Abstract
Antimicrobial immune response is mediated by a signal-transducing sensor, peptidoglycan recognition protein-SA (PGRP-SA), that can recognize non-self molecules. Although several studies have focused on the involvement of Drosophila PGRP-SA in antimicrobial peptide (AMP) expression in response to infections, studies on its role in Tenebrio molitor are lacking. Here, we present a functional analysis of T. molitor PGRP-SA (TmPGRP-SA). In the absence of microbes, TmPGRP-SA was highly expressed in the late-larval fat body, followed by hemocytes, and gut. Interestingly, following Escherichia coli, Staphylococcus aureus, and Candida albicans infections, the mRNA level of TmPGRP-SA was significantly upregulated in both the fat body and gut. TmPGRP-SA silencing had a significant effect on the mortality rates for all the microbes tested. Moreover, TmPGRP-SA is required for regulating the expression of eight AMP genes namely TmTenecin-1, -2, and -4; TmDefensin-1 and -2; TmColeoptericin-1; and TmAttacin-1b and -2 in the fat body in response to E. coli and S. aureus infections. TmPGRP-SA is essential for the transcription of TmTenecin-2, -4; TmDefensin-2; TmColeoptericin-1, -2; and TmAttacin-1a, -1b, and -2 in the gut upon E. coli and C. albicans infections. However, TmPGRP-SA does not regulate AMP expression in the hemocytes. Additionally, TmDorsal isoform X2, a downstream Toll transcription factor, was downregulated in TmPGRP-SA-silenced larval fat body following E. coli and S. aureus challenges, and in the gut following E. coli and C. albicans challenges.
1. Introduction
Innate immunity serves to limit a wide variety of molecules derived from infectious pathogens and neutralizes their virulence through germline-encoded pattern recognition receptors termed pathogen recognition receptors (PRRs). PRRs expressed on host immune cells detect highly conserved microbial surface-derived molecules referred to as pathogen-associated molecular patterns (PAMPs) that are vital for microbial fitness and survival [1,2,3].
Among all the microbe-derived immune elicitors, peptidoglycans (PGNs) are major virulence components of Gram-positive and Gram-negative bacteria, and are structurally divided into two groups, lysine (Lys) type and meso-diaminopimelic acid (DAP) type PGNs [4]. PGNs are unique and well-conserved targets that trigger innate immune responses by binding to members of the PGN recognition protein (PGRP) family in the host. Members of the PGRP family are classified into two types, PGRP-S (short) and PGRP-L (long) [5]. In insects as well as mammals, the short PGRPs and carboxyl-terminal of long PGRPs share conserved amino acids that are homologous to bacteriophage type 2 amidases and T7 lysozymes which are known as amidase type-2 domain or PGRP domain [6]. Early evolutionary separation of insect PGRPs resulted in their categorization into catalytic and non-catalytic PGRPs [7,8]. The former class shares a conserved three-amino-acid structure that cleaves the amide bond between N-acetylmuramic acid (MurNAc) and L-alanine (L-Ala) of PGNs [9,10]. The latter (sensors) bind to PGNs but do not hydrolyze PGNs due to a missing cysteine residue. Instead they activate the intracellular Toll and immune deficiency (IMD) signaling pathways and proteolytic cascades [11,12].
In Drosophila, both types of PGNs are recognized by the PGRPs to subsequently activate the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-dependent signaling cascades. The Toll pathway is activated through recognition of lys-type PGNs which trigger the Toll/Dorsal signaling, whereas the IMD pathway is triggered through detection of DAP-type PGNs which promote the activation of IMD/Relish signaling, and ultimately results in antimicrobial peptides (AMPs) production [13,14]. In this context, 13 PGRP genes from Drosophila including DmPGRP-SA, -SB1, -SB2, -SC1a, -SC1b, -SC2, and -SD (PGRP-S) and DmPGRP-LA, -LB, -LC, -LD, -LE, and -LF (PGRP-L) have been characterized [5].
Phylogenetic studies have inferred that PGRP-S plausibly evolved or originated from a common ancestral gene comprising two introns, and that one (DmPGRP-SB2) or both (DmPGRP-SB1, -SC1a and 1b, -SC2, SD) introns were lost during evolution. In contrast to other PGRP-S, Drosophila PGRP-SA (DmPGRP-SA; 20-kDa) retains both introns in the PGRP domain [15]. Furthermore, the presence of an amino-terminal signal peptide indicates that DmPGRP-SA is an extracellular recognition (secreted) protein expressed in the hemolymph [5]. Further, almost all PGRP-S contain two conserved cysteine residues in close proximity that can be linked by disulfide bridges, which are necessary for the three-dimensional conformation of the PGRP domain. Strikingly, a transition mutation (guanine → adenine) in DmPGRP-SA led to the substitution of one cysteine (Cys80 → Tyr80) that is conserved in <90% of the PGRPs, and affects Toll activation in response to infection with Gram-positive bacteria [16].
A complex of two PRRs, namely DmPGRP-SA and Gram-negative binding protein-1 (DmGNBP-1) is required for recognition of Gram-positive bacteria and for triggering the Toll signaling pathway [17,18,19]. DmGNBP-1 hydrolyzes the PGN resulting in free reducing ends of MurNAc, which are further sensed by DmPGRP-SA [20]. It was previously assumed that DmPGRP-SA lacked enzymatic function and served merely to sense the Lys- and DAP-type PGNs upstream of the intracellular Toll pathway. However, further studies revealed that DmPGRP-SA has an intrinsic L,D-carboxypeptidase activity towards the DAP-type PGNs due to Ser158 and His15 residues [21,22]. Consistent with this finding, recent studies on PGRP-SA from Bombus ignitus, Apis mellifera, and Megachile rotundata revealed preferential binding of PGRP-SA to DAP-type PGNs than to Lys-type PGNs [23].
The mealworm beetle, Tenebrio molitor, presents some advantages when compared to the other invertebrate models (e.g., D. melanogaster) with respect to studying pathogenic infections and immune responses. For instance, the heat tolerance feature of the insect enables it to be maintained at 37 °C, which is an environmental cue for the expression of virulence factors in many pathogens. This makes T. molitor a great model to study host-pathogen interactions [24,25].
In the past decade, several extracellular and intracellular events of the T. molitor Toll signaling pathway have been investigated. In T. molitor, the Lys-type PGNs, DAP-type PGNs, and β-1, 3-glucan are sensed by the TmPGRP-SA/TmGNBP-1 complex and the Gram-negative binding protein 3 (TmGNBP3), respectively, following which they trigger proteolytic cascades which lead to the extracellular cleavage of the Toll endogenous ligand spätzle (TmSpz). The active form of TmSpz binds to the ectodomain of the transmembrane Toll receptors (e.g., TmToll-like receptor 7 [TmToll-7]) resulting in Toll-dependent AMP production [26,27,28,29]. In T. molitor, nine TmSpz ligands including TmSpz-1b, -3, -4, -5, -6, -7, -7a, -7b, 7b, and -like have been identified (unpublished data) [30]. However, the effect of TmPGRP-SA expression on each of the TmSpz genes remains unclear. Concordantly, the intracellular protein cassettes of the Toll pathway comprising TmMyD88 [31], Pelle, and Tube are essential for transducing the signal to TmCactin [32], a positive regulator that interacts with TmCactus, for ensuring the nuclear translocation of TmDorX2 [33] and robust transcription of the TmAMP gene. In agreement with this, earlier immune studies in other invertebrates such as D. melanogaster, Manduca sexta, and Marsupenaeus japonicus have revealed the same mechanism of indirect Toll activation [22,34,35].
In vitro and in vivo studies have primarily addressed the role of TmPGRP-SA as an innate immune recognition molecule that initiates the prophenoloxidase (proPO) cascade as well as the Toll signaling pathway [27,36]. However, the tissue-specific role of TmPGRP-SA in inhibiting the Toll pathway needs to be elucidated. In this study, we sought to understand the functional role of extracellular TmPGRP-SA in the survival and AMP gene expression of T. molitor larvae in response to E. coli, S. aureus, and C. albicans challenges using RNA interference (RNAi). Furthermore, we analyzed the expression pattern of NF-κB genes in T. molitor larvae following TmPGRP-SA silencing and infection with the aforesaid microbes.
2. Results
2.1. Gene Organization, cDNA Analysis, and Phylogenetic Tree of TmPGRP-SA
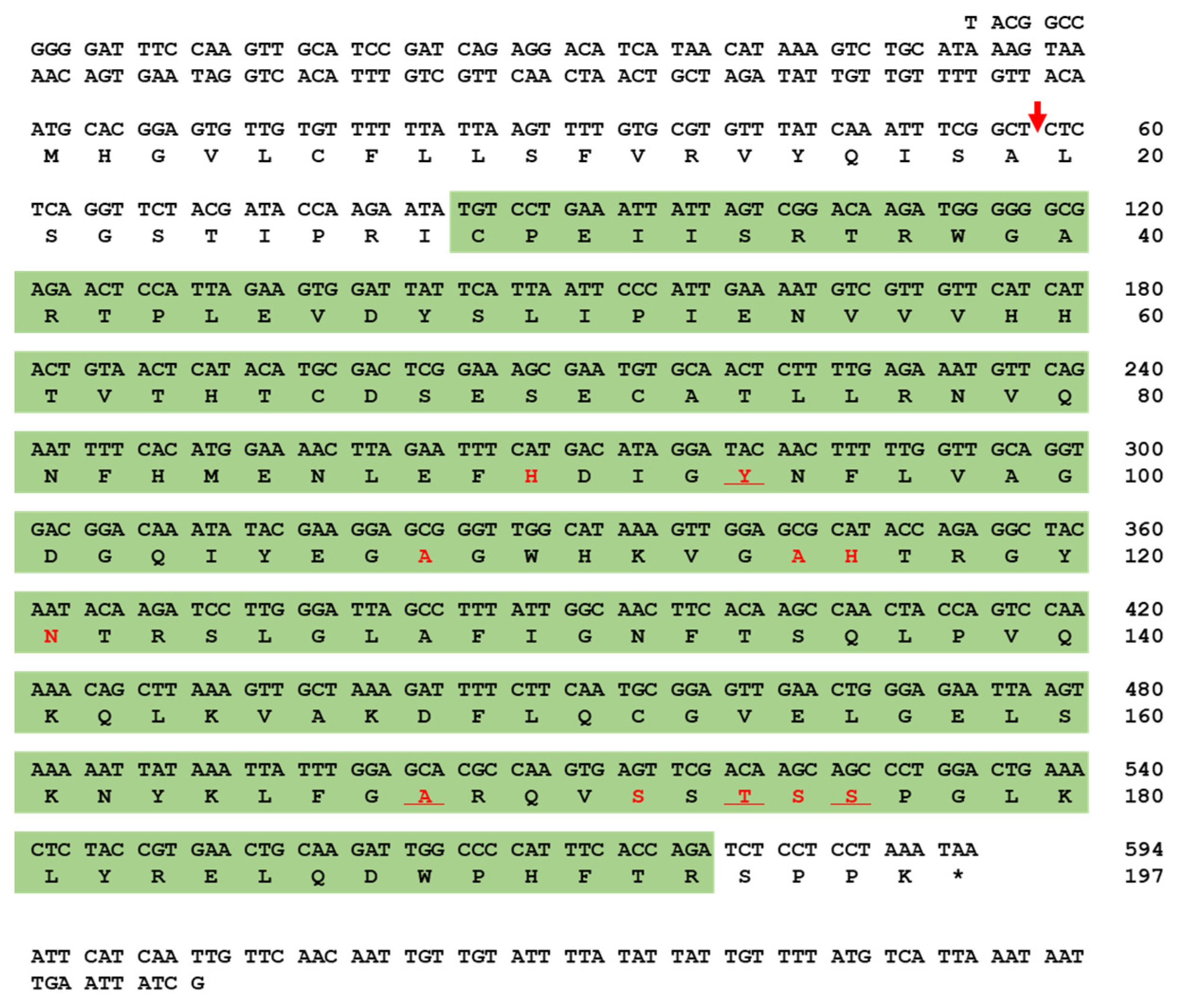
The full-length open reading frame (ORF) of TmPGRP-SA consists of 594 base pairs (bps), and encodes a polypeptide of 197 amino acids (aa). SignalP-5.0 analysis showed that TmPGRP-SA contains a signal peptide at the N-terminal and with cleavage site predicted between the amino acid residues at position 19 and 20. The deduced TmPGRP-SA protein contains a N-acetylmuramoyl-L-alanine amidase/PGRP domain (C29 to R193), based on the predictions from InterProScan 5.0 and BLASTx analyses (Figure 1). SWISS-MODEL and PyMOL visualized the amidase catalytic site (active site) and chemical (substrate) binding site (Figure S2).
