Abstract
Thioredoxin fold proteins (TFPs) form a family of diverse proteins involved in thiol/disulfide exchange in cells from all domains of life. Leptospirillum spp. are bioleaching bacteria naturally exposed to extreme conditions like acidic pH and high concentrations of metals that can contribute to the generation of reactive oxygen species (ROS) and consequently the induction of thiol oxidative damage. Bioinformatic studies have predicted 13 genes that encode for TFP proteins in Leptospirillum spp. We analyzed the participation of individual tfp genes from Leptospirillum sp. CF-1 in the response to oxidative conditions. Genomic context analysis predicted the involvement of these genes in the general thiol-reducing system, cofactor biosynthesis, carbon fixation, cytochrome c biogenesis, signal transduction, and pilus and fimbria assembly. All tfp genes identified were transcriptionally active, although they responded differentially to ferric sulfate and diamide stress. Some of these genes confer oxidative protection to a thioredoxin-deficient Escherichia coli strain by restoring the wild-type phenotype under oxidative stress conditions. These findings contribute to our understanding of the diversity and complexity of thiol/disulfide systems, and of adaptations that emerge in acidophilic microorganisms that allow them to thrive in highly oxidative environments. These findings also give new insights into the physiology of these microorganisms during industrial bioleaching operations.
1. Introduction
Thioredoxin (TRX) fold proteins (TFP) are a family of diverse proteins that possess a common domain consisting of four stranded beta-sheets flanked by three alpha-helices, and the redox-active CXXC motif [1]. The most representative member of this family is thioredoxin, a small disulfide reductase that helps to maintain a reducing cytosolic environment through reduction of oxidized thiols in cytoplasmic proteins [2]. The TRX fold has also been found in a number of other proteins, including the disulfide isomerases (DsbC, DsbD, and DsbG), disulfide oxidases (DsbA and DsbB), glutaredoxin, glutathione S-transferase, glutathione peroxidase, and peroxiredoxin [3].
TFPs share very little sequence homology and can be considered diverse in their functions and structures [4]. Like thioredoxins, many of these proteins are involved in thiol/disulfide exchange reactions, but also in oxidation and reduction reactions, depending on their redox properties. Thus, they play pivotal roles in protein folding and redox control in a number of biological processes. The wide range of redox activities of TFPs is possibly a consequence of modifications to the common scaffold, which result in different properties [5]. Thus, the role of these proteins cannot be directly deduced by the mere identification of a thioredoxin fold as structural feature.
Oxidative stress, caused by reactive oxygen species (ROS), promotes disulfide bond formation in proteins. TFPs are thought to play an important function in response to this condition [4]. In addition to reducing cytoplasmic proteins, thioredoxins are indirectly involved in reducing endogenously- or exogenously-produced inorganic and organic peroxides. In this way, they provide reducing equivalents to thiol-based peroxiredoxins that are responsible for peroxide scavenging activity [6,7].
Thioredoxins also reduce oxidized redox sensors, thus regulating transcriptional activity of genes involved in physiological responses against oxidative stress [8,9]. In addition to thioredoxin and peroxiredoxin, TFPs include disulfide oxidoreductase [10], glutaredoxin [11,12], glutathione S-transferase [13], and glutathione peroxidase [14], all of which play important roles in the defense against oxidative stress. TFPs are thought to protect damaged proteins from inactivation and prevent aggregates of misfolded-protein through the reduction of disulfide-cross links, or through their formation in proper substrate proteins [15]. This latter fact is especially relevant for the folding, stability, and export of secreted proteins.
Leptospirillum spp. is an autotrophic obligate acidophilic bacterium that catalyzes dissimilatory Fe(II) oxidation. It serves as a model bacterium to elucidate the mechanisms used by acidophilic microorganisms to thrive under highly oxidative conditions [16,17,18,19]. Thirteen tfp genes (formerly trx) coding for predicted TFPs have been identified in Leptospirillum spp. [18]. Exposure of Leptospirillum ferriphilum to ROS-generating compounds resulted in a concomitant increase in thioredoxin activity. Interestingly, this activity was significantly higher than that measured in neutrophilic model bacteria. Since thioredoxin activity, measured through 1,4-dithiothreitol (DTT)-induced insulin, is present in most TFPs, the data suggest that proteins containing a TRX fold play a crucial role in the cell physiology and adaptation of Leptospirillum spp. to oxidative conditions. However, the specific contribution of each predicted TFP to the oxidative stress response and fitness in Leptospirillum spp. is still unknown.
The goal of this work was to evaluate the individual participation of 13 tfp genes coding for TRX fold proteins from Leptospirillum sp. strain CF-1 in oxidative stress response. In order to deduce possible functions, we have used an approach that involved bioinformatic examination of the genomic neighborhood of the tfp genes. In addition, the expression patterns of the tfp genes were evaluated in strain CF-1 exposed to oxidative stress conditions. Finally, we established the ability of the genes to confer protection against oxidative stress to Escherichia coli, which is deficient in the thioredoxin system.
2. Results
2.1. In Silico Analysis of the Genomic Context and Prediction of the Roles of the tfp Genes
A previous study demonstrated that the strain Leptospirillum sp. Group II ‘5-way CG’ possesses an extensive and diverse group of TRX fold proteins (TFPs) related to the cellular maintenance of the thiol/disulfide balance [18]. In this work, the genes for the 13 predicted TFPs from the 5-way CG strain were also detected in the genome of strain CF-1, and these predicted proteins were characterized bioinformatically. According to our results, the structural and functional features of these proteins are highly conserved among the two strains. For easier comparison, in this work we kept the numbering of the predicted proteins assigned in a previous paper (Table 1).

Table 1.
Bioinformatic characterization of the predicted thioredoxin fold protein (TFP) of Leptospirillum sp. CF-1.
Currently, a high-quality sequence of the entire genome of the CF-1 strain is available [20] (but not for 5-way CG). Given this, and in order to obtain information on the possible role of tfp genes and their encoded proteins, we analyzed the genomic context of each tfp gene (Figure 1). According to the collected data, TRX fold proteins can be assigned to six functional categories, namely the general thiol-reducing system, cofactor biosynthesis, carbon fixation, cytochrome c biogenesis, signal transduction, and pilus and fimbria assembly (Table 1, Figure 1). A brief description of the major features of each system is provided below.
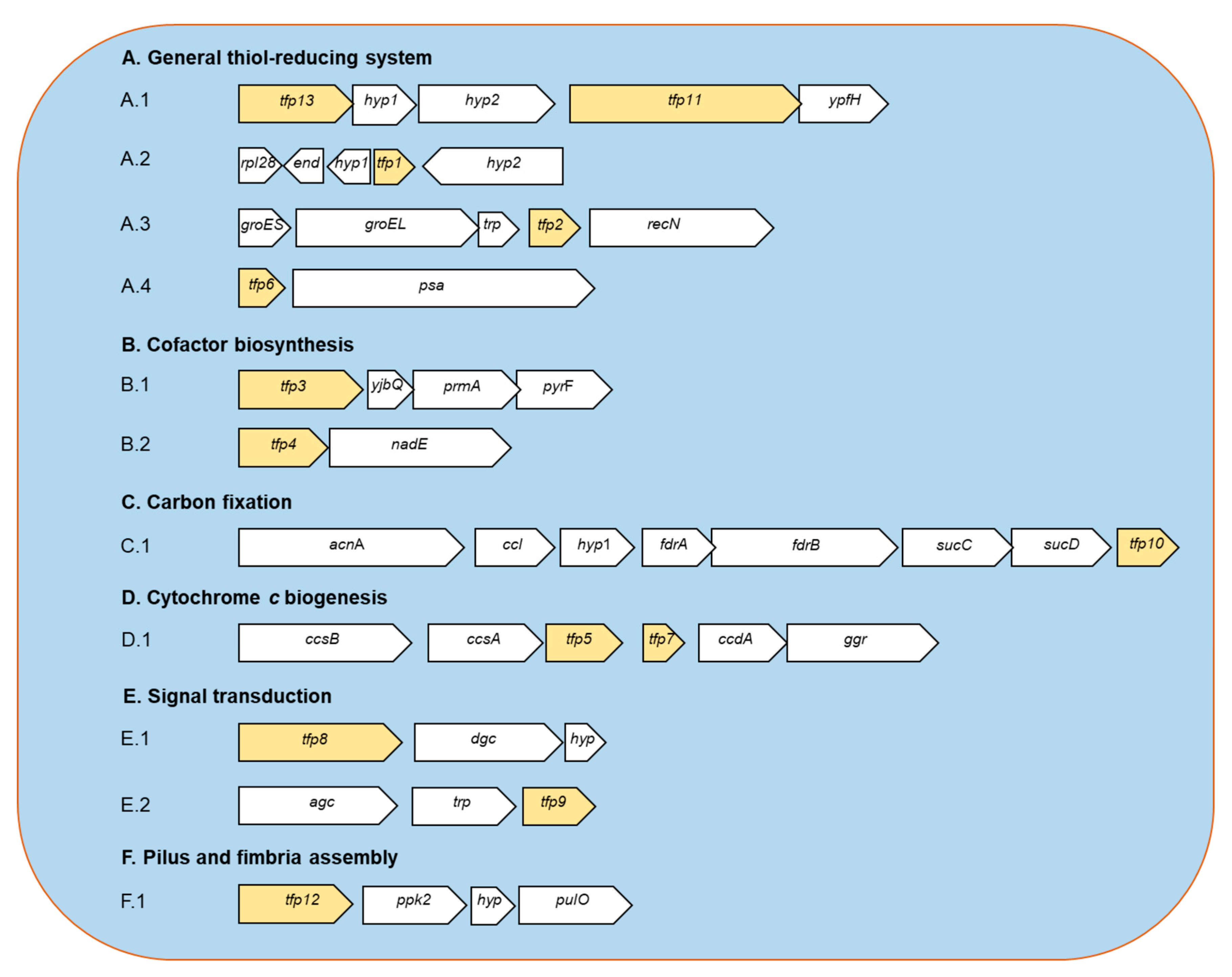
Figure 1.
