Loss of Histone Locus Bodies in the Mature Hemocytes of Larval Lymph Gland Result in Hyperplasia of the Tissue in mxc Mutants of Drosophila
Abstract
1. Introduction
2. Results
2.1. Loss of Function Mutations of mxc and Its Depletion in LG Resulted in Hyperplasia Showing Increased Ratio of Immature Hemocytes in the Larval Tissues
2.2. Hemizygotes for Recessive Lethal But Not Amorphic Mutations of mxc Exhibited Hyperplasia of Lobes in Their Larval LGs
2.3. Depletion of mxc in the Matured Hemocytes of LG Resulted in the Hyperplasia Phenotype of LG
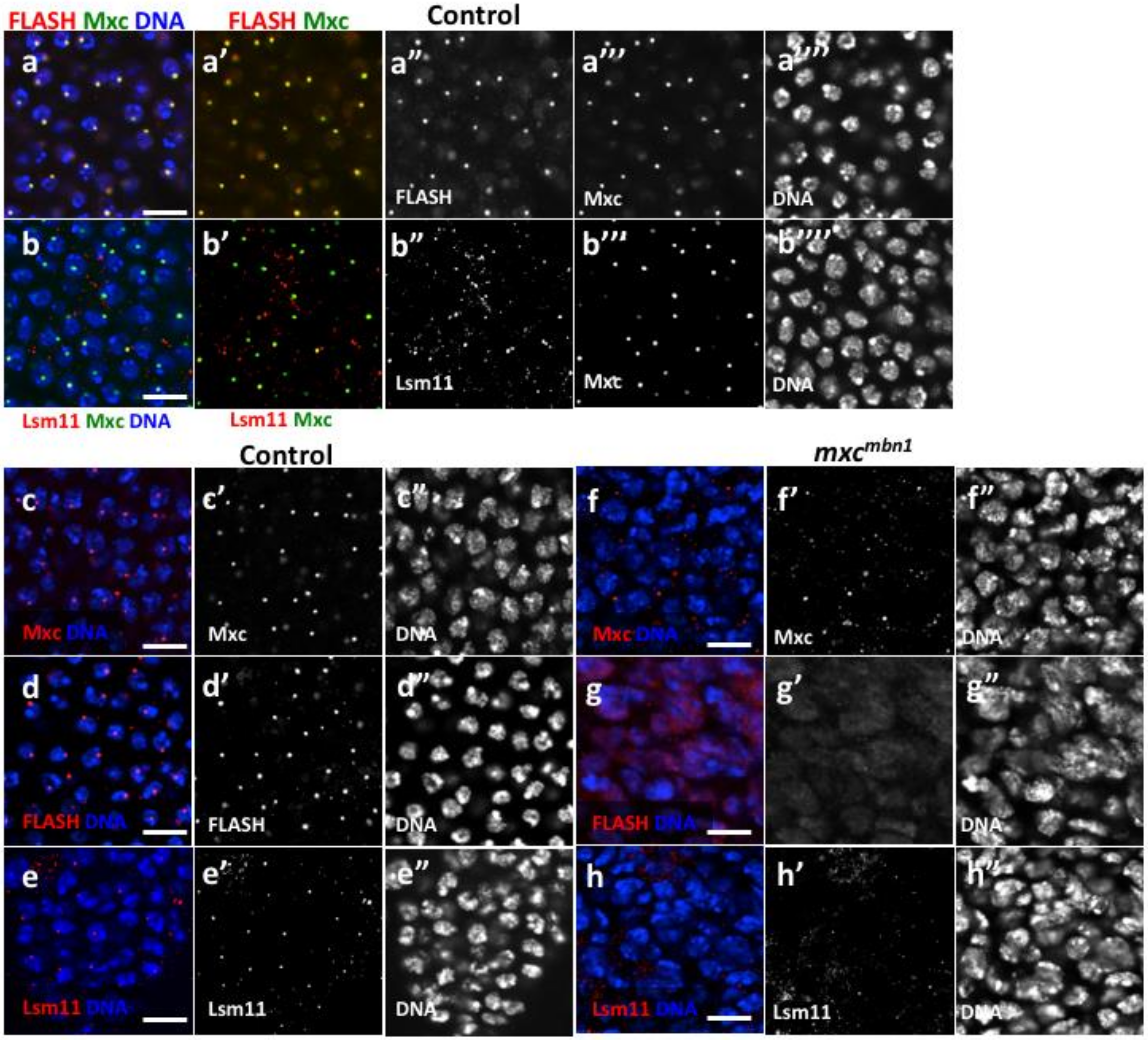
2.4. HLB Formation Was Disrupted in Nuclei of LG Cells From mxcmbn1 Larvae
2.5. Inhibition of HLB Formation in CZ of Larval LGs Reproduced Hyperplasia of the Tissues
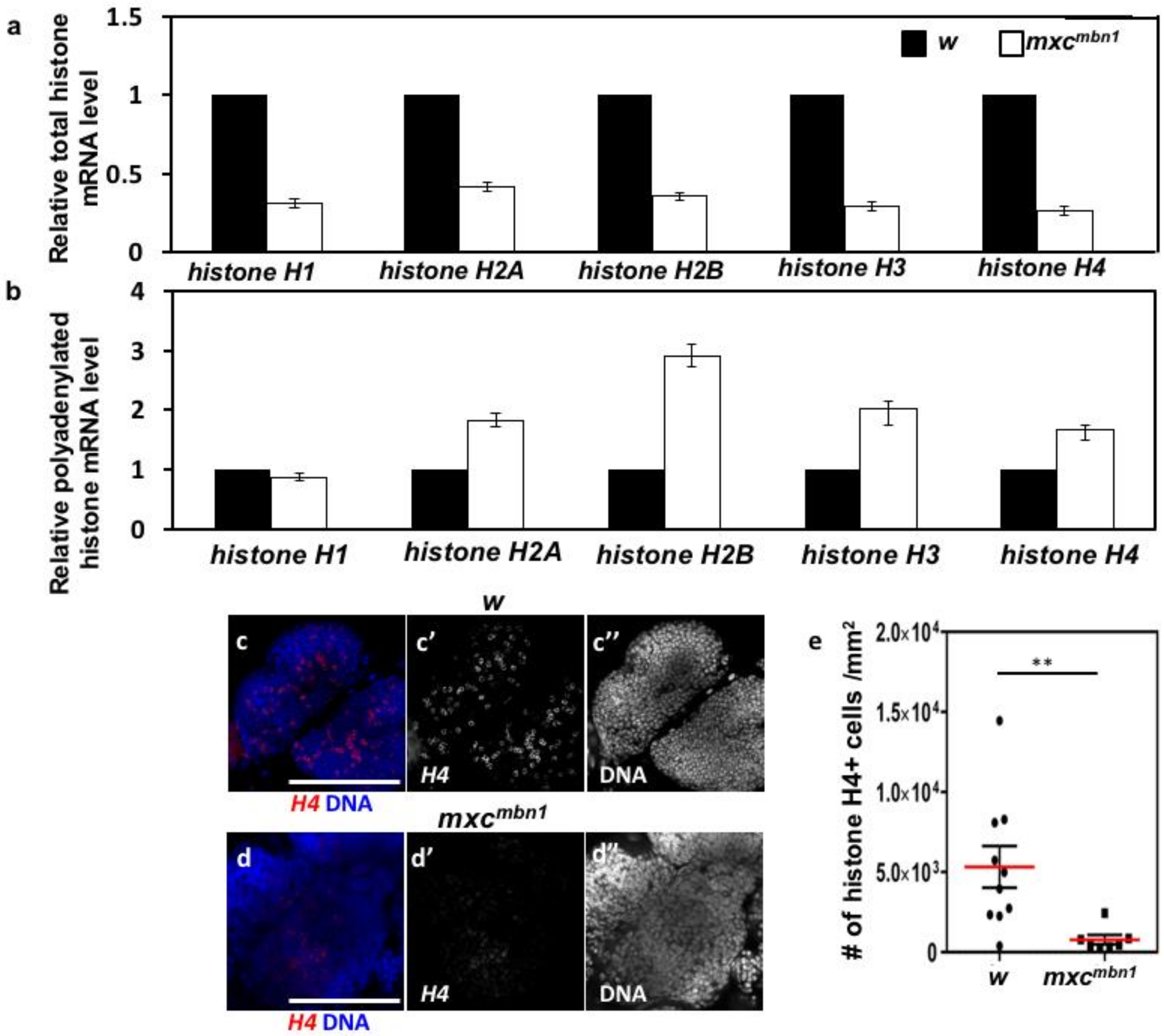
2.6. Reduced mRNA Levels of Canonical Histone mRNAs and Production of Abnormal Polyadenylated mRNAs in mxcmbn1 Larvae
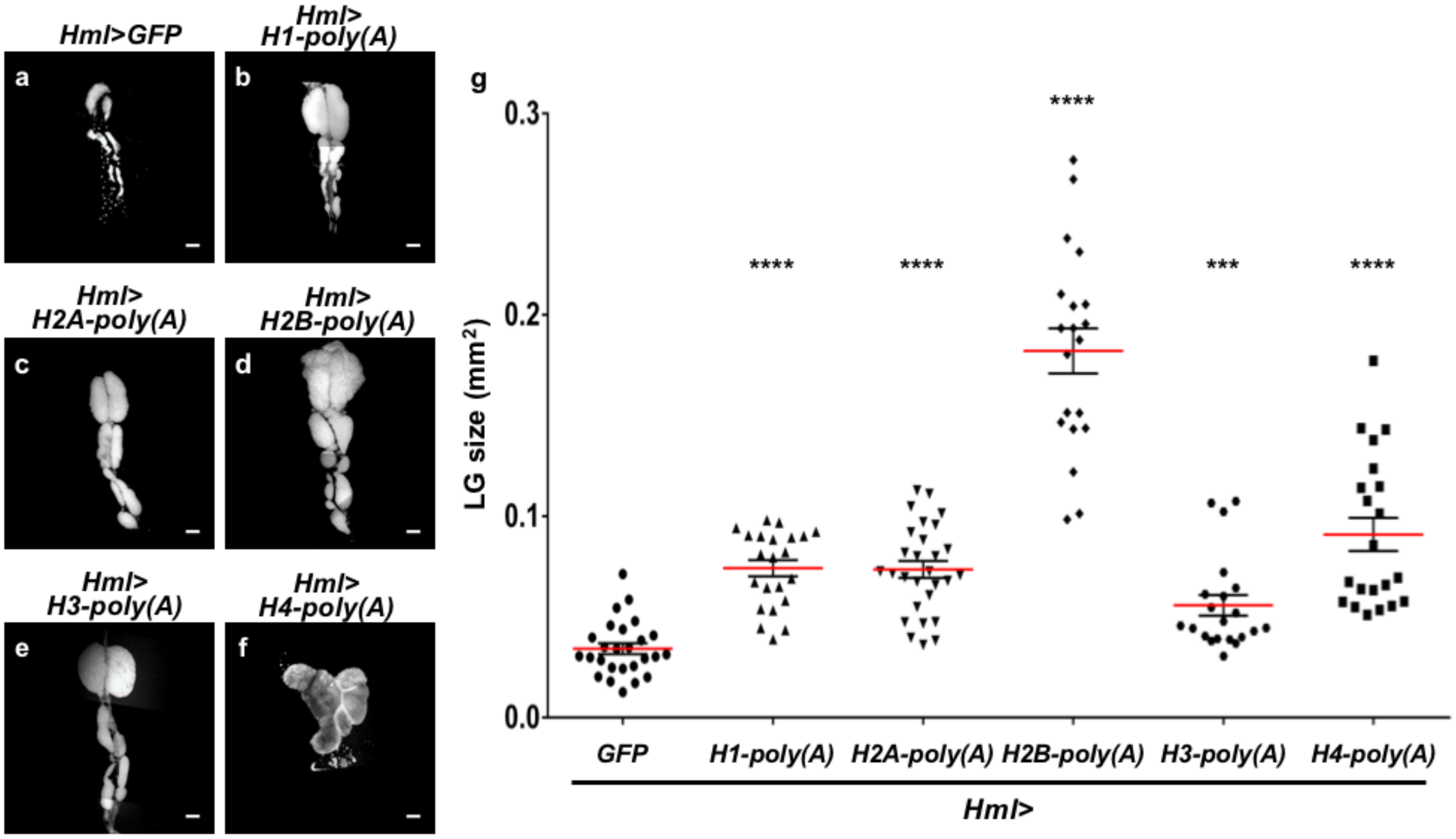
2.7. Ectopic Overexpression of Polyadenylated mRNAs of Canonical Histones in CZ Reproduced the LG Hyperplasia Observed in the mxc Mutants
2.8. Ectopic Expression of the Negative Regulator Adgf-A, Which Suppresses Proliferation of Immature Hemocytes, and Signaling Factors Required for Induction of the Regulator Suppressed Hyperplasia in mxcmbn1
3. Discussion
3.1. Reduction of mxc Expression or Its Function in Mature Hemocytes Resulted in LG Hyperplasia Caused by Over-Proliferation of Undifferentiated Cells in the Tissue
3.2. Mature Hemocyte-Specific Expression of Polyadenylated mRNAs for Canonical Histones Was Responsible for Hyper-Proliferation of Immature Cells in LG Hyperplasia
3.3. Involvement of mxc in the Signaling That Induces Adgf-A Expression, Which Suppresses Excess Proliferation of Immature Cells in LG
4. Materials and Methods
4.1. Drosophila Stocks
4.2. LG Preparation
4.3. qRT-PCR Analysis
4.4. Lymph Gland Immunostaining
4.5. Fluorescence In Situ Hybridization (FISH)
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HLB | Histone locus body |
| LG | Lymph gland |
| CZ | Cortical zone |
| MZ | Medulla zone |
| PSC | Posterior Signaling Center |
