The Encystment-Related MicroRNAs and Its Regulation Molecular Mechanism in Pseudourostyla cristata Revealed by High Throughput Small RNA Sequencing
Abstract
:1. Introduction
2. Results
2.1. Small RNA Populations Collected from P. cristata by HiSeq Technology
2.2. Identification Against Known and Novel miRNAs in P. cristata
2.3. Universally Abundant miRNAs Across the Two Libraries
2.4. Analysis and Validation of DEMs
2.5. DEMs Target Gene Prediction and GO Annotation Analyses
2.6. The DEMs KEGG Functional Annotation Analyses
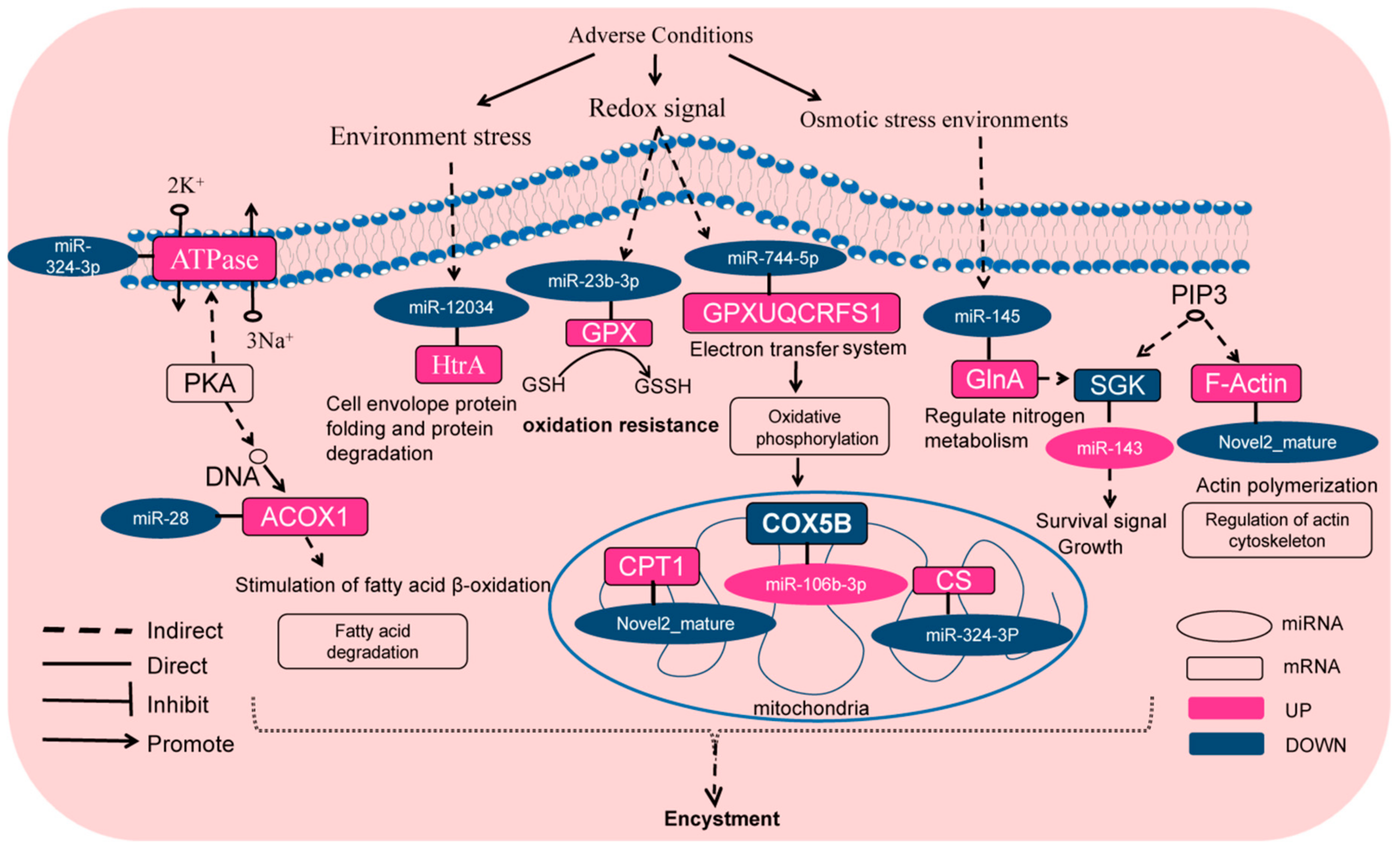
2.7. DEMs Target Genes and Relative KEGG Signaling Pathways Involved in Encystment of P. cristata
3. Discussion
4. Materials and Methods
4.1. Cell Culture and the Encystment Induction
4.2. Sample Collection and RNA Isolation
4.3. Small RNA Library Construction and Sequencing
4.4. Sequence Analysis and Identification of miRNAs
4.5. MiRNA Differential Expression Analysis
4.6. Prediction of miRNA Targets Gene, GO Enrichment, and KEGG Pathway Analysis
4.7. qRT-PCR Analysis
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| miRNAome | microRNAome |
| P. cristata | Pseudourostyla cristata |
| miRNAs | microRNAs |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| yy | vegetative cells |
| bn | dormant cysts |
| DEMs | differentially expressed miRNAs |
| BP | biological process |
| CC | cellular component |
| MF | molecular function |
| clpB | Clp protease ATP-binding subunit ClpB |
| glnA | glutamine synthetase |
| glyS | glycyl-tRNA synthetase alpha chain |
| groEL | chaperonin GroEL |
| HSPA1_8 | heat shock 70kDa protein 8 |
| IPO1 | importin subunit beta-1 |
| PABPC | polyadenylate-binding protein |
| ahpC | alkyl hydroperoxide reductase subunit C |
| RP-L2 | large subunit ribosomal protein L2 |
| TARS | threonyl-tRNA synthetase |
| HSPD1 | chaperonin GroEL |
| dnaK | molecular chaperone DnaK |
| GPX | glutathione peroxidase |
| UQCRFS1 | ubiquinol-cytochrome c reductase iron-sulfur subunit |
| ACOX1 | acyl-CoA oxidase |
| TARS | threonyl-tRNA synthetase |
| UBE1 | ubiquitin-activating enzyme E1 |
| CPT1 | carnitine O-palmitoyltransferase 1 |
| F-Actin | actin beta/gamma 1 |
| HtpG | molecular chaperone HtpG |
| ACSL | long-chain acyl-CoA synthetase |
| SGK | serum / glucocorticoid-regulated kinase |
| rub | rubredoxin-NAD+ reductase |
| COX5B | cytochrome c oxidase subunit 5b |
| asps | aspartyl-tRNA synthetase |
| qRT-PCR | quantitative real-time PCR |
References
- Kaur, H.; Iqbal, S.; Inga, E.; Yawe, D. Encystment and Excystment in Ciliated Protists: Multidimensional Approach. Curr. Sci. 2019, 117, 198. [Google Scholar] [CrossRef]
- Huang, J.; Luo, X.; Bourland, W.A.; Gao, F.; Gao, S. Multigene-based phylogeny of the ciliate families Amphisiellidae and Trachelostylidae (Protozoa: Ciliophora: Hypotrichia). Mol. Phylogenetics Evol. 2016, 101, 101–110. [Google Scholar] [CrossRef]