Predicted genetic context of TFP-encoding genes from Leptospirillum sp. CF-1. A.1. The tfp13 gene codes for a: thioredoxin reductase (TrxB); hyp1: hypothetical protein 1; hyp2: hypothetical protein 2; tfp11: thioredoxin domain-containing protein (YyaL); ypfH: predicted esterase. A.2. rpl28: ribosomal protein L28; end: endonuclease; hyp1: hypothetical protein 1; tfp1: thioredoxin-like protein; hyp2: hypothetical protein 2. A.3. groES: co-chaperone GroES; groEL: chaperone GroEL; trp: TRP-containing small protein; tfp2: thioredoxin-like protein (chaperedoxin); recN: DNA repair protein RecN. A.4. tfp6: thioredoxin-like protein; psa: puromycin- sensitive aminopeptidase. B.1. tfp3: thiol disulfide oxidoreductase; yjbQ: thiamine synthase YjbQ; prmA: ribosomal protein L11 methytransferase PrmA; pyrF: Orotidine 5’-phosphate decarboxylase PyrF. B.2. tfp4: thioredoxin-like protein; nadE: NAD(+) synthetase. C.1. acnA: aconitase A; ccl: citryl-CoA lyase; hyp1: fdrA: fumarate reductase subunit A; frB: fumarate reductase subunit B; sucC: succinyl-CoA synthetase alpha subunit; sucD: succinyl-CoA synthetase beta subunit; tfp10: 2-Cys peroxiredoxin (Prx). D.1. ccsB: cytochrome c-type biogenesis protein CcsB; ccsA: cytochrome c-type biogenesis protein CcsA; tfp5: cytochrome c-type biogenesis protein CcsX; tfp7: thioredoxin-like protein; ccdA: cytochrome c-type biogenesis protein CcdA; ggr: geranylgeranyl reductase. E.1. tfp8: thiol disulfide oxidoreductase; dgc: GGDEF diguanilate cyclase; hyp: hypothetical small membrane protein. E.2. agc: adenylate/guanylate cyclase; trp: tetratricopeptide repeat (TRP)-containing protein; tfp9: thiol disulfide oxidoreductase. F.1. tfp12: cytoplasmic thiol disulfide oxidoreductase; pkk2: polyphosphate kinase 2; hyp: hypothetical protein; pulO: type IV prepilin peptidase.
2.1.1. General Thiol-Reducing System (TFP1-TFP2-TFP6-TFP11-TFP13)
Thioredoxins are considered the primary system for maintenance of the intracellular thiol/disulfide balance. Moreover, in agreement with the previous report [16,18], no candidate genes related to glutathione system were detected in strain CF-1, which reinforces the idea that members of the Leptospirillum genus do not use glutathione as a redox buffer. Bioinformatic predictions suggest that strain CF-1 possesses a Trx system consisting of thioredoxin reductase TrxB (TFP13) and three Trx-like proteins (TFP1, TFP2, and TFP6); each of which has the canonical CXXC motif and is predicted to be located in the cytoplasm of the cell. According to an inspection of neighboring genes, the trxB is located next to genes for two hypothetical proteins, the uncharacterized conserved thioredoxin domain-containing protein YyaL (TFP11) and a putative esterase. The role of TFP11 remains unclear.
In order to gain insights into the possible roles of the three encoded thioredoxins, we compared them phylogenetically to several discrete types of thioredoxins from different sources. The results suggest that thioredoxins 2 and 6 from strain CF-1 are related to the prokaryotic/mitochondrial M-type thioredoxin class [21]. However, they comprise two discrete types, which suggests some degree of functional specialization (Figure S1). The TFP1 encoded protein is a thioredoxin-like protein with a low level of similarity to other characterized thioredoxins, and is grouped with thioredoxins from plants (types F, H, and O). No additional information could be derived from the genetic context of the tfp1 gene. However, specialized functions related to protein folding and turnover could be predicted for TFP2 and TFP6, which have the canonical active site motif WCGPC found in TrxA and TrxC of E. coli, with a similar molecular weight (about 12 kDa) [22].
The GroESL protein complex in E. coli is the best characterized member of the HSP60/HSP10 heat shock chaperonin family and that is formed by chaperone GroEL and its obligate GroES co-chaperone [23]. GroESL is one of the major protein systems involved in folding newly-synthesized cytosolic proteins, and in refolding proteins that have been damaged under stress conditions. Thus, GroESL is believed to play a crucial role in the tolerance to all types of stress. Recent findings have shown that the GroESL system protects E. coli protein substrates during stress in cooperation with the chaperedoxin CnoX (chaperone-redox), which combines holdase activity with the ability to form mixed disulfide complexes with client proteins in an ATP-independent manner to prevent their aggregation [15].
Interestingly, CnoX is a thioredoxin-like protein that not only prevents protein aggregation, but also provides protection against irreversible oxidation by forming disulfide bonds with its substrates, and protecting their sensitive cysteines from over-oxidation. Due to this dual-function, CnoX has been assigned to a new family of proteins called chaperedoxins [15]. The substrates that are bonded to this chaperedoxin can only be transferred to foldase GroESL when intracellular reducing conditions are restored. Thus, this foldase assists the non-covalent folding of the protein in an ATP-dependent manner.
In Leptospirillum sp. CF-1, a putative thioredoxin-like protein (TFP2) gene is co-localized with groEL and groES genes (Figure 1A.3). Interestingly, TFP2 (but no other TFP) is highly similar (55%) to the N-terminal sequence (amino acids 1–66) of the CnoX protein from E. coli (284 aa). No other CnoX-encoding genes were detected in the genome from strain CF-1. Unlike CnoX from E. coli, TFP2 from Leptospirillum CF-1 possesses a CXXC motif and lacks the characteristic tretratricopeptide (TRP) repeat motifs [24]. However, a small open reading frame (ORF) in this genetic cluster that encodes for a predicted TRP-containing protein is located next to the tf2 gene.
The cluster that harbors genes for GroEL, GroES, and the CnoX-like protein also contains a gene that encodes the protein RecN, which is predicted to be involved in the recombination repair of DNA double-strand breaks [25]. The organization of this genetic cluster is in agreement with a role of TFP2 in protein and DNA repair, so it may play a pivotal role in stress response in Leptospirillum sp. The role of TFP2 in Leptospirillum sp. as canonical thioredoxin and/or chaperedoxin should be explored experimentally.
Finally, the TFP6-encoding gene from the strain CF-1 is located close to a gene encoding for the putative puromycin-sensitive aminopeptidase PSA (or endopeptidase N from the M1 class peptidase) [26]. This enzyme is considered the major aminopeptidase in E. coli involved in cytosolic protein degradation, which enables the use of amino acids as nutrients [27]. The aminopeptidase encoded in CF-1 possesses 863 aa and has 35% similarity to the aminopeptidase N from E. coli (879 aa) [28]. The protein of the strain CF-1 has a predicted molecular weight of 97.9 kD, and conserves all the amino acid residues that have been reported as critical for the activity [29].
TFP6 showed a high degree of similarity to thioredoxin TrxC from Rhodobacter capsulatus (59%) and E. coli (54%), suggesting a role as a key antioxidant system through its disulfide reductase activity. Interestingly, TFP6 and aminopeptidase N-encoding genes are contiguous and very close, and could be part of a putative transcriptional unit. A functional relationship between TFP6 and aminopeptidase N linked to protein degradation can also be predicted. Redox-responsive protein degradation has been widely reported in different kinds of cells [30], so the redox status of the cell plays a key role in regulating protein turnover where oxidation has an inactivating proteolytic effect while reduction leads to an activation of proteolysis. Systems that reduce proteases include thioredoxins and glutaredoxins [30]. A relationship between putative thioredoxin TFP6 and aminopetidase N from Leptospirillum sp. CF-1 may be critical in determining redox-responsive enzyme activation and protein turnover.
2.1.2. Cofactor Biosynthesis (TFP3 and TFP4)
The enzymes ThiE and thiamine phosphorylase ThiL catalyze the last two steps of the thiamine (vitamin B1) biosynthetic pathway. Interestingly, an analog (YbjQ) of the thiamine synthase ThiE has been found in many bacteria [31]. YbjQ has no structural similarity to ThiE, although members of the YbjQ protein family from several species have been shown to have thiamin phosphate synthase activity and complement thiE mutants [31].
In Leptospirillum sp. CF-1, visualization of the genomic neighborhood showed that the YbjQ-encoding gene clusters with three other genes that encode orotidine 5′-phosphate decarboxylase, methyltransferase, and thiol disulfide oxidoreductase (TFP3) (Figure 1B.1). According to our inspection, this cluster structure is only conserved in leptopirilli species. The biological significance of this genetic clustering should be studied. However, it may represent a relationship between disulfide oxidoreductase and thiamine biosynthesis. The involvement of the vitamin biosynthesis as part of oxidative response cannot be ruled out.