| PC | Pericardial cells |
| Adgf-A | Adenosine deaminase-related growth factor-A |
References
- McCormack, E.; Bruserud, O.; Gjertsen, B.T. Review: Genetic models of acute myeloid leukaemia. Oncogene 2008, 27, 3765–3779. [Google Scholar]
- Jan, M.; Majeti, R. Clonal evolution of acute leukemia genomes. Oncogene 2013, 32, 135–140. [Google Scholar]
- Ferrando, A.A.; López-Otín, C. Clonal evolution in leukemia. Nat. Med. 2017, 23, 1135–1145. [Google Scholar]
- Drexler, H.G. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia–lymphoma cells. Leukemia 1998, 12, 845–859. [Google Scholar]
- Martín, V.; Valencia, A.; Agirre, X.; Cervera, J.; San Jose-Eneriz, E.; Vilas-Zornoza, A.; Rodriguez-Otero, P.; Sanz, M.A.; Herrera, C.; Torres, A.; et al. Epigenetic regulation of the non-canonical Wnt pathway in acute myeloid leukemia. Cancer Sci. 2010, 101, 425–432. [Google Scholar]
- Paulsson, K. Genomic heterogeneity in acute leukemia. Cytogenet. Genome Res. 2013, 139, 174–180. [Google Scholar]
- Saarinen, S.; Aavikko, M.; Aittomäki, K.; Launonen, V.; Lehtonen, R.; Franssila, K.; Lehtonen, H.J.; Kaasinen, E.; Broderick, P.; Tarkkanen, J.; et al. Exome sequencing reveals germline NPAT mutation as a candidate risk factor for Hodgkin lymphoma. Blood 2011, 118, 493–498. [Google Scholar]
- Borchmann, S.; Engert, A. The genetics of Hodgkin lymphoma: An overview and clinical implications. Curr. Opin. Oncol. 2017, 29, 307–314. [Google Scholar]
- Platzer, M.; Rotman, G.; Bauer, D.; Uziel, T.; Savitsky, K.; Bar-Shira, A.; Gilad, S.; Shiloh, Y.; Rosenthal, A. Ataxia-telangiectasia locus: Sequence analysis of 184 kb of human genomic DNA containing the entire ATM gene. Genome Res. 1997, 7, 592–605. [Google Scholar]
- Miele, A.; Braastad, C.D.; Holmes, W.F.; Mitra, P.; Medina, R.; Xie, R.; Zaidi, S.K.; Ye, X.; Wei, Y.; Harper, J.W.; et al. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 2005, 25, 6140–6153. [Google Scholar]
- Medina, R.; van der Deen, M.; Miele-Chamberland, A.; Xie, R.-L.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S. The HiNF-P/p220NPAT cell cycle signaling pathway controls nonhistone target genes. Cancer Res. 2007, 67, 10334–10342. [Google Scholar]