- Pan, N.; Niu, T.; Bhatti, M.Z.; Zhang, H.; Fan, X.; Ni, B.; Chen, J. Novel insights into molecular mechanisms of Pseudourostyla cristata encystment using comparative transcriptomics. Sci. Rep. 2019, 9, 19109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corliss, J.O.; Esser, S.C. Comments on the role of the cyst in the life cycle and survival of free-living protozoa. Trans. Am. Microsc. Soc. 1974, 578–593. [Google Scholar] [CrossRef]
- Grisvard, J.; Lemullois, M.; Morin, L.; Baroin-Tourancheau, A. Differentially expressed genes during the encystment-excystment cycle of the ciliate Sterkiella histriomuscorum. Eur. J. Protistol. 2008, 44, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Palacios, G.; Martin-Gonzalez, A.; Gutierrez, J.C. Macronuclear DNA demethylation is involved in the encystment process of the ciliate Colpoda inflata. Cell Biol. Int. 1994, 18, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Miao, W.; Xiong, J.; Bowen, J.; Wang, W.; Liu, Y.; Braguinets, O.; Grigull, J.; Pearlman, R.E.; Orias, E.; Gorovsky, M.A. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE 2009, 4, e4429. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Gao, X.; Wang, B.; Chen, F.; Wu, N.; Zhang, Y. Proteomic approach to reveal the proteins associated with encystment of the ciliate Euplotes encysticus. PLoS ONE 2014, 9, e97362. [Google Scholar] [CrossRef]
- Gao, X.; Chen, F.; Niu, T.; Qu, R.; Chen, J. Large-scale identification of encystment-related proteins and genes in Pseudourostyla cristata. Sci. Rep. UK 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Wei, W.; Yan, G.; Shi, T.; Miao, W. Transcriptome analysis reveals the molecular mechanism of resting cyst formation in Colpoda aspera. J. Eukaryot. Microbiol. 2019, 66, 212–220. [Google Scholar] [CrossRef]
- Dhorne-Pollet, S.; Crisci, E.; Mach, N.; Renson, P.; Jaffrezic, F.; Marot, G.; Maroilley, T.; Moroldo, M.; Lecardonnel, J.; Blanc, F.; et al. The miRNA-targeted transcriptome of porcine alveolar macrophages upon infection with porcine reproductive and respiratory syndrome virus. Sci. Rep. 2019, 9, 3160. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Ni, B.; Yang, Z.; Du, B. Ultrastructure of the vegetative and resting cyst in Pseudourostyla cristata (Ciliophora, Hypotrichida). Curr. Zool. 2002, 48, 251–257. [Google Scholar]
- Zhou, W.; Xu, J.; Wang, C.; Shi, D.; Yan, Q. MiR-23b-3p regulates apoptosis and autophagy via suppressing SIRT1 in lens epithelial cells. J. Cell. Biochem. 2019, 120, 19635–19646. [Google Scholar] [CrossRef]