TFP4 is thought to be a membrane-anchored thioredoxin-like protein oriented towards the periplasm similar to the characterized thioredoxin-like protein TlpA protein (41% similarity) from Bradyrhizobium japonicum [32]. TlpA acts as a protein-disulfide reductase that has been shown to be physiologically significant [32]. The tfp4 gene from Leptospirillum CF-1 was found next to the nadE gene (Figure 1B.2), which encodes for a glutamine-dependent NAD(+) cytoplasmic synthetase. NadE catalyzes ATP-dependent amidation of deamido-NAD in the last step of NAD(+) synthesis in both de novo and salvage biosynthetic pathways. The generated NAD(+) is an indispensable and universal redox cofactor involved in a variety of cellular processes. This dinucleotide is also a precursor of NADP+/NADPH, which is a key player in redox homeostasis maintenance in the cell [33].
The NADH:NAD(+) ratio is a significant indicator of the metabolic cellular state. It has been shown that intracellular concentration and the ratio of NAD(+)/NADH in Bacillus subtilis and Mycobacterium tuberculosis is modified in response to stress conditions [34]. Although cytoplasmic regulation of the NAD(+)/NADH ratio in chemolitoautotrophic bacteria is largely unknown, oxidative conditions may have an important effect in modulating the levels of this redox pair, so the synthesis of NAD(+) by the NadE enzyme may to some extent depend on the disulfide membrane reductase TFP4. Thioredoxin-mediated activation has been found in a number of enzymes [35], although redox activation has been not proven for NAD(+) synthetase. Thus, the relationship between this enzyme and disulfide reductase remains to be established.
2.1.3. CO2 Fixation (TFP10)
Members of the Leptospirillum genus are obligate chemolitotrophic and autotrophic bacteria. CO2 fixation in these microorganisms seems to be performed by the reductive tricarboxylic acid (RTCA) cycle instead of the conventional Calvin–Benson cycle [36]. In Leptospirillum ferriphilum DSM14947, the enzymes for the RTCA cycle are encoded in four transcriptional units named ccs (for citryl-CoA synthetase), ccl (for citryl-CoA lyase), for (for 2-oxoglutarate ferredoxin oxidoreductase), and por (for pyruvate ferredoxin oxidoreductase) operons [36].
As in the strain DSM14647, the ccl operon of the strain CF-1 contains genes coding for aconitase A (acnA), citryl-CoA lyase (ccl), fumarate reductase (fdrAB), and succinyl-CoA synthetase (sucCD). Furthermore, this operon contains a gene that encodes for TFP10 that is thought to be a 2-Cys peroxiredoxin (Prx) belonging to the thioredoxin superfamily (Figure 1C.1). The Prx enzyme has been implicated in the reductive detoxification of peroxides [37]. Organic and inorganic peroxides are produced close to the cytoplasmic membrane during respiration and are strong oxidizing compounds that can damage carbon fixation proteins like aconitase [38], impairing the activity of the pathway. The activity of Prx can contribute to the maintenance of the cellular redox environment and consequently to the carbon fixation taking place.
2.1.4. Cytochrome c Biogenesis (TFP5 and TFP7)
Biogenesis of c-type cytochromes involves the covalent attachment of the heme group to two cysteines present in apocytochrome c, and the subsequent folding of apocytochrome c around the heme. With this purpose, the prokaryotes use the pathways called Systems I, II, and III (extensively reviewed in Kranz et al., 2009) [39]. In general, attachment occurs on the external side of the cytoplasmic membrane and involves the formation of thioester bonds between the reduced cysteine residues of the CXXCH cytochrome motif and the alpha carbons of the heme vinyl groups [39]. The CXXCH cysteines are reduced, using a general transmembrane thioredoxin protein such as DsbD and specific thioredoxins such as CcmG of cytochrome c maturation system I or CcsX of cytochrome c synthesis system II. It has been reported that Leptospirillum sp. uses a plethora of c-type cytochromes that contribute to the maintenance of high respiratory rates during ferrous ion oxidation [40].
As noted in a previous report [18], our bioinformatic analysis in this work revealed that the genome of the strain CF-1 contains a cluster with genes that possibly encode for the CcsA, CcsB, CcsX, and CcdA proteins, which belong to System II of cytochrome c biogenesis. In addition to the gene that codes for the thioredoxin fold protein CcsX (TFP5), a second gene that codes for TFP7 was detected in this gene locus and annotated as a thioredoxin-like protein with a predicted membrane location. This protein is thought to contribute with reducing equivalents to restore the reduced state of CcsX or other target proteins.
Finally, it is interesting to note the presence of a geranylgeranyl reductase-encoding gene that may be involved in the structural modification of the hydrophobic side-chain of the heme molecule [41]. Hence, this locus seems to contain a complete set of genes related to the modification and trafficking of the heme group, and the biogenesis of type-c cytochromes. As described above, the participation in this process of two thioredoxin fold proteins (TFP5 and TFP7) was predicted.
2.1.5. Signal Transduction (TFP8 and TFP9)
The second messenger cyclic diguanylate (c-di-GMP) has emerged in recent years as a central regulator for a number of bacterial processes, including biofilm formation, motility, and virulence [42]. c-di-GMP pathways have been reported in different Acidithiobacillus species. However, c-di-GMP effectors and signal transduction networks are still largely uncharacterized in this and other extremophile species. Interestingly, the involvement of c-di-GMP in biofilm formation on elemental sulfur in Acidithiobacillus thiooxidans was recently reported [43]. In Leptospirillum sp. CF-1, the existence of c-di-GMP pathways is suggested by the presence of genes that code for diguanilate cyclases GGDEF (with Gly-Gly-Asp-Glu-Phe at the active site) or GGEEF (with Gly-Gly-Glu-Glu-Phe at the active site), and EAL (Glu-Ala-Leu) or HD-GYP (His-Asp and Gly-Tyr-Pro) domain-containing phosphodiesterases, which are involved in synthesis and degradation of this molecule, respectively [44]. In this context, we were able to detect 12 genes coding non-identical versions of diguanylate cyclase in the genome of the strain CF-1, suggesting a role of these proteins in triggering cellular responses mediated by c-di-GMP under different environmental stimuli.
Similarly, to cyclic diguanylate, cyclic AMP (cAMP) is another important second messenger which is made by from adenosine triphosphate (ATP). In the vast majority of organisms, the formation and degradation of cAMP are mediated respectively by adenylyl cyclases and phophodiesterase, whose activity is controlled by glucose availability [45]. Changes in intracellular cAMP concentration are perceived by the cAMP receptor protein CRP. The cAMP-CRP regulatory coupling is able to respond to distinct external and internal inputs [45].
According to our bioinformatic inspection, TFP8 and TFP9 from the strain CF-1 are cytoplasmic thiol/disulfide oxidoreductases. The genetic context of the respective encoding genes revealed that they are located in two clusters that contain genes for a GGDEF domain-containing diguanylate cyclase (Figure 1E.1) and adenylate cyclase (Figure 1E.2) that may be directly involved in formation of c-di-GMP and cAMP, respectively.
The diguanylate cyclase (DGC) protein, encoded next to tfp8 gene, possesses a transmembrane helix and a high isoelectric point, so this protein is possibly associated with the membrane and exposed to the acidic periplasm of the cell [46]. DGC has been shown to have a role in bacteria as a broad-range environmental sensor on the membrane [47]. The characteristics of the detected DGC from the strain CF-1 are consistent with a predicted role as a sensor. This genetic cluster also contains a gene that encodes for a small conserved protein of unknown function. However, according to bioinformatics, this protein has a predicted location in the cell membrane that also fits a possible role in the signaling pathway.
Disulfide oxidoreductase TFP8 codified in this genetic cluster is a soluble protein that may exert an antioxidant role in response to environmental assaults that generate oxidative stress. Interestingly, c-di-GMP has also been linked to the bacterial response to oxidative conditions [48]. Thus, a link between signaling pathways mediated by c-di-GMP and oxidative damage protection response could be established in the strain CF-1.
The TFP9-encoding gene is located close to genes for adenylate cyclase involved in cyclic AMP synthesis, and tetratricopeptide repeat-containing protein, as described in Figure 1. Tetratricopeptide repeats (TPRs) typically contain 34 amino acids and are found in proteins with a variety of functions [49]. To our knowledge, no relationship has been established between TRP proteins and the cAMP-dependent signaling pathway. However, it has recently been reported that the activity of regulating intracellular cAMP levels by the FimV protein is dependent on its C-terminal TRP domain in Pseudomonas aeruginosa [49]. Whether the activity of thiol-disulfide interchange activity of TFP9 is influenced by adenylate cyclase and TRP-containing protein or vice versa in Leptospirillum spp. is still an open question.
2.1.6. Pilus and Fimbria Assembly (TFP12)
Phosphates can accumulate in cells as polymers of tens to hundreds of phosphate residues linked by inorganic phosphoanhydride bonds called polyphosphates (PolyPs). These polymers are synthesized by polyphosphate kinases (PPK) and degraded by polyphosphatases (PPX) [50]. PolyPs are produced by different Acidithiobacillus strains [51] and are believed to be present in other acidophiles.
There are two families of PPKs, type 1 (PPK1) and type 2 (PPK2). PPK1 from E. coli catalyzes the transfer of the terminal phosphate of ATP to a short polyP chain, but it can also catalyze the reverse reaction [50]. PPK2 is present in Pseudomonas aeruginosa and preferentially catalyzes the phosphorylation of GDP to support the biosynthesis of exopolysaccharides [52]. In the vicinity of tfp12 there is a predicted ppk gene (Figure 1F.1) that encodes for PPK2 (51.74% homology to Meiothermus ruber PKK2). No PKK1 homologue was detected. The other interesting gene nearby in the genome encodes for a type IV prepilin peptidase (PulO) that is involved in the biosynthesis of pilus and secretion of substrates like toxins and enzymes that possess the IV pilin signal peptide [53].