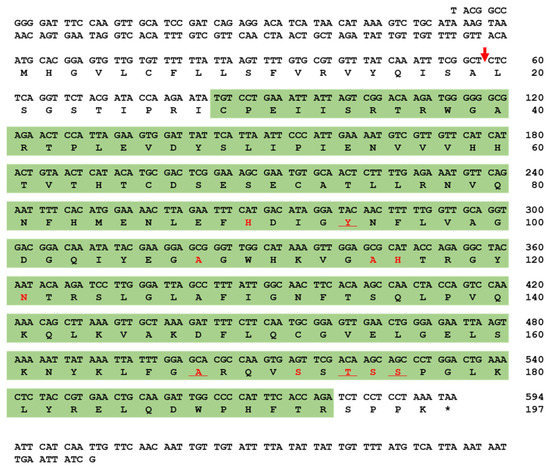
Figure 1.
Nucleotide and deduced amino acid sequence of T. molitor peptidoglycan recognition protein-SA (TmPGRP-SA). The nucleotide and amino acid sequence numbers are shown at the right margin, indicating that full-length open reading frame (ORF) sequence of TmPGRP-SA consists of 594 nucleotides encoding 197 amino acid residues. The predicted signal peptide cleavage site is marked with a red arrow. The amidase/PGRP domain predicted by InterProScan 5.0 is highlighted in the green box. The amidase catalytic site (active site) is underlined and chemical (substrate) binding site is shown in red.
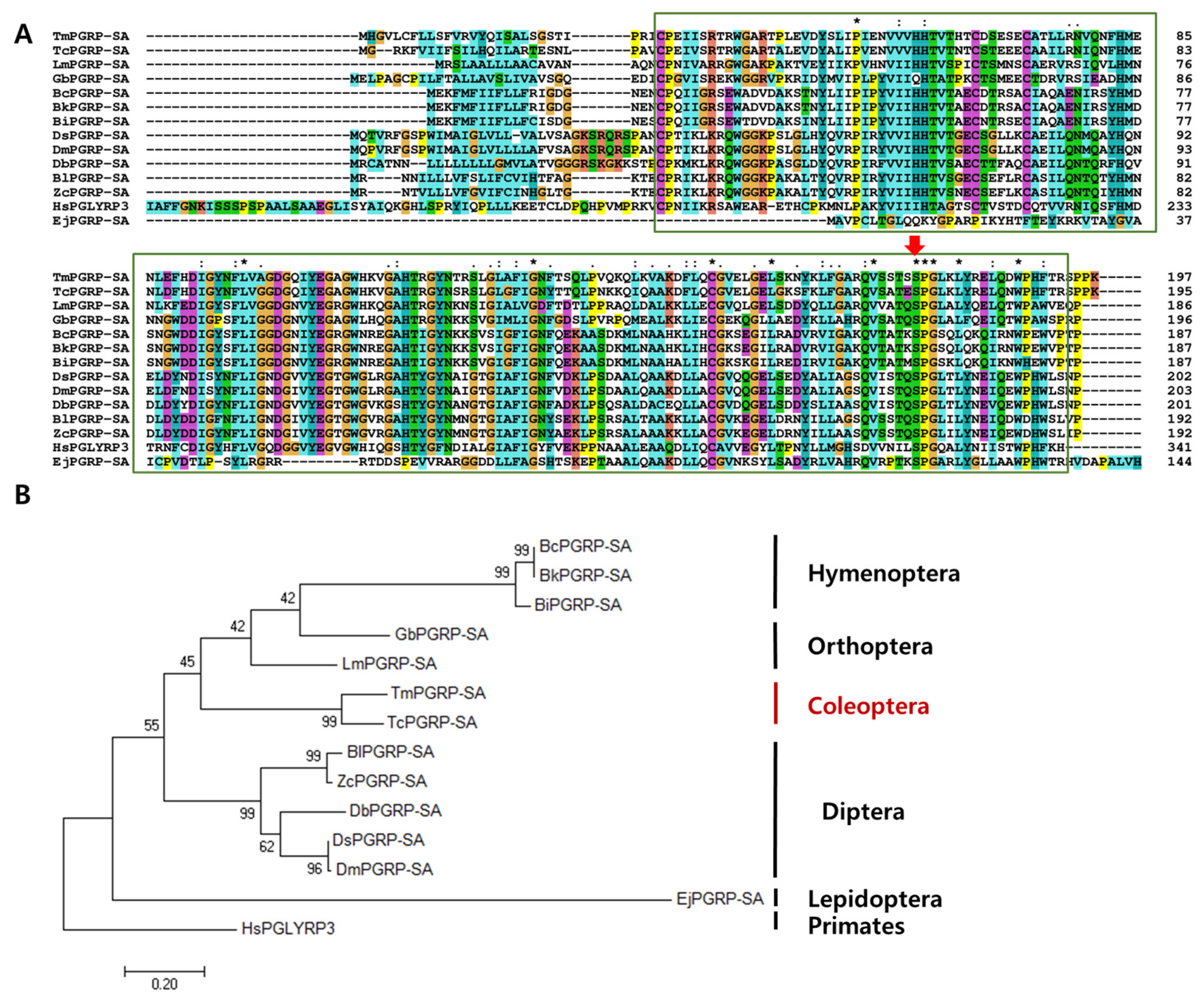
Multiple sequence alignment of TmPGRP-SA and other representative insect proteins revealed the relationship between the residues in the PGRP domains of corresponding sequences. Notably, the most conserved amino acids (displayed with asterisk) include proline, leucine, glycine, cysteine, valine, serine, and tryptophan (Figure 2A). The percent identity based on the full-length ORF of TmPGRP-SA and that from other insects indicated that TmPGRP-SA was highly similar to PGRP-SA from Tribolium castaneum (76% similarity), followed by 50% and 43% identity with the Orthopterans, Locusta migratoria (LmPGEP-SA) and Gryllus bimaculatus (GbPGRP-SA), respectively. Furthermore, a minimum and maximum similarity of TmPGRP-SA with Hymenoptera (37–38%) and Diptera (43–45%) orders was also noted, suggesting that TmPGRP-SA shares higher similarity with Diptera than Hymenoptera. The least similarity was observed with the order Lepidoptera (27%) (Figure S3).
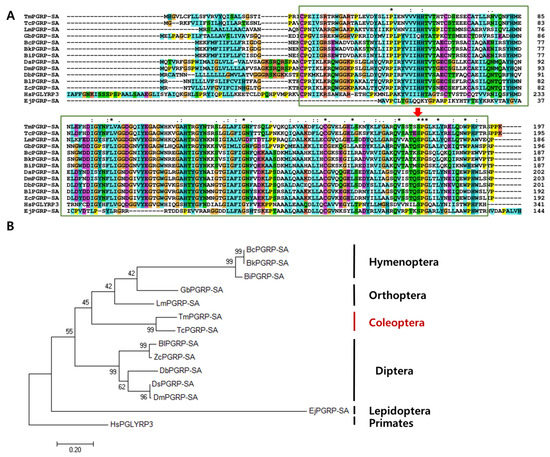
Figure 2.
Phylogenetic analyses and alignment of TmPGRP-SA amino acid sequence with those from other species. The amidase/PGRP domain is marked in green boxes. Asterisk, colon, and period symbols indicate conservation scores between the sequences of representative PGRP-SA proteins according to the Gonnet PAM 250 matrix (‘*’ > ‘:’ > ‘.’) and ‘-’ indicates internal or terminal gaps. The Red Green Blue (RGB) colors are identified based on RGB values (from 0 to 255). Red color shows 229, 51, and 25 RGB values, blue (25, 127, and 229), green (25, 204, and 25), cyan (25, 178, and 178), pink (299, 127, and 127), magenta (204, 76, and 204), yellow (204, 204, and 0), and orange (229, 153, and 76) (A). Red arrow indicates the conserved serine amino acids. Phylogenetic tree constructed using maximum likelihood (ML) method based on the Jones-Taylor-Thornton (JTT) matrix model in MEGA 7.0 (1000 bootstrap replicates). The MT tree were rooted with the protein sequences of TmPGRP-SA (Tenebrio molitor peptidoglycan recognition protein SA), TcPGRP-2 (T. castaneum REDICTED: peptidoglycan recognition protein 2; P_008192927.1), LmPGRP-SA (Locusta migratoria peptidoglycan recognition protein SA; AFD54029.1), BcPGRP-SA (Bombus consobrinus peptidoglycan recognition protein SA; ATL64828.1), BkPGRP-SA (Bombus koreanus peptidoglycan recognition protein SA; ATL64813.1), GbPGRP-SA (Gryllus bimaculatus peptidoglycan recognition protein SA; BBG28438.1), DsPGRP-SA (Drosophila simulans peptidoglycan recognition protein SA; XP_002106687.1), DbPGRP-SA (Drosophila busckii peptidoglycan recognition protein SA; XP_002106687.1), EjPGRP-SA (Eumeta japonica peptidoglycan recognition protein SA; GBP17419.1), DmPGRP-SA (Drosophila melanogaster peptidoglycan recognition protein SA; CAD89124.1), BlPGRP-SA (Bactrocera latifrons peptidoglycan recognition protein SA; JAI23539.1), ZcPGRP-SA (Zeugodacus cucurbitae peptidoglycan recognition protein SA; JAD13283.1), BiPGRP-SA (Bombus ignites peptidoglycan recognition protein SA; ATL64812.1). HsPGLYRP3 (Homo sapiens peptidoglycan recognition protein 3; AAI28116.1) was used as an outgroup (B).
An ML tree was constructed based on the protein sequences of PGRP-SA from twelve representative insect species and one human homolog (outgroup) (Figure 2B). The phylogenetic tree showed that the PGRP-SA isoforms from T. molitor and T. castaneum clustered together, and that TmPGRP-SA had a close evolutionary position with respect to LmPGEP-SA and GbPGRP-SA (Orthoptera). Additionally, all Diptera and Hymenoptera species were grouped into two independent clusters. Phylogenetic analysis of the selected Diptera PGRP-SA protein sequences depicted two separate clusters, one formed by Drosophila (DmPGRP-SA (D. melanogaster), DsPGRP-SA (D. simulans), DbPGRP-SA (D. busckii)) and another by other flies (BlPGRP-SA (B. latifrons), ZcPGRP-SA (Z. cucurbitae)) (Figure 2B).