- Savaskan, N.E.; Ufer, C.; Kuhn, H.; Borchert, A. Molecular biology of glutathione peroxidase 4: From genomic structure to developmental expression and neural function. Biol. Chem. 2007, 388, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.O.; Milbury, P.E.; Blumberg, J.B. Polyphenols in almond skins after blanching modulate plasma biomarkers of oxidative stress in healthy humans. Antioxidants 2019, 8, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aktar, K.; Kafi, A.; Dahiya, R. Association of Gpx1 fluctuation in cell cycle progression. In Vitro Cell. Dev. Biol. Anim. 2019, 55, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, L.; Miao, Y.; Yang, J.; Wang, X.; Wang, C.C.; Feng, J.; Wang, L. Homocysteine causes vascular endothelial dysfunction by disrupting endoplasmic reticulum redox homeostasis. Redox Biol. 2019, 20, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, J.C.; Callejas, S.; Borniquel, S.; Benitez, L.; Martin-Gonzalez, A. Ciliate cryptobiosis: A microbial strategy against environmental starvation. Int. Microbiol. 2001, 4, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, C.; Ji, Y.; Chen, Z.; Kitazato, K.; Xiang, Y.; Zhong, M.; Wang, Q.; Pei, Y.; Ju, H.; Wang, Y. Proteomics analysis of autophagy-deficient Atg7-/-MEFs reveals a close relationship between F-actin and autophagy. Biochem. Biophys. Res. Commun. 2013, 437, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Bai, H.; Jin, Y.; Yang, Z. Exercise combined with administration of EGCG and L-carnitine affects the body weight, visceral fat and CPT1 expression in obese rat. Chin. J. Sports Med. 2012, 31, 331–335. [Google Scholar]
- Zhao, S.; Liu, H.; Liu, Y.; Wu, J.; Wang, C.; Hou, X.; Chen, X.; Yang, G.; Zhao, L.; Che, H.; et al. MiR-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells. Cancer Lett. 2013, 333, 253–260. [Google Scholar] [CrossRef]
- Lang, F.; Bohmer, C.; Palmada, M.; Seebohm, G.; Strutz-Seebohm, N.; Vallon, V. (Patho)physiological significance of the serum-and glucocorticoid-inducible kinase isoforms. Physiol. Rev. 2006, 86, 1151–1178. [Google Scholar] [CrossRef]
- Isikbay, M.; Otto, K.; Kregel, S.; Kach, J.; Cai, Y.; Vander, G.D.; Conzen, S.D.; Szmulewitz, R.Z. Glucocorticoid receptor activity contributes to resistance to androgen-targeted therapy in prostate cancer. Horm. Cancer 2014, 5, 72–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, R.Y.; Zhang, D.; Zou, H.D.; Zuo, X.S.; Zhou, Q.S.; Huang, H. MiR-28 inhibits cardiomyocyte survival through suppressing PDK1/Akt/mTOR signaling. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 1020–1025. [Google Scholar] [CrossRef]