According to our inspection, a complete type IV secretion system is not encoded in the CF-1 genome. Nevertheless, we detected genes in this strain related to the general secretory pathway (Gsp) that are clustered and form a putative operon gsp. Interestingly, the proteins encoded in this operon, which include the GspCDEF, and major pseudopilin GspG (NCBI accession number AKS23819.1 to AKS23827.1) are also necessary for the assembly of the type II secretion system [54]. Since most of the genes for the type IV secretion system do not appear to be present, the pulO gene that encodes a type IV prepilin peptidase and that is next to the TFP12 coding gene may have a role as the peptidase of the type II pilin. Our prediction suggests that the CF-1 genome contains a number of genes coding for possible target proteins of PulO, which typically contain a GFTLIE motif in the N-terminal region [55]. In agreement with this, bioinformatic predictions showed that target proteins include the pseudopilin GspG (AKS23823.1), among others.
In E. coli, periplasmic disulfide oxidase DsbA is required for the correct assembly of type IV fimbria that maintain the disulfide bond at the bundling C-terminal [56]. According to our inspection, TFP12 is a cytoplasmic thiol/disulfide protein, so activity similar to that of DsbA is unlikely. However, it may play a similar role in the CF-1 strain by maintaining proteins in proper redox and folding states for the further assembly of structures similar to those of pili or fimbria. Thus, we propose that protein products from tfp12 and neighboring genes are involved in secretion and assembly of appendages, with TFP12 ensuring protein folding, PKK2 ensuring ATP supply required for transport of secreted proteins, and PulO cleaving signal sequences of necessary proteins (like GspG). TPL12 in Leptospirillum spp. may indirectly mediate many important functions where pilin/fimbria usually play pivotal roles (e.g., adherence to minerals).
2.2. The Expression of tfp Genes from Leptospirillum sp. CF-1 Is Differentially Regulated under Oxidative Stress Conditions
In order to determine the contribution of the predicted tpf genes to redox homeostasis, we determined whether oxidative stress elicitors increase the mRNA levels of each tfp gene. ROS overproduction and oxidative stress induction by exposure to ferric iron in Leptospirillum sp. had been assessed previously [18,19]. Diamide was used because of its ability to induce generalized disulfide stress. Cells were exposed for one hour to oxidative stress conditions with 260 mM ferric iron or 4 mM diamide. Changes in mRNA levels were estimated by real-time PCR and normalized against the 16S rRNA (rrsB) [19].
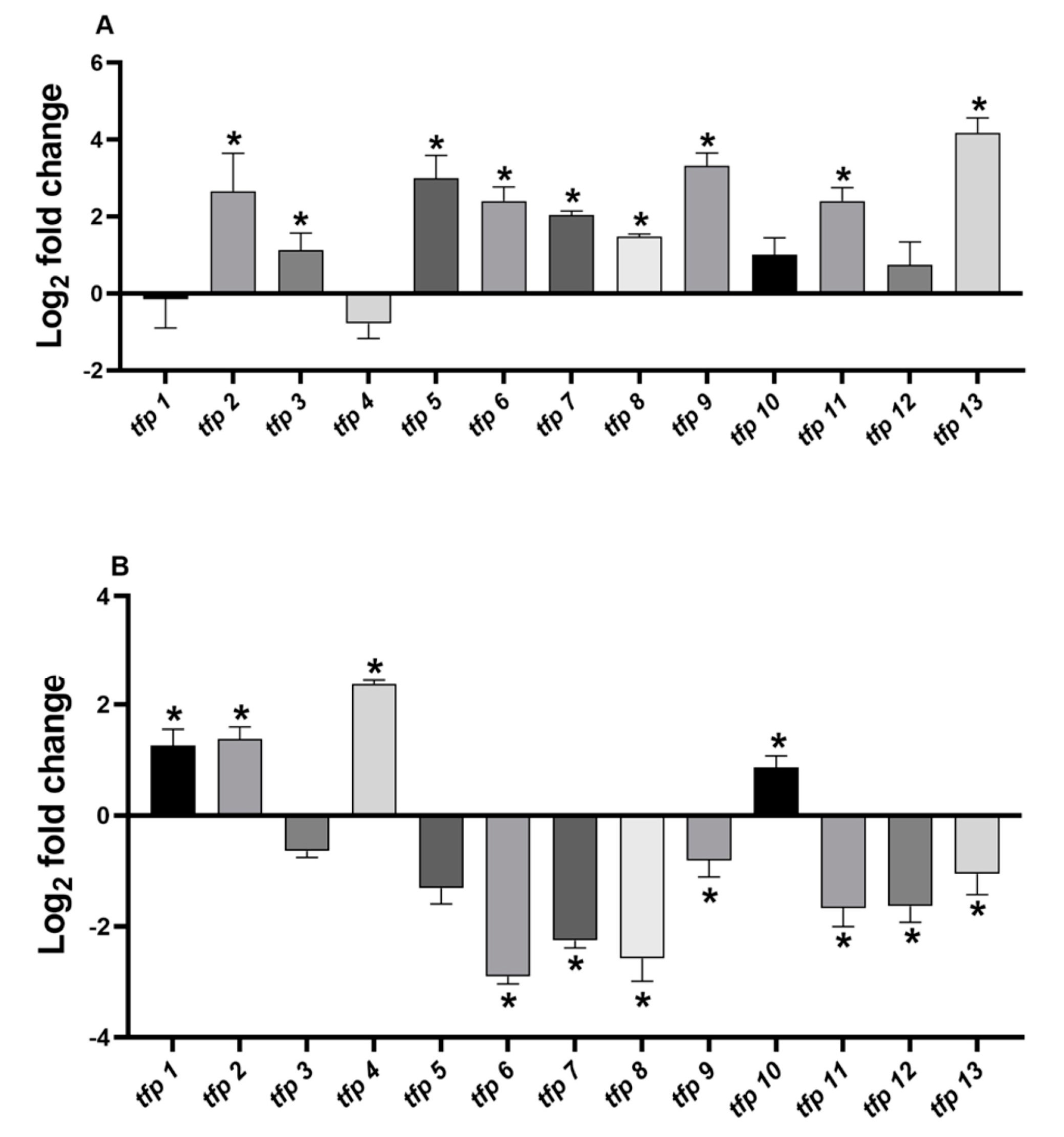
Under ferric stress culture conditions, the genes tfp2, tfp5, tfp9, and tfp13 were upregulated between 7- and 18-fold, while tfp3, tfp6, tfp7, tfp8, and tfp11 showed increases of 2- to 5-fold (Figure 2A, Table S2) compared to control condition (without ferric iron). No significant changes were detected for tfp1, tfp4, tfp10, and tfp12 mRNA levels.
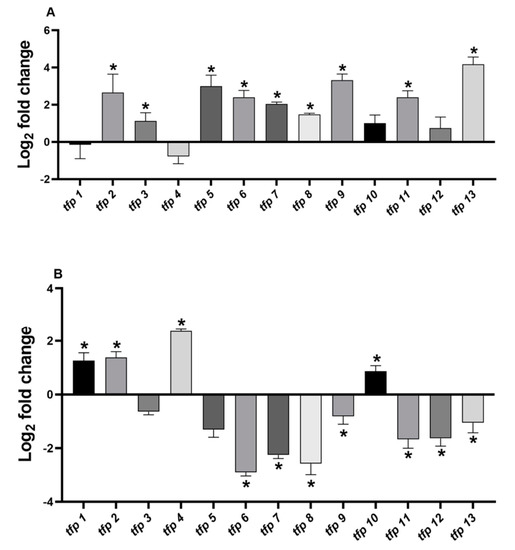
Figure 2.
Relative expression of tfp genes from Leptospirillum sp. CF-1. Cells were stressed with (Fe2(SO4)3) (A) or diamide (B) for 1 h as indicated in Material and Methods. The RNA samples were analyzed in triplicate and ∆Ct results compared to the control 16S RNA gene. Bars represent the average of three independent experiments ± standard deviation. (*): p < 0.05.
The analysis of the relative mRNA levels of tfp genes in response to 4 mM diamide showed that tfp1, tfp2, tfp4 and tfp10 were upregulated between 2- and 5-fold, whereas tfp6, tfp7, tfp8, tfp9, tfp11, tfp12 and tfp13 were downregulated, compared to the results under the control condition (without diamide) (Figure 2B, Table S2). No significant changes in mRNA levels were detected for tfp3 and tfp5.
The results show that under ferric iron stress generalized upregulation of tfp genes took place (9 out of 13), whereas stress with diamide downregulated most tfp genes (7 out 13). The downregulation ranged from −1 to −2.9 log2 fold changes, representing decreases in mRNA levels of 50%–90%. For example, tfp9 and tfp13 showed 10.2- and 18.5-fold change, respectively, in ferric iron stress, but in response to diamide they decreased by almost 50% (Table S2). Remarkably, one group of tfp genes was upregulated solely in response to ferric iron (tfp3, tfp5, tfp6, tfp7, tfp8, tfp9, tfp11, and tfp13), while another group was induced solely in response to diamide (tfp1, tfp4, and tfp10), suggesting that they are responsive to different stress signals. It is notable that tfp2 increased in response to both ferric iron and diamide, which is in agreement with its possible role in preventing protein oxidation and aggregation.