2.2. Developmental and Tissue Distribution of TmPGRP-SA
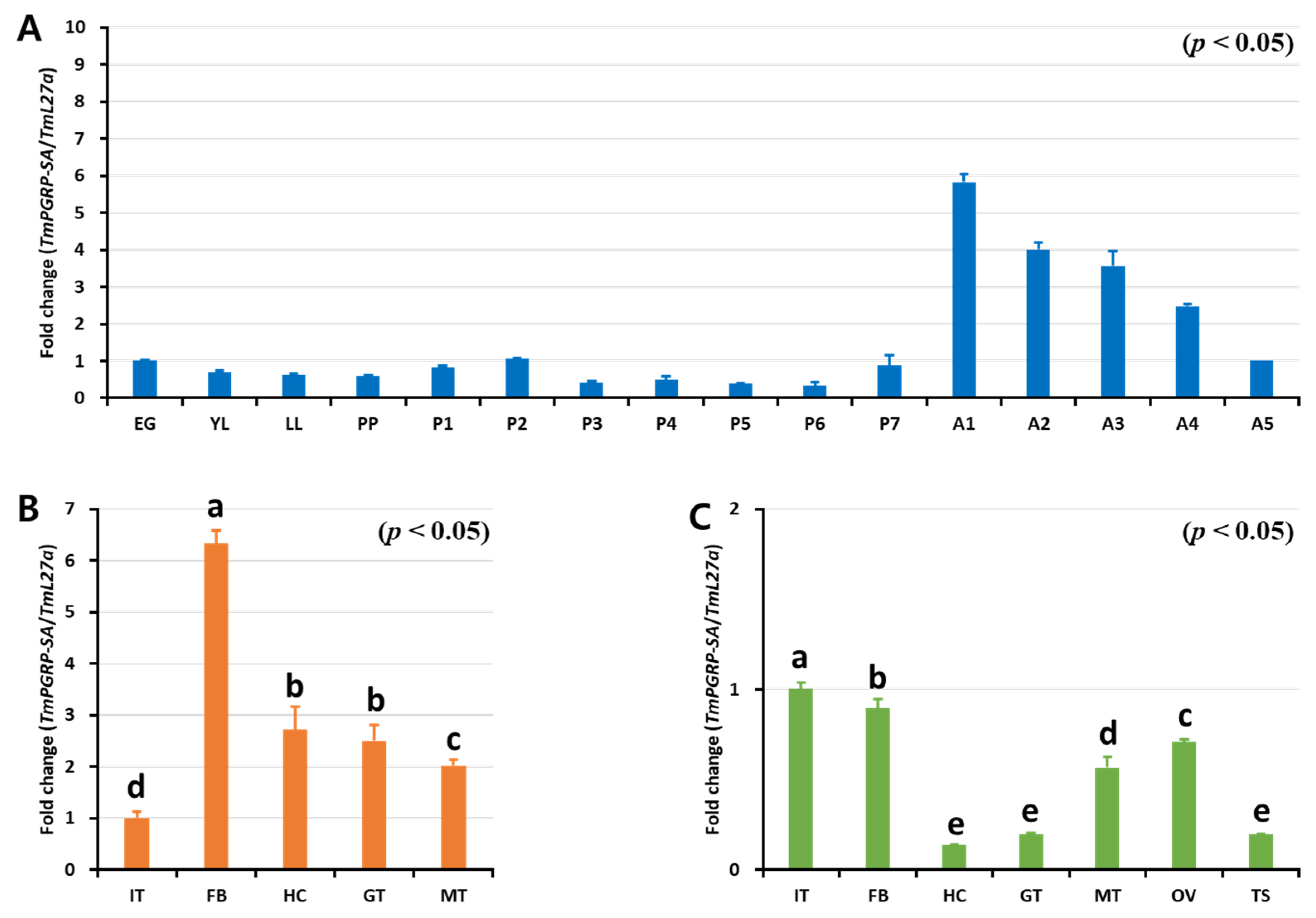
Insect PGRP-Ss (e.g., Drosophila) were previously shown to exhibit a tissue-specific expression profile, with short PGRPs being expressed almost exclusively in the fat body cells, with some expression also being detectable in the epidermal cells, gut, hemolymph, and cuticle [5]. To determine if TmPGRP-SA was active during developmental stages and in the larval or adult tissues, we sought to evaluate its development- and tissue-specific expression using RT-qPCR (Figure 3). Notably, TmPGRP-SA mRNA was detected at all the developmental stages tested. Although TmPGRP-SA expression was hardly detectable in the larval and pupal stages, its expression was highly increased in the adult stages, with the highest expression in the 1-day-old adult, followed by a rapid decline in expression in the older adults (Figure 3A). Further, we found that TmPGRP-SA mRNA was detectable in all the tissues analyzed. TmPGRP-SA showed elevated expression in the larval fat body, followed by hemocytes, gut, and Malpighian tubules. Additionally, we observed the lowest level of TmPGRP-SA mRNA expression in the integument (Figure 3B). RT-qPCR analysis of adult tissues revealed a markedly different pattern of TmPGRP-SA transcript expression in all tissues, with the highest expression detected in the integument and fat body, followed by ovary and Malpighian tubules. TmPGRP-SA mRNA was barely detectable in the adult hemocytes, gut, and testis (Figure 3C).
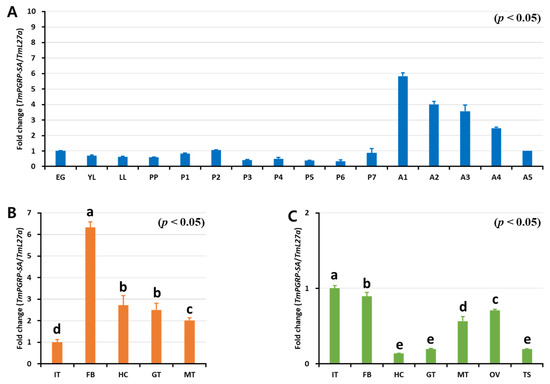
Figure 3.
Expression of TmPGRP-SA in different developmental stages and multiple tissues of late-instar larvae and 5-day-old adults. RT-qPCR transcript analysis of TmPGRP-SA at different T. molitor developmental stages. EG: eggs, YL: young larvae, LL: late-instar larvae, PP: Pre-pupa, P1 – P7: 1 to 7-day-old pupa, and A1–A5: 1 to 5-day-old adults (A). mRNA profile of TmPGRP-SA in late-instar larval tissues (IT: integument, FB: fat body, HC: hemocytes, GT: gut, and MT: Malpighian tubules) (B) and in 5-day-old adult tissues (OV: ovary and TS: testis) using RT-qPCR (C). The results were normalized to T. molitor 60S ribosomal protein L27a (TmL27a). The mean values and SE were obtained from 20 insects per group. Bars in each graph with the same letter are not significantly different from each other (p > 0.05).
2.3. TmPGRP-SA is Upregulated following Microbial Infection in vivo
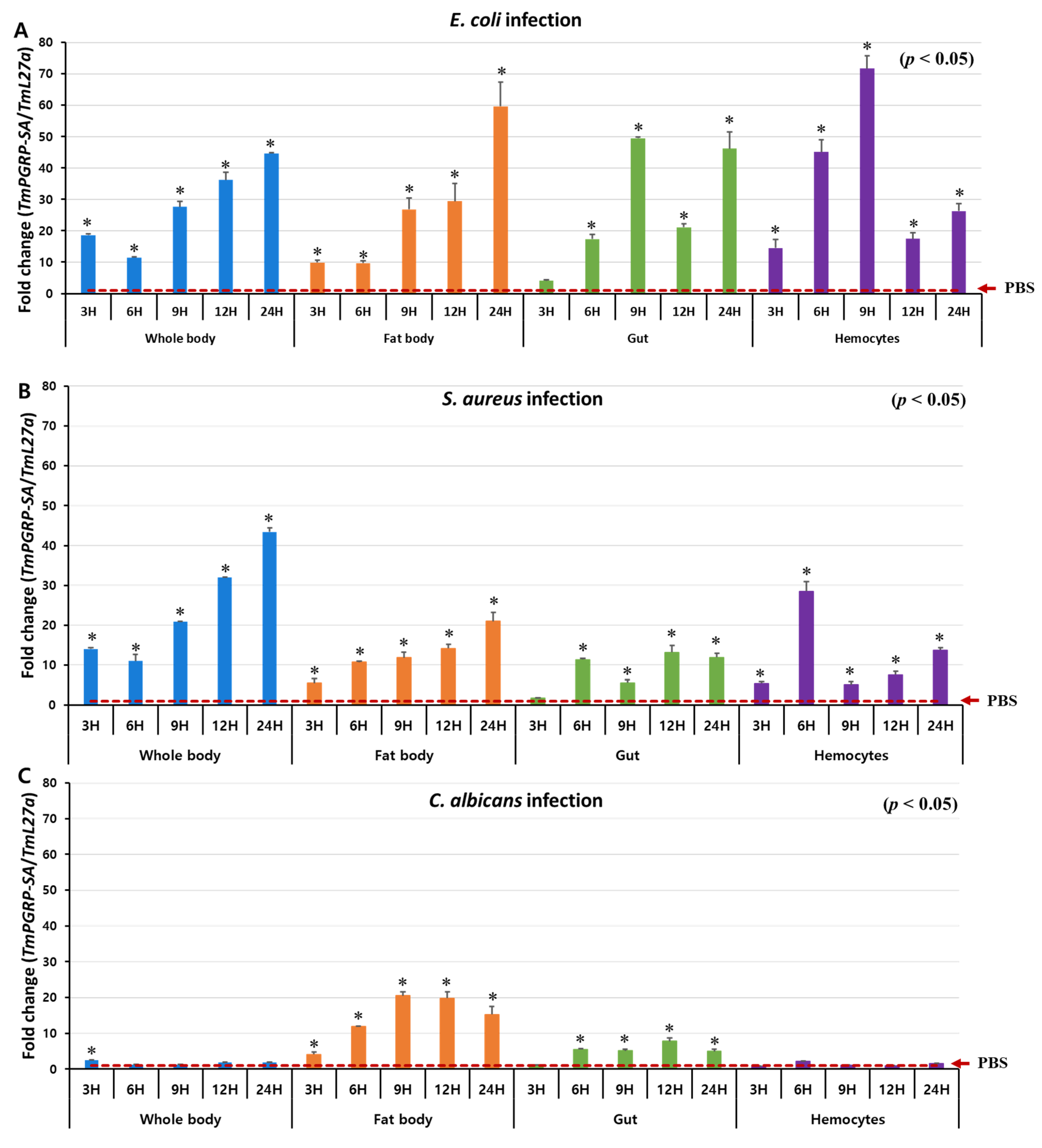
Previous studies have demonstrated that in D. melanogaster, PGRP-SA is involved in Toll-dependent immune defense against Gram-positive bacteria but not against fungal or Gram-negative bacterial infections [17]. Therefore, to investigate the involvement of TmPGRP-SA in promptly detecting various infectious pathogens, we examined the response of T. molitor larvae (whole body and multiple tissues) to infection with E. coli, S. aureus, and C. albicans at specific time points (3, 6, 9, 12, and 24 h post-challenge) (Figure 4). We observed significantly elevated levels of TmPGRP-SA mRNA when E. coli and S. aureus were injected in the whole body of T. molitor larvae (Figure 4A,B). However, TmPGRP-SA expression showed a slight but significant induction at 3 h (or no response) to the C. albicans at the other time points (p < 0.05) (Figure 4C). Upon bacterial infection, a gradual increase in the transcript levels of TmPGRP-SA leading to a 40-fold upregulation in mRNA expression was noted with respect to the PBS-injected control at 24 h post-infection (Figure 4A,B). In the larval fat body of T. molitor, infection-mediated induction of TmPGRP-SA was significantly higher than in the PBS-injected cohorts (p < 0.05) (Figure 4A–C). E. coli and S. aureus challenge moderately increased TmPGRP-SA expression in the fat body, with the highest level observed at 24 h (Figure 4A,B). In the gut, induction of TmPGRP-SA mRNA by E. coli was stronger than in response to S. aureus and C. albicans (Figure 4A–C). Following microbial infections, induction of TmPGRP-SA in the hemocytes varied depending on the type of microbe. Whereas challenge with C. albicans did not induce TmPGRP-SA expression relative to that observed in PBS-injected cohorts (Figure 4C); exposure to the Gram-negative and Gram-positive bacteria, E. coli and S. aureus, respectively, triggered significant upregulation in TmPGRP-SA at the early time points (6 h), but did not persist at a high level at 12 and 24 h post-challenge (Figure 4A,B). Additionally, in agreement with the aforementioned results in hemocytes, high accumulation of TmPGRP-SA in the hemolymph of T. molitor following challenge with lys-type and DAP-type PGNs was previously observed [37]. These findings highlight the role of a bacterial (but not fungal) elicitor, and that of TmPGRP-SA as a sensor in the fat body, gut, and hemocytes of T. molitor larvae against E. coli and S. aureus challenges.
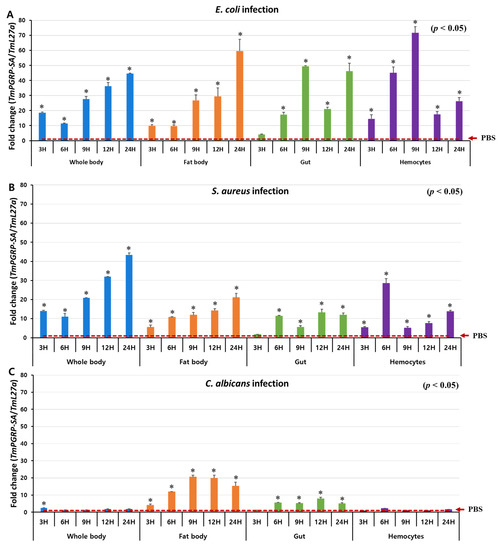
Figure 4.
TmPGRP-SA is differentially induced following microbial challenges. Relative changes in gene expression (mean ± SE) of TmPGRP-SA in the whole body, fat body, gut, and hemocytes of T. molitor (10th–12th instar) larvae challenged with E. coli (A), S. aureus (B), and C. albicans (C) at the indicated time points by RT-qPCR (n = 20 per treatment group per time point). TmPGRP-SA mRNA expression was normalized to the reference gene, T. molitor 60S ribosomal protein L27a (TmL27a), followed by normalization to the PBS-injected control mRNA expression. Statistical significance is denoted with asterisks (p < 0.05).