- Gao, G.; Bracken, A.P.; Burkard, K.; Pasini, D.; Classon, M.; Attwooll, C.; Sagara, M.; Imai, T.; Helin, K.; Zhao, J. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol. Cell. Biol. 2003, 23, 2821–2833. [Google Scholar]
- Ma, T.; Van Tine, B.A.; Wei, Y.; Garrett, M.D.; Nelson, D.; Adams, P.D.; Wang, J.; Qin, J.; Chow, L.T.; Harper, J.W. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000, 14, 2298–2313. [Google Scholar]
- Zhao, J.; Kennedy, B.K.; Lawrence, B.D.; Barbie, D.A.; Matera, A.G.; Fletcher, J.A.; Harlow, E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000, 14, 2283–2297. [Google Scholar]
- Ghule, P.N.; Dominski, Z.; Yang, X.-C.; Marzluff, W.F.; Becker, K.A.; Harper, J.W.; Lian, J.B.; Stein, J.L.; van Wijnen, A.J.; Stein, G.S. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16964–16969. [Google Scholar]
- White, A.E.; Burch, B.D.; Yang, X.-C.; Gasdaska, P.Y.; Dominski, Z.; Marzluff, W.F.; Duronio, R.J. Drosophila histone locus bodies form by hierarchical recruitment of components. J. Cell Biol. 2011, 193, 677–694. [Google Scholar]
- Terzo, E.A.; Lyons, S.M.; Poulton, J.S.; Temple, B.R.S.; Marzluff, W.F.; Duronio, R.J. Distinct self-interaction domains promote Multi Sex Combs accumulation in and formation of the Drosophila histone locus body. Mol. Biol. Cell 2015, 26, 1559–1574. [Google Scholar]
- Santamaría, P.; Randsholt, N.B. Characterization of a region of the X chromosome of Drosophila including multi sex combs (mxc), a Polycomb group gene which also functions as a tumour suppressor. Mol. Gen. Genet. 1995, 246, 282–290. [Google Scholar]
- Saget, O.; Forquignon, F.; Santamaria, P.; Randsholt, N.B. Needs and targets for the multi sex combs gene product in Drosophila melanogaster. Genetics 1998, 149, 1823–1838. [Google Scholar]
- Shrestha, R.; Gateff, E. Ultrastructure and cytochemistry of the cell types in the larval hematopoietic organs and hemolymph of Drosophila melanogaster. Dev. Growth Diff. 1982, 24, 65–82. [Google Scholar]
- Gateff, E. Tumor suppressor and overgrowth suppressor genes of Drosophila melanogaster: Developmental aspects. Int. J. Dev. Biol. 1994, 38, 565–590. [Google Scholar]