It is also interesting to note that ferric iron, a naturally occurring stressor in bioleaching environments, produced a generalized upregulation and a likely activation of TRX fold proteins, including thioredoxin reductase TrxB (TFP13), as well as proteins thought to be involved in protein turnover (TFP2 and TFP6), thiamine biosynthesis (TFP3), biogenesis of cytochrome c, (TFP5 and TFP7), signaling, and disulfide bond isomerization (TFP8 and TFP9). Differences in the expression profile under ferric and diamide stress will be further discussed.
2.3. Tfp Genes from Leptospirillum sp. CF-1 Restored the Response to Oxidative Stress of a Thioredoxin-Deficient Strain of E. coli
Complementation assays were performed to determine whether tfp genes from Leptospirillum sp. CF-1 are functional and capable of conferring antioxidative protection to a ΔtrxAΔtrxC thioredoxin-deficient strain (E. coli JEM-136). According to the expression profile determined by the RT-qPCR experiments, the following tfp genes were analyzed: tfp2 and tfp6 (upregulated in response to ferric ions), tfp1 and tfp10 (upregulated in response to diamide), and tfp12, which was not responsive to ferric ions, but was downregulated under diamide stress. E. coli trxA (reported as constitutively expressed) encoding thioredoxin Trx1 from the wild-type strain K12 was used as a positive control.
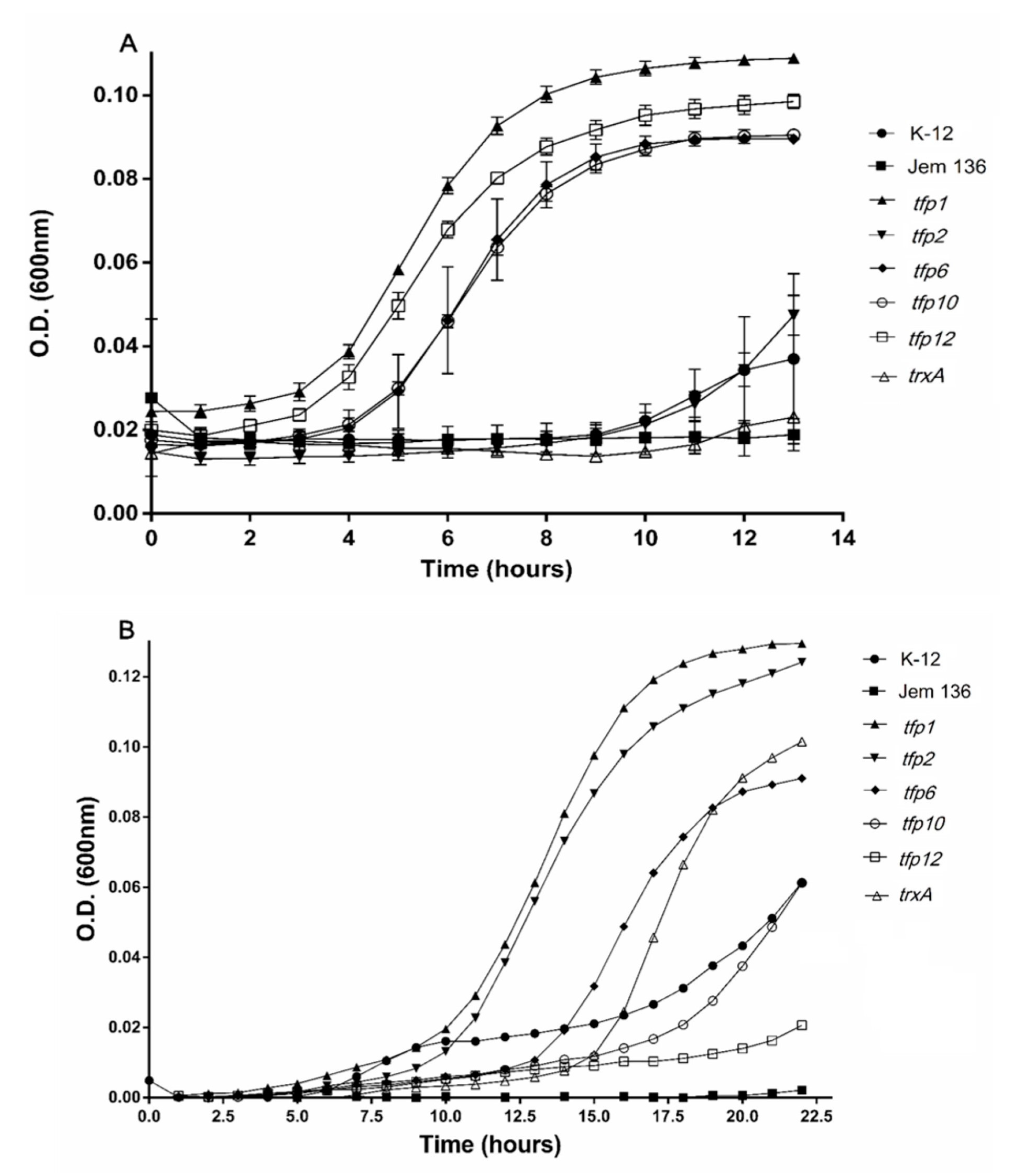
Initially, the susceptibility of E. coli ∆trxAΔtrxC mutant JEM-136 and the wild-type K-12 strains to ferric sulfate and diamide was assayed by determining their minimal inhibitory concentration (MIC). Sub-lethal levels of ferric iron (0.5 mM) and diamide (0.4 mM) (1/2 MIC) were used to evaluate the growth of the E. coli strains. As shown in Figure 3A, the strains transformed with the plasmid-carrying genes tfp1, tfp6, tfp10, and tfp12 had higher growth rates with exposure to ferric ions than the wild-type strain, and the mutant JEM-136 strain transformed with an empty vector. These genes conferred a dramatically more notable protective effect than did trxA from E. coli. Interestingly, not only is the cell growth of strains carrying tfp genes higher than that of the control strains, but also the growth of complemented strains exposed to ferric iron were higher than those of control cells not exposed to the oxidant compound. For example, after 10 h of incubation under a ferric iron-stress condition, cells complemented with tfp1, tfp12, tfp10, or tfp6 (induced with l-arabinose) had growth rates that were 67.5%, 63.7%, 57%, and 54.3% higher, respectively, than those of non-stressed cells (100%) (Table S3). In turn, growth of the mutant JEM-136 or the mutant strain complemented with trxA from E. coli K12 (TrxA-Ec) reached 15.3% and 25.7%, respectively. These data show that the tfp1, tfp12, tfp10, and tfp6 genes from the Leptospirillum sp. strain CF-1 confer more protection to E. coli when exposed to oxidative conditions with ferric iron than does the endogenous gene trxA. As mentioned above, the TrxA protein from E. coli is a thioredoxin directly related to oxidative protection [7,57].
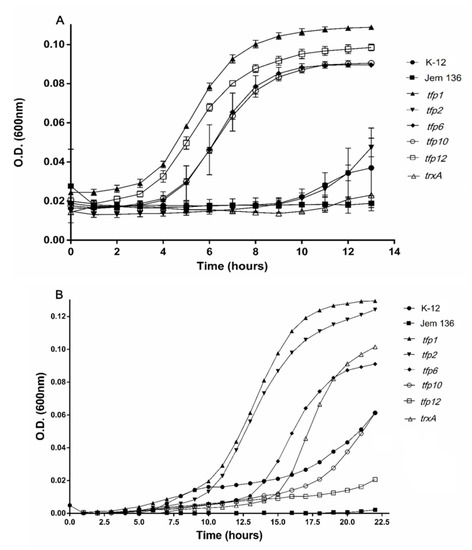
Figure 3.
Growth of Escherichia coli JEM-136 complemented with tfp genes under oxidative stress culture conditions. Expression induction of the tfp gene in E. coli carrying pBADTopo-tfp was carried out with l-arabinose, as described in Materials and Methods. The complemented strain was grown on minimum tryptone yeast extract salts (TYES) medium under stress conditions with 0.5 mM (Fe2(SO4)3) (A) for 15 min, then transferred to a fresh medium, and grown for another 13 h. For diamide stress, the cells were exposed to 0.4 mM diamide and then grown for 20 h (B).
In the case of cultures exposed to diamide, the mutant strain JEM-136 complemented with predicted thioredoxin genes tfp1 or tfp2 had higher growth than the trxA-Ec-complemented strain or the wild-type K-12 strain, suggesting that these genes restore the phenotype to the thioredoxin-deficient strain, thus allowing this mutant to respond even more effectively to oxidative conditions than do wild-type strains (Figure 3B). Notably, after 10 h of incubation, higher percentages of growth recovery than under control condition (100%) were reached by strains complemented with tfp1 (66.5%), tfp2 (41.5%), and tfp6 (10.5%), all of which are predicted to encode actual thioredoxins (Table S3). The strains complemented with the genes tfp10 and tfp12 did not have any noticeable growth stimulating effect under the oxidative condition.
Taken together, these results suggest that tfp genes from the strain CF-1 are functional and capable of restoring, and even improving the capability of E. coli to face stress conditions. In particular, tfp1, tfp2, and tfp6, which possibly encode thioredoxin-like proteins, restore and ameliorate the phenotype to a mutant trx E. coli strain in response to oxidative stress conditions. In addition to these genes, a significant protective effect was conferred by tfp10, which possibly encodes the 2-Cys peroxiredoxin Prx. In addition to their bioinformatic putative functions associated with regulation of the redox state, protein folding and turnover, and peroxide detoxification, these genes have a general protective role that is important under oxidative stress conditions. TFPs may contribute significantly to the fitness of Leptospirillum spp. in the extreme environments in which they inhabit and thrive.