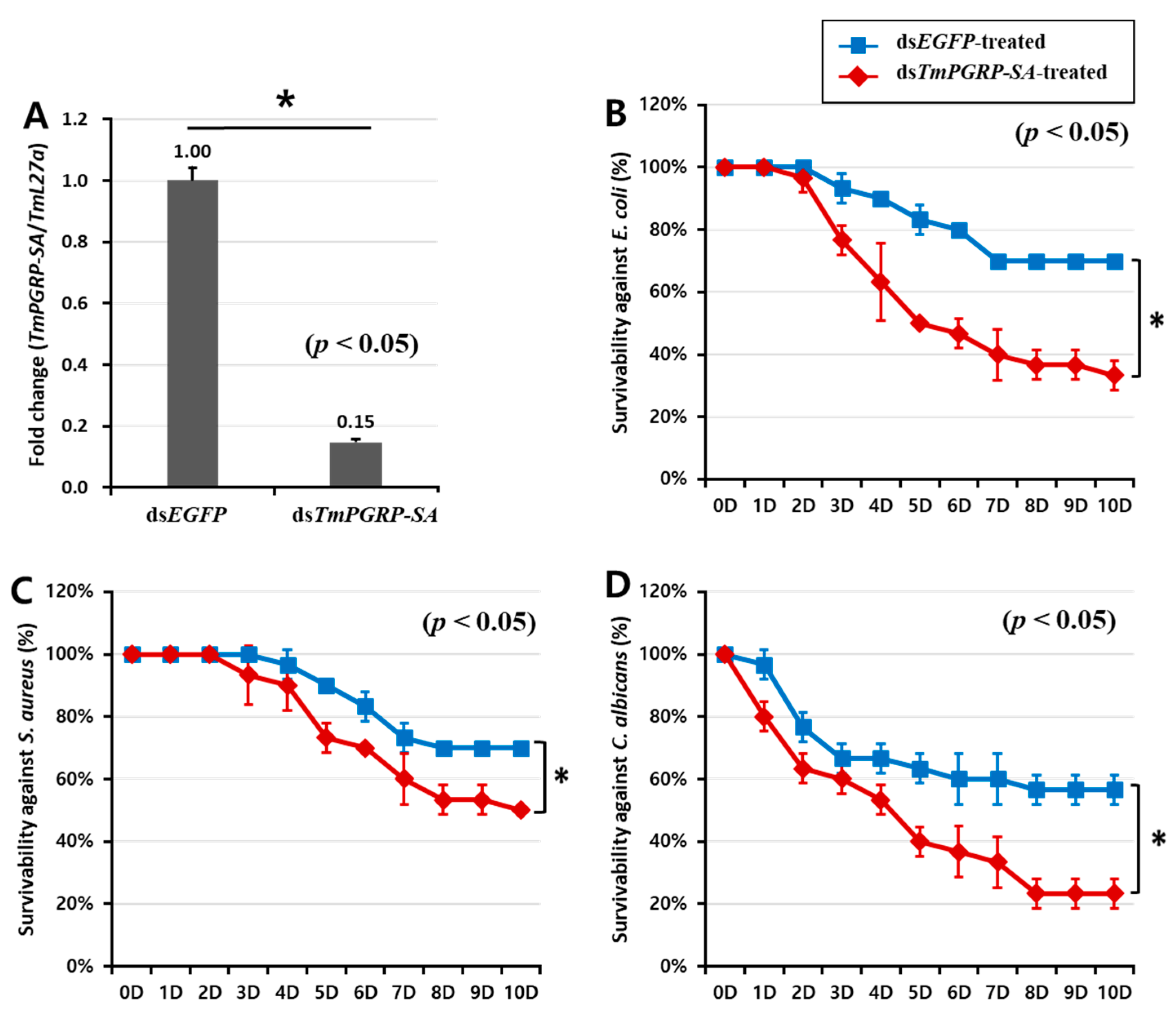
2.4. TmPGRP-SA Silenced Larvae are Susceptible to Microbial Infections
The Drosophila Toll pathway can be activated by Gram-positive Lys-type and Gram-negative DAP-type PGNs that are recognized by DmPGRP-SA [22]. As DmPGRP-SA-silenced flies are more susceptible to Gram-positive bacteria than to Gram-negative bacteria and fungi [17], we investigated whether suppression of TmPGRP-SA followed by E. coli, S. aureus, and C. albicans infections would lead to increased mortality of T. molitor larvae in a 10-day period. As shown Figure 5A, TmPGRP-SA was effectively silenced (85% reduction) on the second day post the dsRNA treatment. The percent survival of TmPGRP-SA-silenced larvae after challenges with E. coli (30%, Figure 5B) and C. albicans (20%, Figure 5D) were significantly compromised compared to that of the control larvae (70% and 60% respectively; p < 0.05). TmPGRP-SA knockdown larvae survived up to 50% compared to 70% survival of dsEGFP-treated control following S. aureus challenge (Figure 5C). These data suggest the prominent role of TmPGRP-SA in activation of immunity against microbial infections. Simultaneously, we assessed the expression of Toll-dependent genes (AMP and NF-κB genes) in TmPGRP-SA-silenced larvae following the microbial challenges.
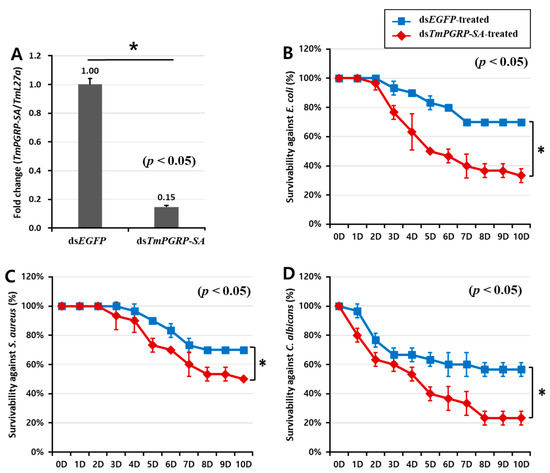
Figure 5.
The pivotal role of TmPGRP-SA in the survival of T. molitor larvae following microbial infections. Knockdown efficiency of TmPGRP-SA in T. molitor larvae (n = 3 per group) quantified by mRNA transcription after TmPGRP-SA depletion compared to dsEGFP controls carried out on the second day post-injection (80%) (A). Lifespan curves of dsEGFP- and dsTmPGRP-SA-injected larvae (n = 10 per group) following immune challenge with E. coli (B), S. aureus (C), and C. albicans (D) over a ten-day period. Results presented are representative of three independent experiments. Asterisks depict significant differences between negative control and TmPGRP-SA-silenced larvae (p < 0.05).
2.5. Induction of AMPs is Regulated by TmPGRP-SA in Immunocompetent Tissues
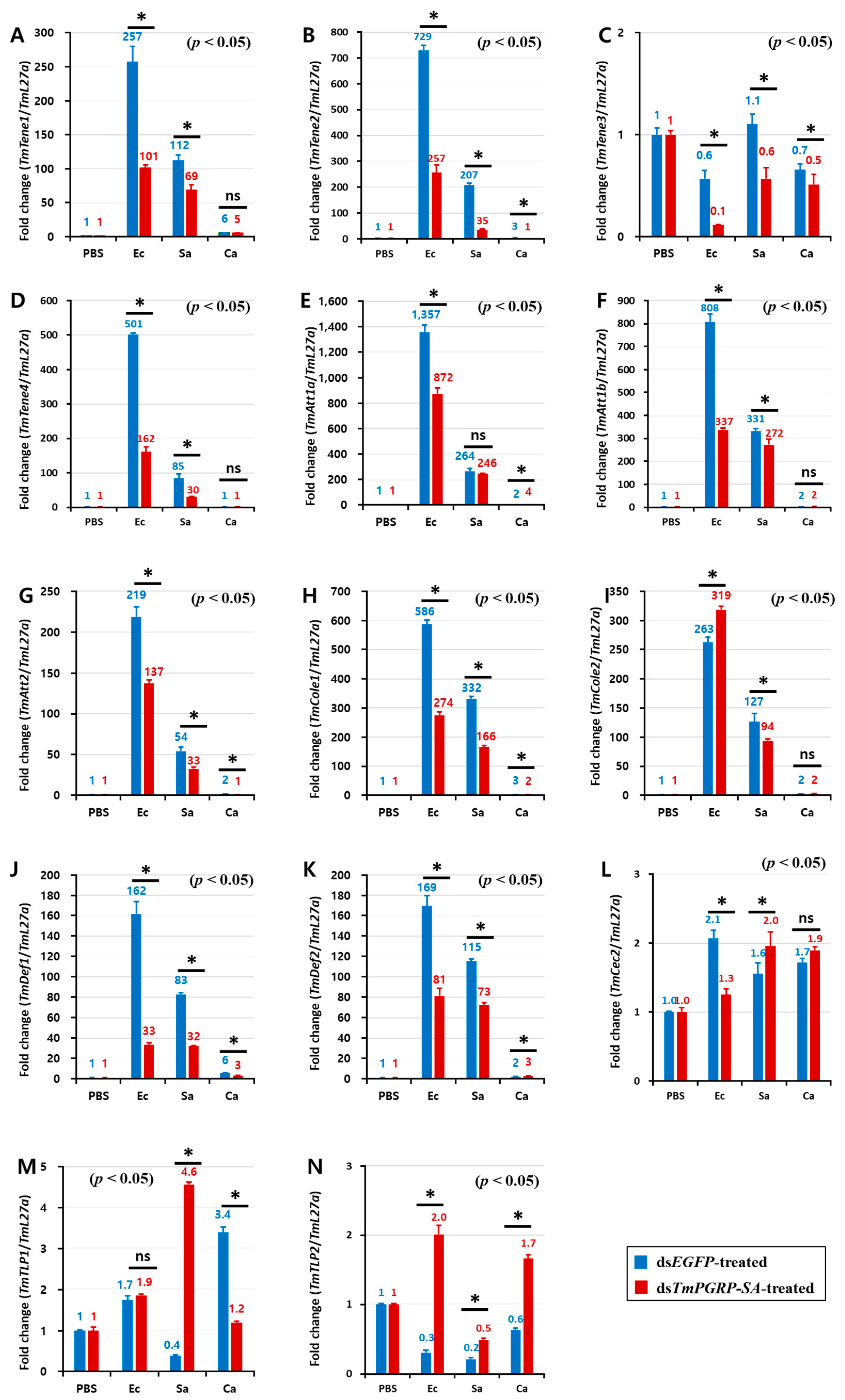
We hypothesized that the survival phenotypes of insects challenged with different microbes may be due to differential expression of AMP genes. To investigate the downstream signal transduction of TmPGRP-SA following infection which leads to NF-κB translocation and induction of immune responses, we determined the expression of 14 AMP genes in larval fat body, gut, and hemocytes of ds TmPGRP-SA- and dsEGFP-treated groups 24 h post-E. coli, -S. aureus, and -C. albicans challenges.
Infection of dsEGFP-treated larvae with E. coli induced the expression of TmTene1 (Figure 6A), TmTene2 (Figure 6B), TmTene4 (Figure 6D), TmAtt1a (Figure 6E), TmAtt1b (Figure 6F), TmAtt2 (Figure 6G), TmCole1 (Figure 6H), TmDef1 (Figure 6J), TmDef2 (Figure 6K), and TmCec2 (Figure 6L) genes in the fat body. It should be noted that a significant downregulation of all these AMPs was observed in E. coli-infected larvae in which TmPGRP-SA was silenced (p < 0.05) (Figure 6). Exposing TmPGRP-SA-depleted cohorts to S. aureus markedly decreased the expression of nine AMP genes, namely TmTene1 (Figure 6A), TmTene2 (Figure 6B), TmTene3 (Figure 6C), TmTene4 (Figure 6D), TmAtt1b (Figure 6F), TmAtt2 (Figure 6G), TmCole1 (Figure 6H), TmCole2 (Figure 6I), TmDef1 (Figure 6J), and TmDef2 (Figure 6K) relative to that in the dsEGFP-treated groups. In contrast, impairment of TmPGRP-SA expression did not affect C. albicans-induced expression of TmTene1 (Figure 6A), TmTene4 (Figure 6D), TmAtt1b (Figure 6F), TmCole2 (Figure 6I), and TmCec2 (Figure 6L). However, TmPGRP-SA silencing slightly downregulated few AMP mRNAs including TmTene2 (Figure 6B), TmAtt2 (Figure 6G), TmCole1 (Figure 6H), TmDef1 (Figure 6J), and TmTLP1 (Figure 6M) in the larval fat body following C. albicans challenge.
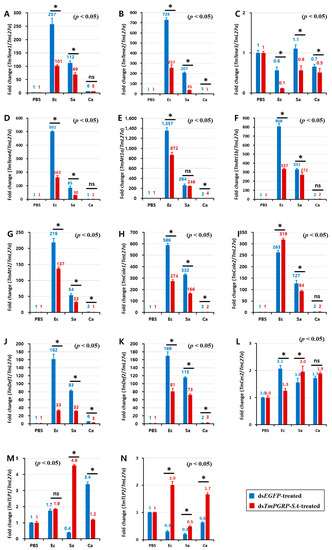
Figure 6.