- Remillieux-Leschelle, N.; Santamaria, P.; Randsholt, N.B. Regulation of larval hematopoiesis in Drosophila melanogaster: A role for the multi sex combs gene. Genetics 2002, 162, 1259–1274. [Google Scholar]
- Araki, M.; Kurihara, M.; Kinoshita, S.; Awane, R.; Sato, T.; Ohkawa, Y.; Inoue, Y.H. Anti-tumor effects of antimicrobial peptides, components of the innate immune system, against hematopoietic tumours in Drosophila mxc mutants. Dis. Model. Mech. 2019, 12, dmm037721. [Google Scholar] [CrossRef]
- Landais, S.; D’Alterio, C.; Jones, D.L. Persistent replicative stress alters polycomb phenotypes and tissue homeostasis in Drosophila melanogaster. Cell Rep. 2014, 7, 859–870. [Google Scholar]
- Tanabe, K.; Awane, R.; Shoda, T.; Yamazoe, K.; Inoue, Y.H. Mutations in mxc tumor-suppressor gene induce chromosome instability in Drosophila male meiosis. Cell Struct. Funct. 2019, 44, 121–135. [Google Scholar]
- Elrod-Erickson, M.; Mishra, S.; Schneider, D. Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol. 2000, 10, 781–784. [Google Scholar]
- Rizki, R.M.; Rizki, T.M. Hemocyte responses to implanted tissues in Drosophila melanogaster larvae. Wilehm Roux Arch. Dev. Biol. 1980, 189, 207–213. [Google Scholar]
- Tepass, U.; Fessler, L.I.; Aziz, A.; Hartenstein, V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 1994, 120, 1829–1837. [Google Scholar]
- Holz, A.; Bossinger, B.; Strasser, T.; Janning, W.; Klapper, R. The two origins of hemocytes in Drosophila. Development 2003, 130, 4955–4962. [Google Scholar]
- Evans, C.J.; Hartenstein, V.; Banerjee, U. Thicker than blood. Developmental Cell 2003, 5, 673–690. [Google Scholar]
- Govind, S. Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 2008, 15, 29–43. [Google Scholar]
- Jung, S.-H.; Evans, C.J.; Uemura, C.; Banerjee, U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development 2005, 132, 2521–2533. [Google Scholar]
- Banerjee, U.; Girard, J.R.; Goins, L.M.; Spratford, C.M. Drosophila as a Genetic Model for Hematopoiesis. Genetics 2019, 211, 367–417. [Google Scholar]