3. Discussion
The reduction of disulfide protein bonds is mediated by a variety of thiol-redox enzymes that perform a rapid and reversible thiol/disulfide exchange. Members of the Leptospirillum genus lack the protein coding genes to generate glutathione and other thiol reductants such as glutaredoxins, or any known low-molecular-weight thiols. However, they have a predicted thioredoxin-based thiol/disulfide system composed of 13 coding genes.
The results presented in this research indicate that tfp genes found in Leptospirillum sp. CF-1 are transcriptionally responsive and their regulation is linked to oxidative stress, although they have differential expression depending on the oxidative agent used. Ferric iron resulted in the significant upregulation of nine tfp genes that are possibly linked to thioredoxins, cytochrome c biogenesis, cofactor biosynthesis, and signal transduction. Leptospirillum CF-1 seems to activate a number of genes encoding thioredoxin fold proteins from both cytoplasmic and membrane compartments to maintain the thiol/disulfide balance. The mRNA levels of the genes tfp1, tfp2, and tfp6, which possibly encode thioredoxins, significantly increased under ferric ion and/or diamide stress, and all were able to restore the growth of the mutant E. coli.
The deduced role of the gene products under oxidative stress is in accordance with the increased thioredoxin activity previously observed in cell extracts of the Leptospirillum strain 5-way CG in response to exposure to iron or diamide [18]. There was also a significant increase in the mRNA level of the gene tfp13, which is predicted to encode the NADPH-dependent thioredoxin reductase, TrxR. The role of this protein in providing reducing power to thioredoxins and peroxidases has been well-described, as has its essential function in the direct reduction of ROS and the acquisition of stress tolerance in a wide range of organisms [58,59]. It may also have an important role in oxidative protection in Leptospirillum spp. The tfp10 and tfp12 genes, which possibly encode the 2-Cys Prx and a predicted cytoplasmic thiol/disulfide protein, were not upregulated when the strain CF-1 was exposed to ferric iron. Nevertheless, they allowed significant growth of the mutant E. coli strain when it was exposed to a sub-lethal concentration of ferric iron, which suggests these proteins have a protective role.
In contrast to the generalized upregulation of genes under ferric exposure, the mRNA levels of tfp genes in Leptospirillum CF-1 exposed to diamide stress indicate that only four genes were upregulated. Interestingly, the genes tfp1 and tfp2, which were upregulated, were also functional in E. coli as they complemented the thioredoxin-deficient strain JEM-136. The results also suggest a discrete involvement of 2-Cys peroxiredoxin (TFP10) in disulfide stress defense. Strikingly, most of genes for thioredoxin fold proteins were not significantly affected by exposure to diamide stress. However, they may confer a significant level of protection to the thioredoxin-deficient E. coli strain. This protection was significantly higher than that conferred by the endogenous trxA gene.
In our previous study [18], the genes tfp1, tfp6, and tfp13 of the strain 5-way CG were significantly upregulated by exposure to diamide, suggesting that this strain is more tolerant and active than the CF-1 strain when exposed to disulfide stress elicitors. A possible explanation to the reduced mRNA levels of these genes in response to diamide exposure is that this results from disulfide stress overcoming cell defense because of a generalized reaction of thiols to diazenecarbonyl diamide derivatives [60]. Consequently, cellular machinery for transcription, protein biosynthesis, and other metabolic pathways are negatively affected, thus impairing the ability of the cell to properly respond to extreme environmental conditions.
Exposure to ferric ions and diamide provides two models of oxidative stress induction [18], although they present noteworthy differences. While diamide triggers disulfide stress, ferric ions have a more direct oxidative effect that leads to their reduction and the consequent generation of ferrous ions, as well as the induction of Fenton chemistry, which in turn facilitates the generation of highly harmful radical species [17]. Additionally, oxidative stress was induced in this work with ferric iron in E. coli by exposure for 15 min and further growth was evaluated in fresh medium. Diamide was added and remained in the culture medium, which is why cells were constantly exposed to the oxidant.
Different stress induction protocols were applied because ferric ions tend to generate precipitates in neutral pH culture medium that reduce their bioavailability and alter the spectrophotometric measurement of growth. Although both agents induce oxidative stress, the mechanisms involved in this induction and the induced cell protection systems obviously differ to some extent. Despite this, the expression of these genes in one condition or another allows us to infer about their participation in the defense against ferric iron and diamide, as well as general redox protection.
An interesting question is why tfp genes from the strain CF-1 restore the wild type phenotype even better than the trxA from E. coli. In a previous work, we reported that crude cell extracts from L. ferriphilum showed higher thioredoxin activity than did B. subtilis and E. coli under either standard or diamide-induced stress conditions [18]. Furthermore, E. coli activity under the control condition was so low that it could not be detected in the extract under the test conditions. The results obtained here also support the view that thioredoxins from Leptospirillum spp. are more active and therefore more effective in dealing with oxidative stress. Highly redundant and complex thioredoxin systems in other microorganisms have been described [61,62]. Whether Leptospirillum spp. possesses high intracellular concentrations and/or high levels of specific Trx protein activity remains to be elucidated.
In addition, since there is no glutathione system, it is likely that thioredoxins from this microorganism play diverse roles in cellular metabolism, some of which may partially overlap with classic functions associated with glutaredoxins or glutathione in other organisms [63]. Finally, it should be noted that we were unable to detect canonical genes for the DsbA/B and DsbC/D systems, which have been widely described in a number of bacteria as participating in the oxidation and isomerization of thiols of extra-cytoplasmatic proteins, respectively [64]. Leptospirillum spp. may contain alternative systems for these functions. Therefore, a deeper characterization of the systems that govern the formation of disulfide bonds and increase stability of proteins at low pH is missing and needs to be assessed in this acidophilic bacterium.
In conclusion, the set of tfp genes that encodes for thioredoxin fold proteins in Leptospirillum sp. CF-1 is an active and functional system. Whether these proteins can successfully replace the role of the lacking glutathione or whether a different low-molecular-weight thiol contributes to redox homeostasis in this bacterium is still an open question. However, according to our results it should be assumed that the thioredoxin-based thiol/disulfide system plays an important role in oxidative stress response and the survival of Leptospirillum spp. under the highly oxidizing conditions imposed by acidic bioleaching environments.
4. Materials and Methods
4.1. Cell Cultures
Leptospirillum strain CF-1 cultures were grown aerobically at 37 °C and 180 rpm as described previously [19]. Bacterial growth was measured by direct microscopic counting using the improved Neubaüer chamber. Escherichia coli strains (DH5α, JEM-136, MG-1655) were grown in Luria Bertani (LB) or tryptone yeast extract salts (TYES) media at 37 °C and 150 rpm. Bacterial growth was estimated by measuring optical density (O.D.) at 600 nm, as indicated.
4.2. Induction of Stress in Leptospirillum sp. CF-1
One liter of Leptospirillum sp. CF-1 culture was grown to late exponential phase. Cells were harvested by centrifugation at 9000× g for 20 min at 15 °C and suspended in 25 mL of 9K-BR medium supplemented with 18.4 g/L ferrous sulfate (Fe2SO4). Cells were exposed to oxidative stress with 260 mM ferric iron (Fe2(SO4)3) or 4 mM diamide for 1 h at 37 °C and 150 rpm. Finally, cells were collected by centrifugation and washed with fresh medium.
4.3. RNA Extraction and cDNA Synthesis
Cells harvested from 1 L of Leptospirillum sp. CF-1 culture were suspended in 400 µL of lysozyme for 2 min (3 mg/mL in Tris EDTA (TE) buffer, pH 8.0. Later, 400 µL of lysis solution (20 mM sodium acetate pH 5.5, 1 mM EDTA, 0.5% SDS) and 400 µL of Trizol reagent (TRIsure Bioline, London, UK) were added. The samples were incubated for 10 min at 65 °C. The aqueous phase, obtained after centrifugation at 16,000× g for 5 min, was chloroform-extracted and ethanol-precipitated. Finally, the pellet was suspended in 30 µL of nuclease-free water. RNA integrity was electrophoretically analyzed. RNA concentration was estimated by spectrophotometry. The cDNA was synthesized using RevertAid reverse transcriptase (Thermo Fisher Scientific, Walthman, MA, USA) following the manufacturer’s instructions.
4.4. RT-qPCR and PCR
The reaction mixture contained 10 µL of 2× Fast SYBR Kapa, 0.4 µL of each primer (10 µM), 2 µL of cDNA, 0.4 µL 50× High ROX, and 6.8 µL of nuclease-free water. The primers are listed in Table S1. The qPCR conditions were an initial denaturation (95 °C for 20 s), followed by 40 cycles of denaturation (95 °C for 3 s), annealing (58 °C for 30 s) and extension (72 °C for 30 s). The reactions were performed in the StepOne Real-Time PCR system (Applied Biosystems, Foster, CA, USA). The relative abundance of the transcripts of the genes was determined using the constitutively expressed rrsB gene (16S rDNA) [19]. The experiments were carried out in triplicate.
4.5. Cloning of tfp Genes in an Expression Vector
The tfp genes were cloned in pBADTopo TA expression vector (Thermo Fisher Scientific, Walthman, MA, USA) to generate pBADTopo-tfp. The cloning was performed using the PCR product obtained from each tfp gene with genomic DNA from Leptospirillum sp. CF-1 as a template and specific primers (Table S1). The ligation product was obtained according to manufacturer’s instructions, and then introduced in E. coli TOP10 using the CaCl2 transformation protocol. To select transformants, the transformation mix was plated on media containing 100 µg/mL ampicillin.