TmPGRP-SA-mediated transcription induction of antimicrobial peptides is lower in the fat body of dsEGFP-injected larvae following bacterial infections. Quantitative RT-PCR was used to measure the expression of fourteen antimicrobial peptide (AMP) genes including TmTenecin-1 (TmTene1, A); TmTenecin-2 (TmTene2, B); TmTenecin-3 (TmTene3, C); TmTenecin-4 (TmTene4, D); TmAttacin-1a (TmAtt1a, E); TmAttacin-1b (TmAtt1b, F); TmAttacin-2 (TmAtt2, G); TmColeptericin-1 (TmCole1, H); TmColeptericin-2 (TmCole2, I); TmDefensin-1 (TmDef1, J); TmDefensin-2 (TmDef2, K); TmCecropin-2 (TmCec2, L); TmTLP-1 (TmTLP1, M); and TmTLP-2 (TmTLP2, N) in the TmPGRP-SA-silenced T. molitor larval fat body (n = 20 per group) in response to E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) challenges. EGFP dsRNA injection served as a negative control, and the mRNA level of the respective AMP genes are presented relative to those for TmL27a as an internal control. Significant differences between dsEGFP- and dsTmPGRP-SA-treated groups are shown with asterisks (p < 0.05); ns = not significant.
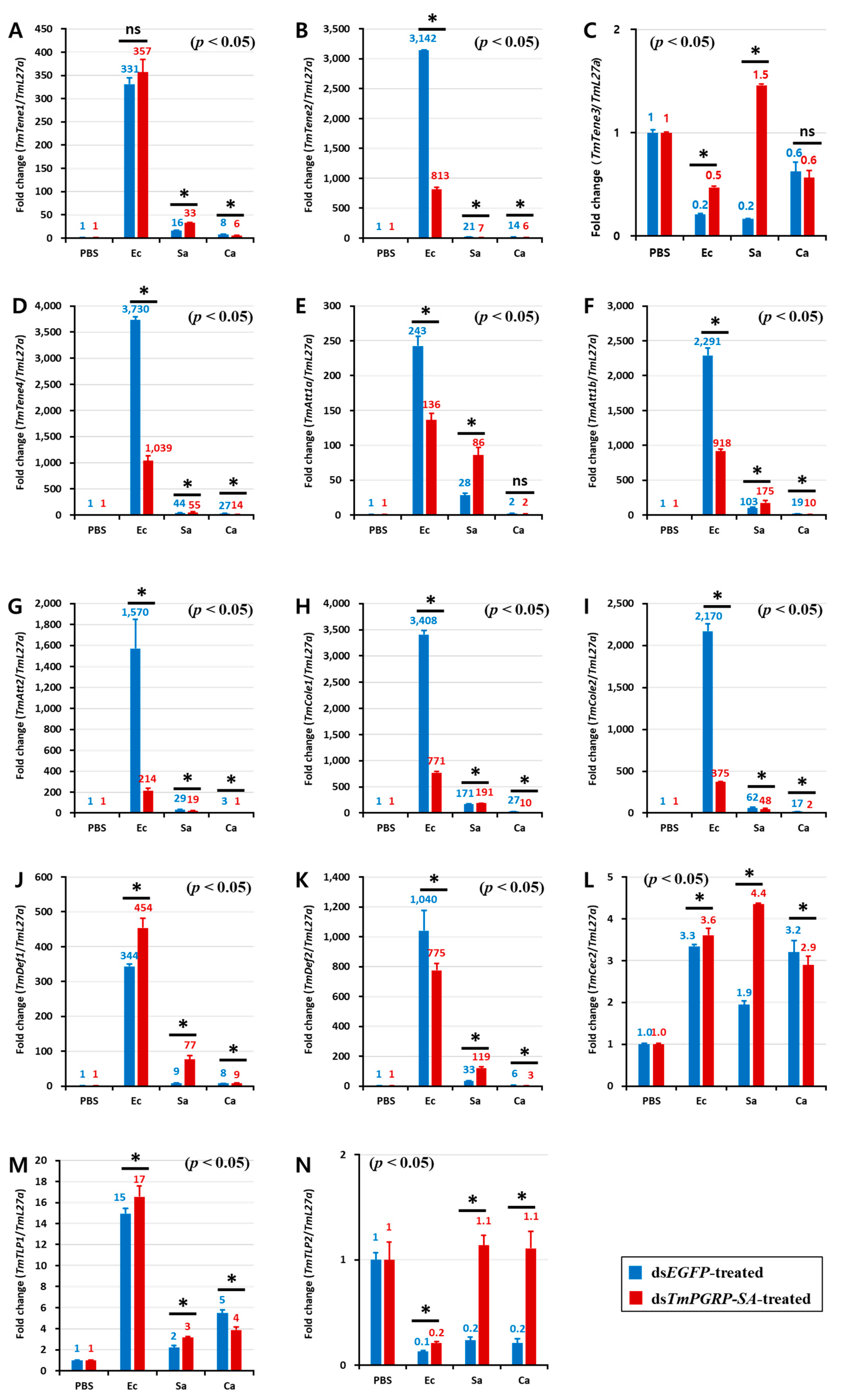
In the gut of the control (dsEGFP-treated) larvae, E. coli triggered a potent transcription of TmTene2 (Figure 7B), TmTene4 (Figure 7D), TmAtt1a (Figure 7E), TmAtt1b (Figure 7F), TmAtt2 (Figure 7G), TmCole1 (Figure 7H), TmCole2 (Figure 7I), and TmDef2 (Figure 7K). However, the gut of dsTmPGRP-SA-silenced larvae showed lower expression of all AMPs except TmTene1 (Figure 7A), TmDef1 (Figure 7J), TmCec2 (Figure 7L), and TmTLP1 (Figure 7M) following challenge with E. coli. In contrast, when infected with S. aureus, induction of almost all AMPs including TmTene1 (Figure 7A), TmTene3 (Figure 7C), TmTene4 (Figure 7D), TmAtt1a (Figure 7E), TmAtt1b (Figure 7F), TmCole1 (Figure 7H), TmDef1 (Figure 7J), TmDef2 (Figure 7K), TmCec2 (Figure 7L), TmTLP1 (Figure 7M), and TmTLP2 (Figure 7N) was significantly higher than that of the control groups in the gut (p < 0.05). Interestingly, C. albicans challenge markedly induced the mRNA levels of 11 AMP genes in the gut of dsEGFP-treated control larvae, whereas these transcripts were decreased in dsTmPGRP-SA-treated larvae (Figure 7). These results indicated that the gut responds to infections and that immune responses to E. coli and C. albicans are conveyed by TmPGRP-SA.
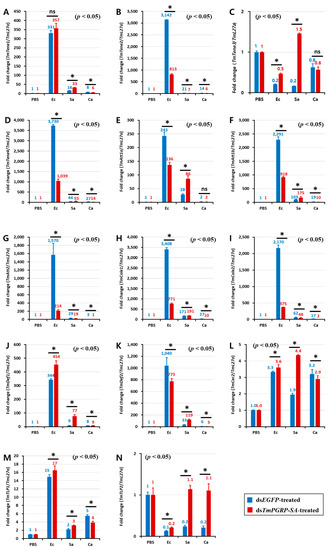
Figure 7.
Expression of TmPGRP-SA is required for the induction of antimicrobial peptides in the gut. Effect of TmPGRP-SA reduction mediated by double-stranded RNA on infection-dependent expression of fourteen AMPs namely TmTenecin-1 (TmTene1, A), TmTenecin-2 (TmTene2, B), TmTenecin-3 (TmTene3, C), TmTenecin-4 (TmTene4, D), TmAttacin-1a (TmAtt1a, E), TmAttacin-1b (TmAtt1b, F), TmAttacin-2 (TmAtt2, G), TmColeptericin-1 (TmCole1, H), TmColeptericin-2 (TmCole2, I), TmDefensin-1 (TmDef1, J), TmDefensin-2 (TmDef2, K), TmCecropin-2 (TmCec2, L), TmTLP-1 (TmTLP1, M), and TmTLP-2 (TmTLP2, N) in the larval gut of T. molitor against microbial challenge with E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) were quantified relative to L27a at 24 h post-infection. ‘*’ indicates significant difference between dsTmPGRP-SA and dsEGFP-treated group (p < 0.05); ns = not significant.
Unlike lower expression of AMPs observed in the hemocytes following microbial challenges, antimicrobial responses were strong in both fat body and gut (Figure S4). In this context, no differences in TmTene1 (Figure S4A) and TmAtt1a (Figure S4E) transcription were found between dsEGFP- and dsTmPGRP-SA-treated larvae against all microbes tested. On the other hand, a mild downregulation of TmTene2 (Figure S4B) was noticed following TmPGRP-SA silencing upon E. coli-, S. aureus-, and C. albicans-infection in the hemocytes. Altogether, TmPGRP-SA does not appear to affect AMP regulation in the hemocytes.
2.6. Effect of TmPGRP-SA Knockdown on NF-κB Gene Expression
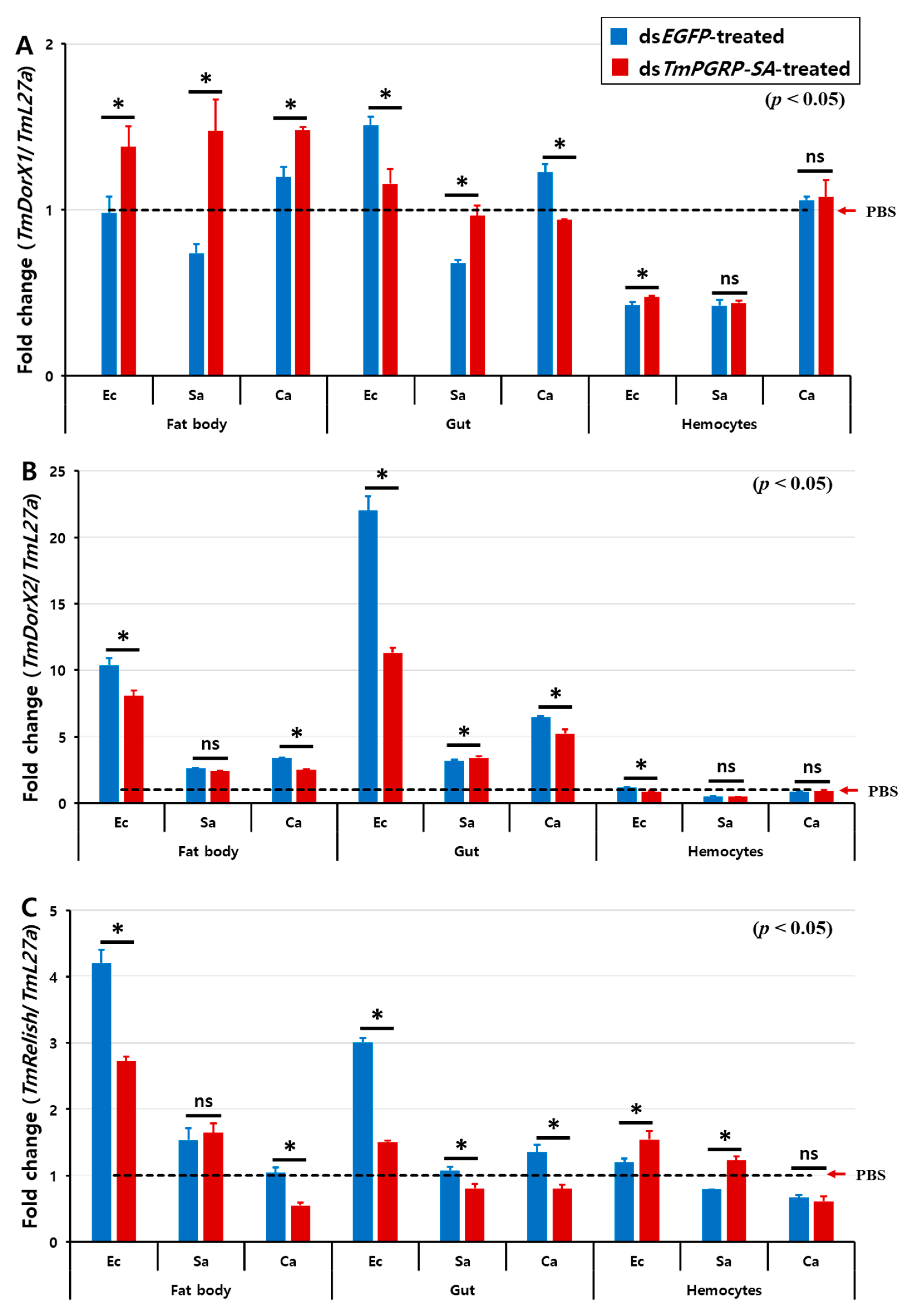
The above data demonstrated that as a receptor TmPGRP-SA plays an important role in the recognition of bacterial pathogens such as E. coli, S. aureus, and C. albicans through the induction of AMP gene expression. Thus, we asked whether the depletion of TmPGRP-SA would impair the mRNA expression of T. molitor transcription proteins, including TmRelish, TmDorX1, and TmDorX2 in the fat body, gut, and hemocytes following E. coli, S. aureus, and C. albicans infections.