- Morais, S.; Knoll-Gellida, A.; André, M.; Barthe, C.; Babin, P.J. Conserved expression of alternative splicing variants of peroxisomalacyl-CoA oxidase 1 in vertebrates and developmental and nutritional regulation in fish. Physiol. Genom. 2007, 28, 239–252. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Qiu, Y.; Zhang, Q.F.; Li, D. Chlorogenic acid and caffeine in combination inhibit fat accumulation by regulating hepatic lipid metabolism-related enzymes in mice. Br. J. Nutr. 2014, 112, 1034–1040. [Google Scholar] [CrossRef] [Green Version]
- Ryckelynck, M.; Giege, R.; Frugier, M. tRNAs and tRNA mimics as cornerstones of aminoacyl-tRNA synthetase regulations. Biochimie 2005, 87, 835–845. [Google Scholar] [CrossRef]
- Wang, M.; Yang, S. Research progress on the structure, function and pathogenic mechanism of glycyl-tRNA synthetase. Chin. J. Anat. 2016, 39, 116–119. [Google Scholar]
- Crumbley, C.; Wang, Y.; Banerjee, S.; Burris, T.P. Regulation of expression of citrate synthase by the retinoic acid receptor-related orphan receptor alpha (RORalpha). PLoS ONE 2012, 7, e33804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, J.R.; Donini, S.; Kappock, T.J. An active site-tail interaction in the structure of hexahistidine-tagged Thermoplasma acidophilum citrate synthase. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 1292–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, T.; Xi, J. Identification of COX5B as a novel biomarker in high-grade glioma patients. Onco Targets Ther. 2017, 10, 5463–5470. [Google Scholar] [CrossRef] [Green Version]
- Hodge, M.R.; Singh, K.; Cumsky, M.G. Upstream activation and repression elements control transcription of the yeast COX5b gene. Mol. Cell. Biol. 1990, 10, 5510–5520. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.; Stewart, R.C. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998, 117, 723–731. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.I.; Carozza, M.; Klein, M.; Nantermet, P.; Luk, D.; Crowl, R.M. Human HtrA, an evolutionarily conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J. Biol. Chem. 1998, 273, 34406–34412. [Google Scholar] [CrossRef] [Green Version]
- Fitriani, D.; Rohman, M.S.; Prijambada, I.D. Proteolytic activity of recombinant DegP from Chromohalobacter salexigens BKL5. Electron. J. Biotechnol. 2017, 29, 7–12. [Google Scholar] [CrossRef]
- Ohashi, Y.; Kaneko, S.J.; Cupples, T.E.; Young, S.R. Ubiquinol cytochrome c reductase (UQCRFS1) gene amplification in primary breast cancer core biopsy samples. Gynecol. Oncol. 2004, 93, 54–58. [Google Scholar] [CrossRef]
- Murray, D.S.; Chinnam, N.; Tonthat, N.K.; Whitfill, T.; Wray, L.J.; Fisher, S.H.; Schumacher, M.A. Structures of the bacillus subtilis glutamine synthetase dodecamer reveal large intersubunit catalytic conformational changes linked to a unique feedback inhibition mechanism. J. Biol. Chem. 2013, 288, 35801–35811. [Google Scholar] [CrossRef] [Green Version]