- Krzemień, J.; Dubois, L.; Makki, R.; Meister, M.; Vincent, A.; Crozatier, M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature 2007, 446, 325–328. [Google Scholar]
- Mandal, L.; Martinez-Agosto, J.A.; Evans, C.J.; Hartenstein, V.; Banerjee, U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature 2007, 446, 320–324. [Google Scholar]
- Zurovec, M.; Dolezal, T.; Gazi, M.; Pavlova, E.; Bryant, P.J. Adenosine deaminase-related growth factors stimulate cell proliferation in Drosophila by depleting extracellular adenosine. Proc. Natl. Acad. Sci. USA 2002, 99, 4403–4408. [Google Scholar]
- Mondal, B.C.; Mukherjee, T.; Mandal, L.; Evans, C.J.; Sinenko, S.A.; Martinez-Agosto, J.A.; Banerjee, U. Interaction between differentiating cell- and niche-derived signals in hematopoietic progenitor maintenance. Cell 2011, 147, 1589–1600. [Google Scholar]
- White, A.E.; Leslie, M.E.; Calvi, B.R.; Marzluff, W.F.; Duronio, R.J. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol. Biol. Cell 2007, 18, 2491–2502. [Google Scholar]
- Battle, D.J.; Doudna, J.A. The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA 2001, 7, 123–132. [Google Scholar]
- Rajendra, T.K.; Praveen, K.; Matera, A.G. Genetic analysis of nuclear bodies: From nondeterministic chaos to deterministic order. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 365–374. [Google Scholar]
- Duronio, R.J.; Marzluff, W.F. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 2017, 14, 726–738. [Google Scholar]
- Dominski, Z.; Yang, X.; Marzluff, W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 2005, 123, 37–48.R. [Google Scholar]
- Godfrey, A.C.; Kupsco, J.M.; Burch, B.D.; Zimmerman, R.M.; Dominski, Z.; Marzluff, W.F.; Duronio, R.J. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 2006, 12, 396–409. [Google Scholar]
- Barcaroli, D.; Bongiorno-Borbone, L.; Terrinoni, A.; Hofmann, T.G.; Rossi, M.; Knight, R.A.; Matera, A.G.; Melino, G.; and De Laurenzi, V. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. USA 2006, 103, 14808–14812. [Google Scholar]