The effect of TmPGRP-SA knockdown on the induction of TmDorX2 and TmRelish, encoding Toll- and IMD-pathway mediated transcription proteins, respectively, by E. coli and C. albicans showed a similar tendency in both fat body and gut (Figure 8). In TmPGRP-SA-silenced larvae, the expression of TmDorX2 and TmRelish were significantly decreased in both the fat body and gut following E. coli and C. albicans challenges (Figure 8B,C) unlike in the case of TmDorX1, which was upregulated in the fat body in TmPGRP-SA-depleted group following challenges with all microbes (Figure 8A). Although, TmDorX1 expression in the gut appeared to be downregulated following E. coli and C. albicans challenges (Figure 8A). Of note, expression of NF-κB genes were insensitive to regulation by TmPGRP-SA in the hemocytes (Figure 8A–C). Overall, our data highlight that loss of TmPGRP-SA caused significant downregulation of TmDorX2 and TmRelish after E. coli and C. albicans infections in both the immune-related organs, fat body, and gut.
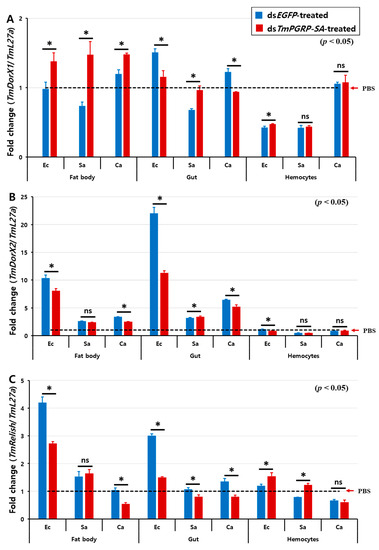
Figure 8.
Relative transcript level of three NF-κB transcription factors in TmPGRP-SA knockdown insects following microbial infection. The average fold-change of TmRelish (A), TmDorX1 (B), and TmDorX2 (C) in TmPGRP-SA-silenced larvae following challenge with E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) was quantified relative to that of L27a at 24 h post-challenge. dsEGFP served as a negative control. Bars represent mean ± SE of three independent experiments. ‘*’ indicates significant difference (p < 0.05); ns = not significant.
3. Discussion
In the present study, we identified the same PGRP-SA from T. molitor that was previously identified by Lee and his colleagues (accession number: AB219970.1), and it showed a high degree of high identity with Drosophila PGRP-SA [36]. Like all known PGRPs, TmPGRP-SA possesses the same conserved domain and a disulfide bond between cysteine residues which is essential for protein stability. Remarkably, the DmPGRP-SA contains a signal peptide (N-terminus) and a single PGRP domain. The cleaved PGRP-SA has a single PGRP domain, indicating that it is a secreted protein [5,21]. In agreement with Drosophila studies, the N-terminal residues of TmPGRP-SA correspond to the putative signal peptide and contains one PGRP domain. Given that the PGRP domains of both insects and mammals share conserved amino acids that are homologous to N-acetylmuramoyl-L-alanine amidase (T7 lysozymes), we found only one tyrosine (Y) residue of three conserved amino acids (histidine, tyrosine, and lysine) in the active site of TmPGRP-SA, suggesting that like in Drosophila, TmPGRP-SA may not have amidase activity [6,36,38]. However, further studies in DmPGRP-SA have revealed that a serine residue (Ser158) is a key amino acid for carboxypeptidase activity toward DAP-type PGNs and Toll activation, which is highly conserved in PGRPs that lack amidase catalytic activity [21]. Furthermore, PGRP-SA from B. ignitus, A. mellifera, and M. rotundata had stronger interaction with DAP-type PGNs than with lys-type PGNs [23]. Consistent with these findings, TmPGRP-SA possesses the conserved serine residue in its active site (threonine174, serine175, serine176), suggesting that TmPGRP-SA may have carboxypeptidase activity. Similar to PGRP-SA in D. melanogaster, A. mellifera, and B. ignites, our results show that TmPGRP-SA has four conserved cysteine residues (Cys29, Cys66, Cys72, and Cys151) that may be linked by disulfide bonds. However, a crystal structure of TmPGRP-SA is needed to elucidate the conformation [21,23,39]. Phylogenetic studies highlighted that Hymenoptera diverged from Diptera and Lepidoptera 300 million years ago and this divergence was after the evolution of Coleoptera [40]. Results from the present study show that Hymenoptera insects had fewer similarities with Coleopteran than Dipteran and this difference may be the result of the evolutionary process as the clade of Hymenoptera are more ancient than Diptera [23].
Studies over the past decade have demonstrated that insect short PGRPs are constitutively present in the hemolymph and cuticle proteins, and are expressed mainly in the fat body and gut, and to a lesser extent in the hemocytes [41,42]. In this context, BiPGRP-S was expressed at a high level in the fat body and relatively lower level in the epidermis in B. ignitus worker bees [43]. Furthermore, in the Chinese oak silkworm, Antheraea pernyi, the tissue distribution of PGRP-SA (ApPGRP-SA) was found widely in the different immune-related tissues including the fat body, midgut, epidermis, and hemocytes [44]. Consistent with previous studies, TmPGRP-SA transcripts were highly expressed in the larval fat body, followed by hemocytes and gut.
Innate immune response is triggered by signal-transducing sensors, the peptidoglycan recognition proteins (PGRPs), that recognize non-self molecules, such as the lys-type PGNs of Gram-positive bacteria, DAP-type PGNs of Gram-negative bacteria, and β-1,3-glucans of fungi [45]. Invertebrate PGRPs, like those in some mammals, have shown multivalent binding properties to a variety of pathogenic molecules [46,47]. In the oyster, Crassostrea gigas, PGRPS2 displays high binding affinity to lipopolysaccharide (LPS), PGN, mannan, E. coli, S. aureus, and the fungus Yarrowia lipolytica [48], whereas, ApPGRP-SA can detect S. aureus, M. luteus, E. coli, and C. albicans [46]. Our results showed that the transcriptional level of TmPGRP-SA was significantly upregulated following challenges with E. coli, S. aureus, and C. albicans in both fat body and gut, which was in agreement with expression level of ApPGRP-SA in response to E. coli, Gram-positive bacteria and fungi in the same tissues [44]. Likewise, T. castaneum PGRP-SA (TcPGRP-SA) mRNA expression was significantly increased following E. coli and Micrococcus luteus infections in 3-day-old pupae [49]. Contrary to E. coli- and S. aureus-infected larvae, TmPGRP-SA transcription was not affected by C. albicans infection in the larval hemocytes. This is consistent with high accumulation of TmPGRP-SA in the hemolymph of T. molitor against lys-type and DAP-type PGNs [37]. Taken together, bacterial challenges induce TmPGRP-SA in the larval fat body, gut, and hemocytes of T. molitor, while fungal infection (C. albicans) upregulates its expression only in the fat body and gut.
Monitoring of TmPGRP-SA-silenced larvae following infection with the microbes showed that TmPGRP-SA contributed to survival. Of note, survival of the dsTmPGRP-SA-treated larvae was significantly affected by infection with all the microbes, however they succumbed more to E. coli and C. albicans infection than to S. aureus. It is known that when challenged with non-self molecules derived from pathogens, immunocompetent tissues initiate complex mechanisms to synthesize AMPs and trigger innate immunity. Therefore, expression and regulation of the AMP genes in the fat body, gut, and hemocytes were analyzed.
Comparative studies conducted previously on the two main Toll pathway components revealed that the expression level of TmTene1, TmTene4, TmDef1, TmDef2, TmCole1, TmAtt1b, and TmCec2 were significantly decreased in the fat body of E. coli-challenged T. molitor following silencing of TmDorX2 and TmCactin [32,33]. Our results, similarly, showed significant downregulation of these genes in the fat body of dsTmPGRP-SA-treated larvae upon E. coli challenge. This result was also confirmed by measuring the fold-change of TmDorX2 expression in the TmPGRP-SA-silenced larval fat body following E. coli infection. Notably, the induction of TmAttacin family is regulated by TmPGRP-SA and TmDorX2 in the fat body of E. coli-infected larvae [33]. Furthermore, following infection with S. aureus the mRNA expression of six AMPs, namely TmTene1, TmTene4, TmDef1, TmDef2, TmCole2, and TmAtt1b was suppressed in the fat body of TmPGRP-SA- TmDorX2-, and TmCactin-silenced groups [32,33]. However, fat body of TmPGRP-SA- and TmDorX2-depleted larvae merely control C. albicans-induced transcription of TmDef1 [33]. Moreover, we propose that TmPGRP-SA is a positive regulator in the fat body, and controls the expression levels of TmTene1, TmTene2, TmTene4, TmDef1, TmDef2, TmCole1, TmAtt1b, and TmAtt2 (8 AMPs) upon E. coli and S. aureus infections (Figure 9).
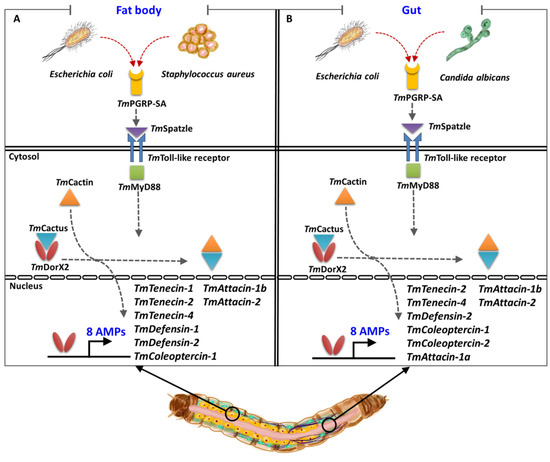
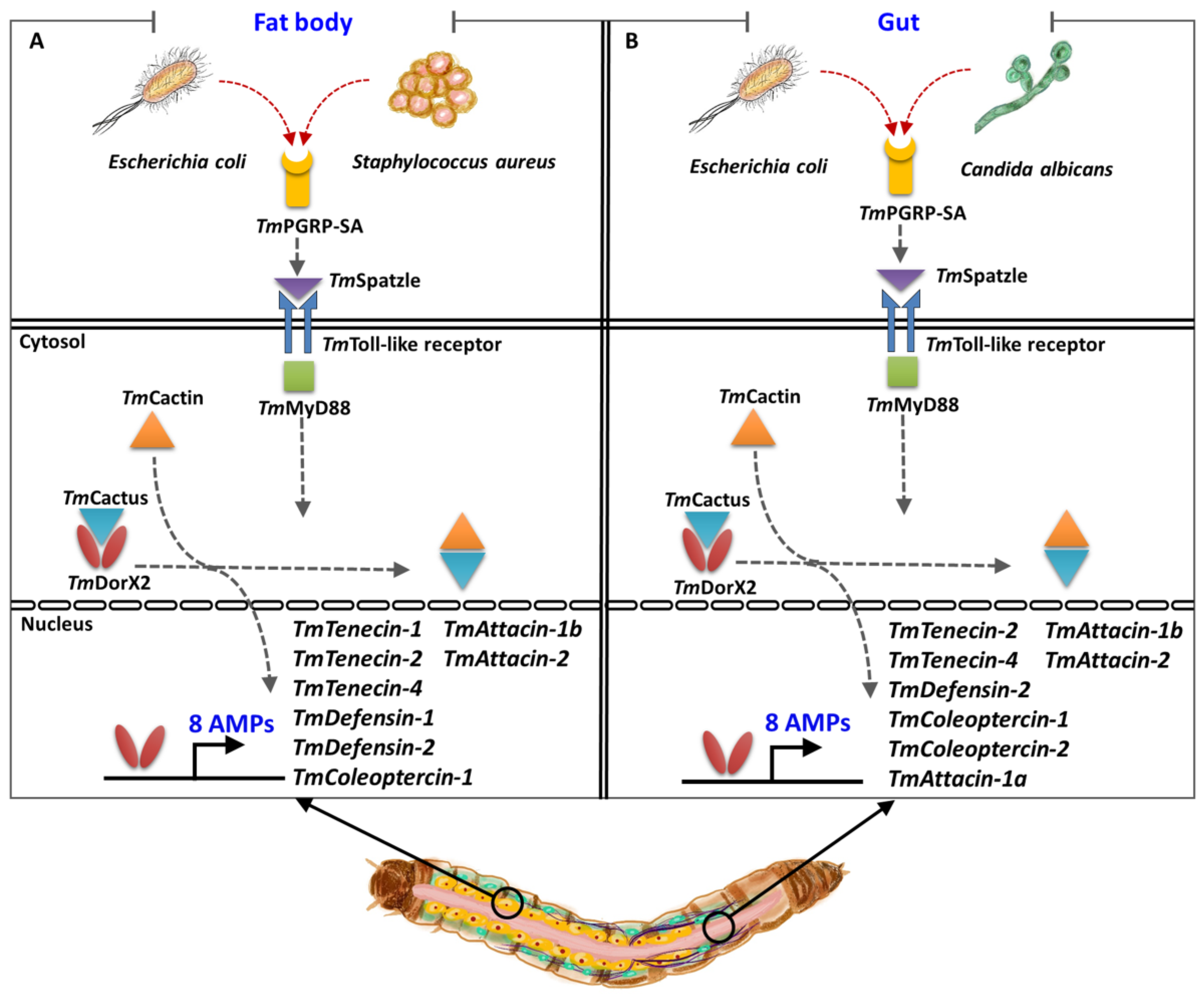
Figure 9.