4.6. Determination of Minimal Inhibitory Concentration
E. coli cultures grown overnight in minimum TYES medium were adjusted to McFarland standard 0.5, equivalent to 1.5 × 108 cfu/mL. A range of concentrations was prepared for ferric sulfate and diamide solutions, and 100 µL were transferred to the first well of a 96-well microtitre plate and then serially diluted 1:1 with minimum TYES medium. Further, 50 µL of a bacterial culture were added. Cultures without oxidative stress elicitors, and TYES medium without cell culture were included as controls. The microplates were incubated overnight at 37˚C for 18 h and the absorbance at 600 nm was determined in an Elisa reader (Thermo Fisher Scientific Multiskan FC Model, Walthman, MA, USA).
4.7. Functional Assays of tfp Genes from the Strain CF-1 in E. coli
The recombinant plasmid pBADTopo-tfp gene was introduced into the E. coli strain JEM-136 (ΔtrxAΔtrxC) by chemical transformation as indicated previously. To induce tfp-gene expression, 100 µL of an overnight culture of E. coli transformant were diluted to 10 mL of TYES minimum medium. Then, cells were grown at 37 °C to an O.D.600 = 0.3, l-arabinose was added to a final concentration of 0.2% w/v, and the culture was incubated for another 5 h at 37 °C. Cultures treated with arabinose were diluted to obtain an O.D.600 = 0.1, and then transferred to a 96-well plate and exposed to 0.4 mM diamide or 0.5 mM ferric iron as oxidative-stress elicitors [18]. For diamide stress, each well had 190 µL of TYES culture medium, 10 µL diamide 8 mM, and 2 µL of cell suspension. The growth was followed by measuring absorbance at 600 nm for indicated times. For ferric iron stress, cells were exposed for only 15 min to 0.5 mM Fe2(SO4)3 dissolved in a salt medium containing 0.5 g/L 7H2O·MgSO4, 0.5 g/L CaCl2·2H2O, 4 g/L NaCl, and 20 g/L glucose. The exposure to ferric ion was limited to avoid iron precipitation of ferric ions, which tends to occur spontaneously due to the neutral pH of the culture medium. Incubation was at 37 °C and 150 rpm, and ferric iron was removed by successive washing with a salt medium. Finally, cells were suspended in 200 µL of TYES medium and cellular growth was followed as indicated above.
4.8. Sequence Alignments and Motif Search
Amino acid sequences of enzymes predicted to possess thioredoxin fold motifs were obtained from the NCBI database. Direct comparisons were made using bioinformatic resources available in Expasy (https://www.expasy.org/). Multiple sequence alignments were performed using CLUSTALW and BLASTP. Putative motif and domains were determined using NCBI-CDD, Motif, and Motif Finder. The subcellular location was determined using ngLOC, CELLO v2.5, PSORTb, and Signal-P. Transmembrane domains were predicted with TMHMM v2.0.
4.9. Statistical Analysis
The data were analyzed through the unpaired student t-test using GraphPad Prism 8.0. A value of p < 0.05 was considered as a limit of significance.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/5/1880/s1.
Author Contributions
Conceptualization: G.L. and O.O. Funding acquisition: G.L. Formal analysis: G.L., D.G., R.C. Investigation: D.G., P.Á., M.R., J.N., and R.C. G.L., D.G., and O.O. prepared the manuscript. All authors have read and agree to the published version of the manuscript.
Funding
This work was supported by Fondecyt Grants 1170799, 1150834 and 1190552 from the government of Chile to G.L and O.O., and Dicyt-Usach to G.L. and R.C. P.A. and M.R. were funded by 02 1743LJ/02 1744LJ POSTDOC and Basal project USA1799/1555/1498 from USACH.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Martin, J. Thioredoxin-a fold for all reasons. Structure 1995, 3, 245–250. [Google Scholar] [CrossRef]
- Meyer, Y.; Buchanan, B.; Vignols, F.; Reichheld, J. Thioredoxins and Glutaredoxins: Unifying elements in redox biology. Ann. Rev. Gen. 2009, 43, 335–367. [Google Scholar] [CrossRef]
- Atkinson, H.; Babbitt, P. An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput. Biol. 2009, 5, e1000541. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin superfamily in oxidative protein folding. Antioxid. Redox Signal. 2014, 21, 457–470. [Google Scholar] [CrossRef]
- Ren, G.; Stephan, D.; Xu, Z.; Zheng, Y.; Tang, D.; Harrison, R.S.; Kurz, M.; Jarrott, R.; Shouldice, S.R.; Hiniker, A.; et al. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J. Biol. Chem. 2009, 284, 10150–10159. [Google Scholar] [CrossRef]
- Netto, L.E.; Antunes, F. The Roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol. Cells 2016, 39, 65–71. [Google Scholar] [PubMed]
- Feng, X.; Sun, W.; Kong, L.; Gao, H. Distinct roles of Shewanella oneidensis thioredoxin in regulation of cellular responses to hydrogen and organic peroxides. App. Environ. Microbiol. 2019, 85, e01700-19. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Neto, J.F.; Negretto, C.C.; Netto, L.E. Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a cys-based redox sensor regulated by thioredoxin. PLoS ONE 2012, 7, e47090. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Nishiyama, Y.; Hisabori, T.; Hihara, Y. Correction: Identification of OmpR-family response regulators interacting with thioredoxin in the cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE 2015, 10, e0124571. [Google Scholar] [CrossRef] [PubMed]
- Khairnar, N.P.; Joe, M.H.; Misra, H.S.; Lim, S.Y.; Kim, D.H. FrnE, a cadmium-inducible protein in Deinococcus radiodurans, is characterized as a disulfide isomerase chaperone in vitro and for its role in oxidative stress tolerance in vivo. J. Bacteriol. 2013, 195, 2880–2886. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.; Baker, B.D.; Munson, R.S., Jr. Overlapping and complementary oxidative stress defense mechanisms in nontypeable Haemophilus influenzae. J. Bacteriol. 2015, 197, 277–285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, L.B.; Tang, L.; Ying, S.H.; Feng, M.G. Regulative roles of glutathione reductase and four glutaredoxins in glutathione redox, antioxidant activity, and iron homeostasis of Beauveria bassiana. Appl. Microbiol. Biotechnol. 2016, 100, 5907–5917. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.M.; Selim, S.; Klöck, G.; AbdElgawad, H. Sensitivity of two green microalgae to copper stress: Growth, oxidative and antioxidants analyses. Ecotoxicol. Environ. Saf. 2017, 144, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Atichartpongkul, S.; Vattanaviboon, P.; Wisitkamol, R.; Jaroensuk, J.; Mongkolsuk, S.; Fuangthong, M. Regulation of organic hydroperoxide stress response by two OhrR homologs in Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0161982. [Google Scholar] [CrossRef] [PubMed]
- Goemans, C.V.; Vertommen, D.; Agrebi, R.; Collet, J.F. CnoX is a chaperedoxin: A holdase that protects its substrates from irreversible oxidation. Mol. Cell 2018, 70, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, J.P.; Moya, F.; Covarrubias, P.; Shmaryahu, A.; Levicán, G.; Holmes, D.; Quatrini, R. Comparative genomics of the oxidative stress response in bioleaching microorganisms. Hydrometallurgy 2012, 127–128, 162–167. [Google Scholar] [CrossRef]
- Ferrer, A.; Orellana, O.; Levicán, G. Oxidative stress and metal tolerance. In Acidophiles: Life in Extremely Acidic Environments, 1st ed.; Johnson, D.B., Quatrini, R., Eds.; Horizon Scientific Press: Norfolk, UK, 2016; pp. 63–76. [Google Scholar]
- Norambuena, J.; Flores, R.; Cárdenas, J.P.; Quatrini, R.; Chávez, R.; Levicán, G. Thiol/disulfide system plays a crucial role in redox protection in the acidophilic iron-oxidizing bacterium Leptospirillum ferriphilum. PLoS ONE 2012, 7, e44576. [Google Scholar] [CrossRef] [PubMed]
- Zapata, C.; Paillavil, B.; Chávez, R.; Álamos, P.; Levicán, G. Cytochrome c peroxidase (CcP) is a molecular determinant of the oxidative stress response in the extreme acidophilic Leptospirillum sp. CF-1. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Ferrer, A.; Bunk, B.; Spröer, C.; Biedendieck, R.; Valdés, N.; Jahn, M.; Jahn, D.; Orellana, O.; Levicán, G. Complete genome sequence of the bioleaching bacterium Leptospirillum sp. group II strain CF-1. J. Biotechnol. 2016, 222, 21–22. [Google Scholar] [CrossRef]
- Lemaire, S.D.; Collin, V.; Keryer, E.; Quesada, A.; Miginiac-Maslow, M. Characterization of thioredoxin y, a new type of thioredoxin identified in the genome of Chlamydomonas reinhardtii. FEBS Lett. 2003, 543, 87–92. [Google Scholar] [CrossRef]
- Mora-García, S.; Rodríguez-Suárez, R.; Wolosiuk, R.A. Role of electrostatic interactions on the affinity of thioredoxin for target proteins. Recognition of chloroplast fructose-1, 6-bisphosphatase by mutant Escherichia coli thioredoxins. J. Biol. Chem. 1998, 273, 16273–16280. [Google Scholar] [CrossRef] [PubMed]