- Salzler, H.R.; Tatomer, D.C.; Malek, P.Y.; McDaniel, S.L.; Orlando, A.N.; Marzluff, W.F.; Duronio, R.J. A sequence in the Drosophila H3-H4 Promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev. Cell 2013, 24, 623–634. [Google Scholar]
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008, 9, 843–854. [Google Scholar]
- Bos, J.L. ras oncogenes in human cancer: A review. Cancer Res. 1989, 49, 4682–4689. [Google Scholar]
- Nakamura, T. NUP98 fusion in human leukemia: Dysregulation of the nuclear pore and homeodomain proteins. Int. J. Hematol. 2005, 82, 21–27. [Google Scholar]
- Ley, T.J.; Mardis, E.R.; Ding, L.; Fulton, B.; McLellan, M.D.; Chen, K.; Dooling, D.; Dunford-Shore, B.H.; McGrath, S.; Hickenbotham, M.; et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2008, 456, 66–72. [Google Scholar]
- Delgado, M.D.; León, J. Myc roles in hematopoiesis and leukemia. Genes Cancer 2010, 1, 605–616. [Google Scholar]
- Xu-Monette, Z.Y.; Medeiros, L.J.; Li, Y.; Orlowski, R.Z.; Andreeff, M.; Bueso-Ramos, C.E.; Greiner, T.C.; McDonnell, T.J.; Young, K.H. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood 2012, 119, 3668–3683. [Google Scholar]
- Jaffe, E.S.; Harris, N.L.; Stein, H.; Isaacson, P.G. Classification of lymphoid neoplasms: The microscope as a tool for disease discovery. Blood 2008, 112, 4384–4399. [Google Scholar]
- Boulet, M.; Miller, M.; Vandel, L.; Walter, L. From Drosophila blood cells to human leukemia. In Drosophila Models for Human Diseases; Yamaguchi, M., Ed.; Springer Nature: Singapore, 2018; pp. 195–214. [Google Scholar]
- Mondal, B.C.; Shim, J.; Evans, C.J.; Banerjee, U. Pvr expression regulators in equilibrium signal control and maintenance of Drosophila blood progenitors. Elife 2014, 3, e03626. [Google Scholar]
- Kalamarz, M.E.; Paddibhatla, I.; Nadar, C.; Govind, S. Sumoylation is tumor-suppressive and confers proliferative quiescence to hematopoietic progenitors in Drosophila melanogaster larvae. Biol. Open 2012, 1, 161–172. [Google Scholar]
- Sorrentino, R.P.; Carton, Y.; Govind, S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev. Biol. 2002, 243, 65–80. [Google Scholar]
- Liu, J.-L.; Murphy, C.; Buszczak, M.; Clatterbuck, S.; Goodman, R.; Gall, J.G. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006, 172, 875–884. [Google Scholar]