A simplified schematic representation of the key role of TmPGRP-SA in regulating antimicrobial peptide (AMP) expression in the larval fat body (A) and gut (B) following infection with E. coli, S. aureus, and C. albicans.
In the gut, silencing of TmPGRP-SA and TmDorX2 following E. coli infection can suppress TmTene2, TmTene4, TmDef2, TmCole1, TmCole2, TmAtt1a, TmAtt1b, and TmAtt2 transcriptions (8 AMPs); while C. albicans infection can impair TmTene1, TmTene2, TmTene4, TmDef1, TmDef2, TmCole1, TmCole2, TmAtt1b, TmAtt2, TmCec2, and TmTLP1 expression (11 AMPs) [33]. In addition, knockdown of TmPGRP-SA and TmDorX2 significantly downregulates TmCole2 and TmAtt2 response to S. aureus challenge [33]. Intriguingly, E. coli- and C. albicans-mediated induction of TmDorX1, TmDorX2, and TmRelish was regulated by TmPGRP-SA in the gut. Collectively, TmPGRP-SA plays a key role in regulating of AMP gene expression in the fat body in response to E. coli and S. aureus, whereas TmPGRP-SA is required for the transcription of AMPs in the gut in response to E. coli and C. albicans (Figure 9). We should also mention that TmPGRP-SA is not a main regulator of AMPs in the hemocytes. The above results demonstrate that the mortality observed in T. molitor larvae following TmPGRP-SA knockdown may be attributable to impairment or abolishment of TmPGRP-SA-mediated signal transduction and antimicrobial defense.
4. Materials and Methods
4.1. Rearing Stock of T. Molitor
T. molitor stocks were reared on an artificial diet at 27 ± 1 °C, 60 ± 5% relative humidity, and under conditions of darkness. Young larvae were fed an artificial diet consisting of 170 g wheat flour, 0.5 g chloramphenicol, 20 g roasted soy flour, 0.5 g sorbic acid, 0.5 mL propionic acid, 10 g soy protein, and 100 g wheat bran in 200 mL of distilled water, autoclaved at 121 °C for 20 min. Healthy and fed 10th–12th instar T. molitor larvae (approximately 2.4 cm) were used for all the experiments. The experimental units maintained in an insectary with artificial diet during all experiments.
4.2. Bioinformatics Analysis for Identification and Sequence Characterization of TmPGRP-SA
To identify TmPGRP-SA, a local-tblastn analysis was carried out using the protein sequence of T. castaneum PGRP-SA (TcPGRP-SA) (GenBank: P_008192927.1) as the query. InterProScan 5.0 [50] and BLASTx [51] were performed to predict the functional domain of TmPGRP-SA by using the deduced amino acid sequence as a template. SignalP-5.0 was used to predict the signal peptide [52]. SWISS-MODEL [53] and PyMOL [54] were used to illustrate the TmPGRP-SA protein structure. To obtain the sequence similarity between TmPGRP-SA protein and its orthologs, multiple protein sequences listed in Table 1 were aligned using ClustalX2 [55]. A maximum likelihood (ML) tree (JTT matrix-based model) [56] was generated based on the protein sequences with 1000 bootstrap replicates using MEGA 7.0 program [57]. Homo sapiens peptidoglycan recognition protein 3 (HsPGRP3) was used as an outgroup in the phylogenetic studies.

Table 1.
The accession number of PGRP-SA proteins used for bioinformatic analysis of this study.
4.3. Gene Expression Analysis of TmPGRP-SA in Multiple Tissues and in Different Developmental Stages
To determine the spatial expression profile of TmPGRP-SA during development, samples were collected from egg (EG), young larvae (YL; 10th–12th instar), late-instar larvae (LL; 19th–20th instar), pre-pupae (PP), 1 to 7-day-old pupae (P1–P7), and 1- to 5-day-old adults (n = 20 per each stage). In addition to TmPGRP-SA developmental profiling, the tissue-specific expression pattern of TmPGRP-SA mRNA was examined using multiple larval and adult tissues of T. molitor including the integument, fat body, hemocytes, gut, Malpighian tubules, ovary, and testis.
4.4. RNA Isolation and cDNA Synthesis
Guanidine thiocyanate-based RNA lysis buffer was used to extract the total RNA from the samples following the modified LogSpin RNA isolation method as described in a previous study [33,58]. First-strand cDNA was synthesized using 2 μg of total RNA and oligo-(dT)12–18 primers in a reaction volume of 20 μL. The reaction was incubated at 42 °C for 5 min. The resultant cDNA was added to the AccuPower® RT PreMix (Bioneer, Daejeon, Korea) kit and then incubated at 72 °C for 1 h. The synthesized cDNAs were stored at −20 °C until use.
4.5. Quantitative Reverse-Transcription PCR (RT-qPCR) Analysis
cDNA was diluted (1:20 with DNase/RNase free water) to establish standard curves for determining empirically optimal template concentration and the primer efficiency. The RT-qPCR reaction mix (20 µL) included 10 µL AccuPower® 2X GreenStar qPCR Master Mix (Bioneer, Daejeon, Korea), 3 µL of 1:20 diluted cDNA template, and 2 µL of designed primers (TmPGRP-SA-qPCR-Fw and TmPGRP-SA-qPCR-Rv) (Figure S1; Table 2). Specific primers against TmPGRP-SA, as well as against the T. molitor housekeeping gene (60 S ribosomal protein L27a), were designed using Primer 3.0 plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi); primer sequences are presented in Table 2. For each sample, duplicate reactions in a total volume of 20 µL were performed with the following thermal program: 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s and amplification at 60 °C for 30 s. the relative mRNA expression levels were evaluated using the comparative CT method (2−ΔΔCT method) [59].

Table 2.
Sequences of the primers used in this study.
4.6. Microbial Strains
The following microorganisms were obtained from the American Type Culture Collection (ATCC) and used in this study: Escherichia coli strain K12, Staphylococcus aureus strain RN4220, and Candida albicans strain AUMC 13529. E. coli and S. aureus were grown aerobically in Luria-Bertani (LB) broth, while C. albicans was cultured in Sabouraud Dextrose broth at 37 °C overnight under continuous shaking. Overnight cultures were centrifuged at 3500 rpm for 15 min at room temperature (around 25 °C), and washed and diluted in phosphate-buffered saline (PBS; pH 7.0). Finally, the cell concentrations (based on OD600 measurements) were adjusted to 1 × 106 cells/μL for E. coli and S. aureus, and 5 × 104 cells/μL for C. albicans.
4.7. TmPGRP-SA mRNA Expression Following Microbial Challenges
To elucidate the effect of microbial strains on TmPGRP-SA mRNA expression in T. molitor, the 10th–12th instar larvae (n = 80 per time point) were divided into the following four subgroups: three groups of larvae (n = 20 per group) were challenged with 1 µL E. coli (1 × 106 cells/µL), S. aureus (1 × 106 cells/µL), and C. albicans (5 × 104 cells/µL), respectively, while the remaining 20 larvae were injected with PBS as control. The experimental groups were maintained in the insectary under identical rearing conditions and then the immune tissues were dissected at 3, 6, 9, 12, and 24 h post-injection. The fat body, hemocytes, and gut were collected for subsequent RNA extraction, cDNA synthesis, and RT-qPCR analyses as described above.
4.8. Preparation of Double-Stranded TmPGRP-SA
The SnapDragon-Long dsRNA design software (https://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl) was used to design gene-specific primers (Table 2). For TmPGRP-SA, we synthesized double-stranded RNA (dsRNA) designing forward and reverse primers; the T7 promoter sequence was added to their 5’ ends. Primers (dsTmPGRP-SA_Fw and dsTmPGRP-SA_Rv) were used to amplify 316 bp amplicons from T. molitor cDNA using AccuPower® Pfu PCR PreMix under the following cycling conditions: 95 °C for 2 min, 30 cycles of 95 °C for 20 s, 56 °C for 30 s, and final extension at 72 °C for 5 min (Figure S1; Table 2). For control, the DNA template was cloned from the enhanced green fluorescent protein (EGFP) gene present within the EGFP-C1 vector, and tailed with T7 promotor using dsEGFP_Fw and dsEGFP_Rv primers (Table 2). The amplicons were purified with AccuPrep® PCR Purification Kit (Bioneer). Next, the dsRNA was transcribed in vitro using the EZTM T7 High Yield in vitro Transcription Kit (Enzynomics, Deajeon, Korea), as per the manufacturer’s protocol.
4.9. T. molitor Survival Bioassay
Survival bioassays were performed by silencing TmPGRP-SA in young larvae of T. molitor followed by microbial infections. A volume of 1 μL (1 μg) of the gene of interest and control, synthesized as previously described, was injected into 10th–12th instar larvae. At least 10–15 larvae were injected with dsTmPGRP-SA and dsEGFP per treatment, and the experiments were repeated three times to obtain a total of 45 RNAi-injected larvae per group. Then, three surviving larvae per group were used to confirm the knockdown efficiency of the target gene on the second day after injection. Subsequently, dsRNA-injected larvae (n = 10 per group) were infected with E. coli (1 × 106 cells/larva), S. aureus (1 × 106 cells/larva), and C. albicans (5 × 104 cells/larva), and survivors were monitored every day for a 10-day period.
4.10. Analysis of the Effect of TmPGRP-SA Silencing on AMP and NF-κB Gene Expression Post-Microbial Challenge
The survival analyses conducted above underline the importance of TmPGRP-SA in T. molitor fitness. These assay results also raise the question about the cause of the reduced lifespan observed in TmPGRP-SA knockdown larvae infected with microbes. It is plausible that low expression of AMP genes could lead to the notable mortality rates. To test this hypothesis, we examined the gene expression profiles of 14 AMPs, including TmTenecin-1 (TmTene1), TmTenecin-2 (TmTene2), TmTenecin-3 (TmTene3), TmTenecin-4 (TmTene4), TmAttacin-1a (TmAtt1a), TmAttacin-1b (TmAtt1b), TmAttacin-2 (TmAtt2), TmDefensin-1 (TmDef1), TmDefensin-2 (TmDef2), TmColeoptericin-1 (TmCole1), TmColeoptericin-2 (TmCole2), TmCecropin-2 (TmCec2), TmThaumatin-like protein-1 (TmTLP1), and TmThaumatin-like protein-2 (TmTLP2) in the TmPGRP-SA-silenced larvae following the microbial challenges. Furthermore, the fold-changes in mRNA of three T. molitor transcription factors, namely TmRelish, TmDorsal X1 isoform (TmDorX1), and TmDorsal X2 isoform (TmDorX2) were also evaluated (Table 2). DsEGFP was used as a negative control, and PBS served as a wound control. Sample tissues (fat body, gut, and hemocytes) were collected 24 h post-challenge and then processed for cDNA synthesis, and RT-qPCR analysis using AMP-specific primers (Table 2).
4.11. Statistical Analysis
Three independent biological replicates were used for all experiments. Values were reported as mean ± standard error (SE). Differences between groups were analyzed using one-way statistical analysis of variance (ANOVA) and Tukey’s test; p values < 0.05 were considered significant. The results of the mortality assay were analyzed using the Kaplan-Meier plot (log-rank Chi-square test) in Excel (http://www.real-statistics.com/survival-analysis/kaplan-meier-procedure/real-statistics-kaplan-meier/).
5. Conclusions
T. molitor peptidoglycan recognition protein-SA (TmPGRP-SA) has multivalent binding properties to Gram-negative and Gram-positive bacteria (E. coli and S. aureus), and fungi (C. albicans). TmPGRP-SA is a positive regulator in the fat body and gut, and regulates the expression of eight of fourteen AMP genes. Invading microbes are sensed by TmPGRP-SA which then likely transduces the signal to TmDorX2, which as a transcription factor to initiate the antimicrobial defense response.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/6/2113/s1.