- Castanié-Cornet, M.P.; Bruel, N.; Genevaux, P. Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys. Acta 2014, 1843, 1442–1456. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wilson, M.A. Escherichia coli thioredoxin-like protein YbbN contains an atypical tetratricopeptide repeat motif and is a negative regulator of GroEL. J. Biol. Chem. 2011, 286, 19459–19469. [Google Scholar] [CrossRef] [PubMed]
- Uranga, L.A.; Reyes, E.D.; Patidar, P.L.; Redman, L.N.; Lusetti, S.L. The cohesin-like RecN protein stimulates RecA-mediated recombinational repair of DNA double-strand breaks. Nat. Commun. 2017, 8, 15282. [Google Scholar] [CrossRef]
- Gumpena, R.; Kishor, C.; Ganji, R.J.; Jain, N.; Addlagatta, A. Glu121-Lys319 salt bridge between catalytic and N-terminal domains is pivotal for the activity and stability of Escherichia coli aminopeptidase N. Protein Sci. 2012, 21, 727–736. [Google Scholar] [CrossRef]
- Chandu, D.; Nandi, D. PepN is the major aminopeptidase in Escherichia coli: Insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology 2003, 149, 3437–3447. [Google Scholar] [CrossRef]
- Foglino, M.; Gharbi, S.; Lazdunski, A. Nucleotide sequence of the pepN gene encoding aminopeptidase N of Escherichia coli. Gene 1986, 49, 303–309. [Google Scholar] [CrossRef]
- Ito, K.; Nakajima, Y.; Onohara, Y.; Takeo, M.; Nakashima, K.; Matsubara, F.; Ito, T.; Yoshimoto, T. Crystal structure of aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli and conformational change of methionine 260 involved in substrate recognition. J. Biol. Chem. 2006, 281, 33664–33676. [Google Scholar] [CrossRef]
- Lockwood, T.D. Redox control of protein degradation. Antioxid. Redox Signal. 2000, 2, 851–878. [Google Scholar] [CrossRef]
- Morett, E.; Saab-Rincón, G.; Olvera, L.; Olvera, M.; Flores, H.; Grande, R. Sensitive genome-wide screen for low secondary enzymatic activities: The YjbQ family shows thiamin phosphate synthase activity. J. Mol. Biol. 2008, 376, 839–853. [Google Scholar] [CrossRef]
- Mohorko, E.; Abicht, H.K.; Bühler, D.; Glockshuber, R.; Hennecke, H.; Fischer, H.M. Thioredoxin-like protein TlpA from Bradyrhizobium japonicum is a reductant for the copper metallochaperone ScoI. FEBS Lett. 2012, 586, 4094–4099. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Wang, R.S.; Handy, D.E.; Loscalzo, J. NAD(H) and NADP(H) Redox couples and cellular energy metabolism. Antioxid. Redox Signal. 2018, 28, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Iqbal, I.K.; Kumar, A. Imaging the NADH:NAD+ homeostasis for understanding the metabolic response of Mycobacterium to physiologically relevant stresses. Front. Cell Infect. Microbiol. 2016, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, M.; Florencio, F.J. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. USA 2003, 100, 16107–16112. [Google Scholar] [CrossRef] [PubMed]
- Levicán, G.; Ugalde, J.A.; Ehrenfeld, N.; Maass, A.; Parada, P. Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: Predictions and validations. BMC Genom. 2008, 9, 581–599. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Baier, M.; Horling, F.; Kahmann, U.; Harris, G.; Schürmann, P.; Dietz, K.J. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc. Natl. Acad. Sci. USA 2002, 99, 5738–5743. [Google Scholar] [CrossRef]
- Tretter, L.; Adam-Vizi, V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J. Neurosci. 2000, 20, 8972–8979. [Google Scholar] [CrossRef]
- Kranz, R.G.; Richard-Fogal, C.; Taylor, J.S.; Frawley, E.R. Cytochrome c biogenesis: Mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 2009, 73, 510–528. [Google Scholar] [CrossRef]
- Levicán, G.; Gómez, M.J.; Chávez, R.; Orellana, O.; Moreno-Paz, M.; Parro, V. Comparative genomic analysis reveals novel facts about Leptospirillum spp. cytochromes. J. Mol. Microbiol. Biotechnol. 2012, 22, 94–104. [Google Scholar] [CrossRef]
- Lübben, M.; Morand, K. Novel prenylated hemes as cofactors of cytochrome oxidases. Archaea have modified hemes A and O. J. Biol. Chem. 1994, 269, 21473–21479. [Google Scholar]
- Opoku-Temeng, C.; Sintim, H.O. Targeting c-di-GMP signaling, biofilm formation, and bacterial motility with small molecules. Methods Mol. Biol. 2017, 1657, 419–430. [Google Scholar] [PubMed]
- Díaz, M.; Castro, M.; Copaja, S.; Guiliani, N. Biofilm formation by the acidophile bacterium Acidithiobacillus thiooxidans involves c-di-GMP pathway and pel exopolysaccharide. Genes (Basel) 2018, 9, E113. [Google Scholar] [CrossRef] [PubMed]
- Cruz, D.P.; Huertas, M.G.; Lozano, M.; Zárate, L.; Zambrano, M.M. Comparative analysis of diguanylate cyclase and phosphodiesterase genes in Klebsiella pneumoniae. BMC Microbiol. 2012, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Green, J.; Stapleton, M.R.; Smith, L.J.; Artymiuk, P.J.; Kahramanoglou, C.; Hunt, D.M.; Buxton, R.S. Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: Adaptation for different ecological niches. Curr. Opin. Microbiol. 2014, 18, 1–7. [Google Scholar] [CrossRef]
- Chi, A.; Valenzuela, L.; Beard, S.; Mackey, A.J.; Shabanowitz, J.; Hunt, D.F.; Jerez, C.A. Periplasmic proteins of the extremophile Acidithiobacillus ferrooxidans: A high throughput proteomics analysis. Mol. Cell Proteom. 2007, 6, 2239–2251. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, J.; Zhang, N.; Yuan, W.; Lin, Z.; Huang, W. Characterization and analysis of a novel diguanylate cyclase PA0847 from Pseudomonas aeruginosa PAO1. Infect. Drug Resist. 2019, 12, 655–665. [Google Scholar] [CrossRef]
- Huang, C.J.; Wang, Z.C.; Huang, H.Y.; Huang, H.D.; Peng, H.L. YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43. PLoS ONE 2013, 8, e66740. [Google Scholar] [CrossRef]
- Buensuceso, R.N.; Nguyen, Y.; Zhang, K.; Daniel-Ivad, M.; Sugiman-Marangos, S.N.; Fleetwood, A.D.; Zhulin, I.B.; Junop, M.S.; Howell, P.L.; Burrows, L.L. The conserved tetratricopeptide repeat-containing c-terminal domain of Pseudomonas aeruginosa FimV is required for its cyclic amp-dependent and -independent functions. J. Bacteriol. 2016, 198, 2263–2274. [Google Scholar] [CrossRef]
- Kornberg, A.; Rao, N.N.; Ault-Riché, D. Inorganic polyphosphate: A molecule of many functions. Annu. Rev. Biochem. 1999, 68, 89–125. [Google Scholar] [CrossRef]
- Orell, A.; Navarro, C.A.; Rivero, M.; Aguilar, J.S.; Jerez, C.A. Inorganic polyphosphates in extremophiles and their possible functions. Extremophiles 2012, 16, 573–583. [Google Scholar] [CrossRef]
- Ishige, K.; Zhang, H.; Kornberg, A. Polyphosphate kinase (PPK2), a potent polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. USA 2002, 99, 16684–16688. [Google Scholar] [CrossRef] [PubMed]
- LaPointe, C.F.; Taylor, R.K. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 2000, 275, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Lewenza, S.; Gardy, J.L.; Brinkman, F.S.; Hancock, R.E. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 2005, 15, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Z.; Donnenberg, M.S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol. Microbiol. 1996, 21, 787–797. [Google Scholar] [CrossRef]
- Prieto-Alamo, M.J.; Jurado, J.; Gallardo-Madueno, R.; Monje-Casas, F.; Holmgren, A.; Pueyo, C. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 2000, 275, 13398–13405. [Google Scholar] [CrossRef]
- Cha, J.Y.; Kim, J.Y.; Jung, I.J.; Kim, M.R.; Melencion, A.; Alam, S.S.; Yun, D.J.; Lee, S.Y.; Kim, M.G.; Kim, W.Y. NADPH-dependent thioredoxin reductase A (NTRA) confers elevated tolerance to oxidative stress and drought. Plant Physiol. Biochem. 2014, 80, 184–191. [Google Scholar] [CrossRef] [PubMed]
- McCarver, A.C.; Lessner, F.H.; Soroeta, J.M.; Lessner, D.J. Methanosarcina acetivorans utilizes a single NADPH-dependent thioredoxin system and contains additional thioredoxin homologues with distinct functions. Microbiology 2017, 163, 62–74. [Google Scholar] [CrossRef]
- Kosower, N.S.; Kosower, E.M. Diamide: An oxidant probe for thiols. Methods Enzymol. 1995, 251, 123–133. [Google Scholar]
- Reott, M.A.; Parker, A.C.; Rocha, E.R.; Smith, C.J. Thioredoxins in redox maintenance and survival during oxidative stress of Bacteroides fragilis. J. Bacteriol. 2009, 191, 3384–3391. [Google Scholar] [CrossRef]
- McCarver, A.C.; Lessner, D.J. Molecular characterization of the thioredoxin system from Methanosarcina acetivorans. FEBS J. 2014, 281, 4598–4611. [Google Scholar] [CrossRef] [PubMed]
- Berndt, C.; Lillig, C.H. Glutathione, glutaredoxins, and iron. Antioxid. Redox Signal. 2017, 27, 1235–1251. [Google Scholar] [CrossRef] [PubMed]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).