- Nelson, D.M.; Ye, X.; Hall, C.; Santos, H.; Ma, T.; Kao, G.D.; Yen, T.J.; Harper, J.W.; Adams, P.D. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 2002, 22, 7459–7472. [Google Scholar]
- Cheng, G.H.; Nandi, A.; Clerk, S.; Skoultchi, A.I. Different 3′-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc. Natl. Acad. Sci. USA 1989, 86, 7002–7006. [Google Scholar]
- Mullen, T.E.; Marzluff, W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008, 22, 50–65. [Google Scholar]
- Parker, R.; Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127. [Google Scholar]
- Fuke, H.; Ohno, M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res. 2008, 36, 1037–1049. [Google Scholar]
- Cheng, H.; Dufu, K.; Lee, C.-S.; Hsu, J.L.; Dias, A.; Reed, R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 2006, 127, 1389–1400. [Google Scholar]
- Ohno, M.; Segref, A.; Bachi, A.; Wilm, M.; Mattaj, I.W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 2000, 101, 187–198. [Google Scholar]
- Belch, Y.; Yang, J.; Liu, Y.; Malkaram, S.A.; Liu, R.; Riethoven, J.J.; Ladunga, I. Weakly positioned nucleosomes enhance the transcriptional competency of chromatin. Plos ONE 2010, 5, e12984. [Google Scholar]
- Celona, B.; Weiner, A.; Felice, F.D.; Mancuso, F.M.; Cesarini, E.; Rossi, R.L.; Gregory, L.; Baban, D.; Rossetti, G.; Grianti, P.; et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. Plos Biol. 2011, 9, e1001086. [Google Scholar]
- Clemente-Ruiz, M.; Prado, F. Chromatin assembly controls replication fork stability. EMBO Rep. 2009, 10, 790–796. [Google Scholar]
- Ozawa, N.; Furuhashi, H.; Masuko, K.; Numao, E.; Makino, T.; Yano, T.; Kurata, S. Organ identity specification factor WGE localizes to the histone locus body and regulates histone expression to ensure genomic stability in Drosophila. Genes Cells 2016, 21, 442–456. [Google Scholar]
- Oka, S.; Hirai, J.; Yasukawa, T.; Nakahara, Y.; Inoue, Y.H. A correlation of reactive oxygen species accumulation by depletion of superoxide dismutases with age-dependent impairment in the nervous system and muscles of Drosophila adults. Biogerontology 2015, 16, 485–501. [Google Scholar]