Author Contributions
Y.S.H. and Y.H.J. conceived and designed the experiments; M.K., T.T.E., and Y.M.B. performed the experiments; M.K. analyzed the data; Y.S.H. contributed reagents/materials/analysis tools; M.K. wrote the manuscript; Y.S.H. and Y.H.J. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning (Grant No. NRF-2019R1I1A3A01057848).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Janeway, C.A. Innate immunity: The virtues of a nonclonal system of recognition. Cell 1997, 91, 295–298. [Google Scholar] [CrossRef]
- Nürnberger, T.; Brunner, F. Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 2002, 5, 318–324. [Google Scholar] [CrossRef]
- Kusumoto, S.; Fukase, K.; Shiba, T. Key structures of bacterial peptidoglycan and lipopolysaccharide triggering the innate immune system of higher animals: Chemical synthesis and functional studies. Proc. Jpn. Acad. Ser. B 2010, 86, 322–337. [Google Scholar] [CrossRef]
- Werner, T.; Liu, G.; Kang, D.; Ekengren, S.; Steiner, H.; Hultmark, D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13772–13777. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-M.; Li, X.; Cocklin, R.R.; Wang, M.; Wang, M.; Fukase, K.; Inamura, S.; Kusumoto, S.; Gupta, D.; Dziarski, R. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 2003, 278, 49044–49052. [Google Scholar] [CrossRef] [PubMed]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006, 7, 232. [Google Scholar] [CrossRef]
- Montaño, A.M.; Tsujino, F.; Takahata, N.; Satta, Y. Evolutionary origin of peptidoglycan recognition proteins in vertebrate innate immune system. BMC Evol. Biol. 2011, 11, 79. [Google Scholar] [CrossRef]
- Mellroth, P.; Karlsson, J.; Steiner, H. A scavenger function for a DrosophilaPeptidoglycan recognition protein. J. Biol. Chem. 2003, 278, 7059–7064. [Google Scholar] [CrossRef]
- Guan, R.; Roychowdhury, A.; Ember, B.; Kumar, S.; Boons, G.-J.; Mariuzza, R.A. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17168–17173. [Google Scholar] [CrossRef]
- Guan, R.; Mariuzza, R.A. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007, 15, 127–134. [Google Scholar] [CrossRef]
- Wang, Q.; Ren, M.; Liu, X.; Xia, H.; Chen, K. Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 2019, 106, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Kleino, A.; Silverman, N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 2014, 42, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Valanne, S.; Wang, J.-H.; Rämet, M. The Drosophila toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef]
- Werner, T. Peptidoglycan Recognition Proteins in Drosophila Melanogaster. Ph.D. Thesis, Umeå Centrum för Molekylär Patogenes (UCMP Medicinska fakulteten), Umea, Sweden, 2004. [Google Scholar]
- Michel, T.; Reichhart, J.-M.; Hoffmann, J.A.; Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756. [Google Scholar] [CrossRef]
- Gobert, V.; Gottar, M.; Matskevich, A.A.; Rutschmann, S.; Royet, J.; Belvin, M.; Hoffmann, J.A.; Ferrandon, D. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 2003, 302, 2126–2130. [Google Scholar] [CrossRef]
- Pili-Floury, S.; Leulier, F.; Takahashi, K.; Saigo, K.; Samain, E.; Ueda, R.; Lemaitre, B. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 2004, 279, 12848–12853. [Google Scholar] [CrossRef]
- Wang, L.; Weber, A.N.; Atilano, M.L.; Filipe, S.R.; Gay, N.J.; Ligoxygakis, P. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 2006, 25, 5005–5014. [Google Scholar] [CrossRef]
- Filipe, S.R.; Tomasz, A.; Ligoxygakis, P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005, 6, 327–333. [Google Scholar] [CrossRef]
- Chang, C.-I.; Pili-Floury, S.; Hervé, M.; Parquet, C.; Chelliah, Y.; Lemaitre, B.; Mengin-Lecreulx, D.; Deisenhofer, J. A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual L, D-carboxypeptidase activity. PLoS Biol. 2004, 2, e277. [Google Scholar] [CrossRef]
- Leulier, F.; Parquet, C.; Pili-Floury, S.; Ryu, J.-H.; Caroff, M.; Lee, W.-J.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003, 4, 478. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, X.; Huang, J.; Chen, M.; An, J. Structural Insights into the Preferential Binding of PGRP-SAs from Bumblebees and Honeybees to Dap-Type Peptidoglycans Rather than Lys-Type Peptidoglycans. J. Immunol. 2019, 202, 249–259. [Google Scholar] [CrossRef]
- Canteri de Souza, P.; Custódio Caloni, C.; Wilson, D.; Sergio Almeida, R. An invertebrate host to study fungal infections, mycotoxins and antifungal drugs: Tenebrio molitor. J. Fungi 2018, 4, 125. [Google Scholar] [CrossRef]
- de Souza, P.C.; Morey, A.T.; Castanheira, G.M.; Bocate, K.P.; Panagio, L.A.; Ito, F.A.; Furlaneto, M.C.; Yamada-Ogatta, S.F.; Costa, I.N.; Mora-Montes, H.M. Tenebrio molitor (Coleoptera: Tenebrionidae) as an alternative host to study fungal infections. J. Microbiol. Methods 2015, 118, 182–186. [Google Scholar] [CrossRef]
- Park, S.; Jo, Y.H.; Park, K.B.; Ko, H.J.; Kim, C.E.; Bae, Y.M.; Kim, B.; Jun, S.A.; Bang, I.S.; Lee, Y.S. TmToll-7 plays a crucial role in innate immune responses against Gram-negative bacteria by regulating 5 AMP genes in Tenebrio molitor. Front. Immunol. 2019, 10, 310. [Google Scholar] [CrossRef]
- Park, J.-W.; Kim, C.-H.; Kim, J.-H.; Je, B.-R.; Roh, K.-B.; Kim, S.-J.; Lee, H.-H.; Ryu, J.-H.; Lim, J.-H.; Oh, B.-H. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc. Natl. Acad. Sci. USA 2007, 104, 6602–6607. [Google Scholar] [CrossRef]
- Yang, Y.T.; Lee, M.R.; Lee, S.J.; Kim, S.; Nai, Y.S.; Kim, J.S. Tenebrio molitor Gram-negative-binding protein 3 (TmGNBP3) is essential for inducing downstream antifungal Tenecin 1 gene expression against infection with Beauveria bassiana JEF-007. Insect Sci. 2018, 25, 969–977. [Google Scholar] [CrossRef]
- Yagi, Y.; Nishida, Y.; Ip, Y.T. Functional analysis of Toll-related genes in Drosophila. Dev. Growth Differ. 2010, 52, 771–783. [Google Scholar] [CrossRef]
- Edosa, T.T.; Jo, Y.H.; Keshavarz, M.; Bae, Y.M.; Kim, D.H.; Lee, Y.S.; Han, Y.S. TmSpz6 Is Essential for Regulating the Immune Response to Escherichia Coli and Staphylococcus Aureus Infection in Tenebrio Molitor. Insects 2020, 11, 105. [Google Scholar] [CrossRef]
- Patnaik, B.B.; Patnaik, H.H.; Seo, G.W.; Jo, Y.H.; Lee, Y.S.; Lee, B.L.; Han, Y.S. Gene structure, cDNA characterization and RNAi-based functional analysis of a myeloid differentiation factor 88 homolog in Tenebrio molitor larvae exposed to Staphylococcus aureus infection. Dev. Comp. Immunol. 2014, 46, 208–221. [Google Scholar] [CrossRef]
- Jo, Y.H.; Kim, Y.J.; Park, K.B.; Seong, J.H.; Kim, S.G.; Park, S.; Noh, M.Y.; Lee, Y.S.; Han, Y.S. TmCactin plays an important role in Gram-negative and-positive bacterial infection by regulating expression of 7 AMP genes in Tenebrio molitor. Sci. Rep. 2017, 7, 46459. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Jo, Y.H.; Park, K.B.; Ko, H.J.; Edosa, T.T.; Lee, Y.S.; Han, Y.S. Tm DorX2 positively regulates antimicrobial peptides in Tenebrio molitor gut, fat body, and hemocytes in response to bacterial and fungal infection. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-J.; Xu, S.; He, Z.-H.; Shi, X.-Z.; Zhao, X.-F.; Wang, J.-X. Activation of toll pathway is different between kuruma shrimp and Drosophila. Front. Immunol. 2017, 8, 1151. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Jiang, H.; Kanost, M.R. Proteolytic activation and function of the cytokine Spätzle in the innate immune response of a lepidopteran insect, Manduca sexta. FEBS J. 2010, 277, 148–162. [Google Scholar] [CrossRef]
- Park, J.W.; Je, B.-R.; Piao, S.; Inamura, S.; Fujimoto, Y.; Fukase, K.; Kusumoto, S.; Söderhäll, K.; Ha, N.-C.; Lee, B.L. A synthetic peptidoglycan fragment as a competitive inhibitor of the melanization cascade. J. Biol. Chem. 2006, 281, 7747–7755. [Google Scholar] [CrossRef]
- Yu, Y.; Park, J.-W.; Kwon, H.-M.; Hwang, H.-O.; Jang, I.-H.; Masuda, A.; Kurokawa, K.; Nakayama, H.; Lee, W.-J.; Dohmae, N. Diversity of innate immune recognition mechanism for bacterial polymeric meso-diaminopimelic acid-type peptidoglycan in insects. J. Biol. Chem. 2010, 285, 32937–32945. [Google Scholar] [CrossRef]
- Kikkawa, H.S.; Ueda, T.; Suzuki, S.-I.; Yasuda, J. Characterization of the catalytic activity of the γ-phage lysin, PlyG, specific for Bacillus anthracis. FEMS Microbiol. Lett. 2008, 286, 236–240. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Zhao, X.; Naeem, M.; An, J. Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae). Protein Sci. 2018, 27, 893–897. [Google Scholar] [CrossRef]
- Consortium, H.G.S. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931. [Google Scholar]
- Dziarski, R. Peptidoglycan recognition proteins (PGRPs). Mol. Immunol. 2004, 40, 877–886. [Google Scholar] [CrossRef]
- Kang, D.; Liu, G.; Lundström, A.; Gelius, E.; Steiner, H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA 1998, 95, 10078–10082. [Google Scholar] [CrossRef]
- You, H.; Wan, H.; Li, J.; Jin, B.R. Molecular cloning and characterization of a short peptidoglycan recognition protein (PGRP-S) with antibacterial activity from the bumblebee Bombus ignitus. Dev. Comp. Immunol. 2010, 34, 977–985. [Google Scholar] [CrossRef]
- Dai, L.-S.; Qian, C.; Wang, L.; Wei, G.-Q.; Liu, Q.-N.; Sun, Y.; Zhang, C.-F.; Li, J.; Liu, D.-R.; Zhu, B.-J. Characterization and function of a short peptidoglycan recognition protein from the Chinese oak silkworm, Antheraea pernyi. J. Asia Pac. Entomol. 2015, 18, 701–707. [Google Scholar] [CrossRef]
- Kurata, S. Peptidoglycan recognition proteins in Drosophila immunity. Dev. Comp. Immunol. 2014, 42, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, X.; Cai, S.; Zhang, S.; Luo, H.; Wu, C.; Zhang, R.; Zhang, J. A novel peptidoglycan recognition protein involved in the prophenoloxidase activation system and antimicrobial peptide production in Antheraea pernyi. Dev. Comp. Immunol. 2018, 86, 78–85. [Google Scholar] [CrossRef]
- Sharma, P.; Dube, D.; Sinha, M.; Yadav, S.; Kaur, P.; Sharma, S.; Singh, T.P. Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: Structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid. PLoS ONE 2013, 8, e53756. [Google Scholar] [CrossRef]
- Yang, C.; Wang, L.; Jia, Z.; Yi, Q.; Xu, Q.; Wang, W.; Gong, C.; Liu, C.; Song, L. Two short peptidoglycan recognition proteins from Crassostrea gigas with similar structure exhibited different PAMP binding activity. Dev. Comp. Immunol. 2017, 70, 9–18. [Google Scholar] [CrossRef]
- Koyama, H.; Kato, D.; Minakuchi, C.; Tanaka, T.; Yokoi, K.; Miura, K. Peptidoglycan recognition protein genes and their roles in the innate immune pathways of the red flour beetle, Tribolium castaneum. J. Invertebr. Pathol. 2015, 132, 86–100. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Mount, D.W. Using the basic local alignment search tool (BLAST). Cold Spring Harbor Protocols 2007, 2007, pdb.top17. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucl. Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Yaffe, H.; Buxdorf, K.; Shapira, I.; Ein-Gedi, S.; Zvi, M.M.-B.; Fridman, E.; Moshelion, M.; Levy, M. LogSpin: A simple, economical and fast method for RNA isolation from infected or healthy plants and other eukaryotic tissues. BMC Res. Notes 2012, 5, 45. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C T method. Nat. Protocols 2008, 3, 1101. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).