- Pennetier, D.; Oyallon, J.; Morin-Poulard, I.; Dejean, S.; Vincent, A.; Crozatier, M. Size control of the Drosophila hematopoietic niche by bone morphogenetic protein signaling reveals parallels with mammals. Proc. Natl. Acad. Sci. USA 2012, 109, 3389–3394. [Google Scholar]
- Pinnola, A.; Naumova, N.; Shah, M.; Tulin, A.V. Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J. Biol. Chem. 2007, 282, 32511–32519. [Google Scholar]
- Feng, W.; Hale, C.J.; Over, R.S.; Cokus, S.J.; Jacobsen, S.J.; Michaels, S.D. Large-scale heterochromatin remodeling linked to overreplication-associated DNA damage. Pro. Natl. Acad. Sci. USA 2017, 114, 406–411. [Google Scholar]
- Sullivan, K.D.; Steiniger, M.; Marzluff, W.F. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell 2009, 34, 322–332. [Google Scholar]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurihara, M.; Komatsu, K.; Awane, R.; Inoue, Y.H. Loss of Histone Locus Bodies in the Mature Hemocytes of Larval Lymph Gland Result in Hyperplasia of the Tissue in mxc Mutants of Drosophila. Int. J. Mol. Sci. 2020, 21, 1586. https://doi.org/10.3390/ijms21051586
Kurihara M, Komatsu K, Awane R, Inoue YH. Loss of Histone Locus Bodies in the Mature Hemocytes of Larval Lymph Gland Result in Hyperplasia of the Tissue in mxc Mutants of Drosophila. International Journal of Molecular Sciences. 2020; 21(5):1586. https://doi.org/10.3390/ijms21051586
Chicago/Turabian StyleKurihara, Masanori, Kouyou Komatsu, Rie Awane, and Yoshihiro H. Inoue. 2020. "Loss of Histone Locus Bodies in the Mature Hemocytes of Larval Lymph Gland Result in Hyperplasia of the Tissue in mxc Mutants of Drosophila" International Journal of Molecular Sciences 21, no. 5: 1586. https://doi.org/10.3390/ijms21051586
APA StyleKurihara, M., Komatsu, K., Awane, R., & Inoue, Y. H. (2020). Loss of Histone Locus Bodies in the Mature Hemocytes of Larval Lymph Gland Result in Hyperplasia of the Tissue in mxc Mutants of Drosophila. International Journal of Molecular Sciences, 21(5), 1586. https://doi.org/10.3390/ijms21051586