- Rose, C.F.; Verkhratsky, A.; Parpura, V. Astrocyte glutamine synthetase: Pivotal in health and disease. Biochem. Soc. Trans. 2013, 41, 1518–1524. [Google Scholar] [CrossRef]
- Liu, H.; Sun, W.; Tan, B.; Chi, S.; Dong, X.; Yang, Q. Molecular cloning and expression of hepatopancreas glutamine synthetase in the Pacific white shrimp, Litopenaeus vannamei, induced by acute hypo-osmotic stress. Aquaculture 2012, 362, 80–87. [Google Scholar] [CrossRef]
- Silvia, G.; Abel Antonio, U.; Francisco, V.; Georgina, H. Ammonia efflux rates and free amino acid levels in Litopenaeus vannamei postlarvae during sudden salinity changes. Aquaculture 2004, 233, 573–581. [Google Scholar] [CrossRef]
- Lu, Z.; Qin, Z.; Sarath Babu, V.; Ye, C.; Su, G.; Li, J.; Yang, G.; Shen, H.; Pan, G.; Lin, L. Expression and functional characterization of glutamine synthetase from giant freshwater prawn (Macrobrachium rosenbergii) under osmotic stress. Aquac. Res. 2019, 50, 2635–2645. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003, 31, 439–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2004, 25, 4–10. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Denman, R.B. Using Rnafold to predict the activity of small catalytic RNAs. Biotechniques 1993, 15, 1090–1095. [Google Scholar] [PubMed]
- Sun, J.; Wang, S.; Li, C.; Ren, Y.; Wang, J. Novel expression profiles of microRNAs suggest that specific miRNAs regulate gene expression for the sexual maturation of female Schistosoma japonicum after pairing. Parasites Vectors 2014, 7, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tino, P. Basic properties and information theory of Audic-Claverie statistic for analyzing cDNA arrays. BMC Bioinform. 2009, 10, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]








| Sample | Raw Reads | Reads Trimmed Length | Reads Trimmed Q20 | Reads Trimmed N | Clean Reads | Clean Reads Uniq |
|---|---|---|---|---|---|---|
| bn | 33,344,602 | 28,940,389 | 28,913,796 | 28,853,904 | 28,853,904 | 1,570,219 |
| yy | 31,646,444 | 30,456,518 | 30,434,734 | 30,368,742 | 30,368,742 | 844,821 |
| Sample | Annotation Type | Number of Total | % of Total | Number of Uniq | % of Uniq |
|---|---|---|---|---|---|
| yy | rRNA | 2504 | 0.01% | 670 | 0.08% |
| tRNA | 270 | 0.00% | 124 | 0.01% | |
| snRNA | 748 | 0.00% | 397 | 0.05% | |
| Cis-reg | 722 | 0.00% | 391 | 0.05% | |
| other_Rfam_RNA | 2593 | 0.01% | 764 | 0.09% | |
| gene | 703,473 | 2.32% | 31,193 | 3.69% | |
| repeat | 870,399 | 2.87% | 38,129 | 4.51% | |
| known_miRNA | 178,799 | 0.59% | 2186 | 0.26% | |
| unannotation | 28,609,234 | 94.21% | 770,967 | 91.26% | |
| bn | rRNA | 16234 | 0.06% | 889 | 0.06% |
| tRNA | 775 | 0.00% | 169 | 0.01% | |
| snRNA | 6346 | 0.02% | 693 | 0.04% | |
| Cis-reg | 3169 | 0.01% | 686 | 0.04% | |
| other_Rfam_RNA | 7899 | 0.03% | 1366 | 0.09% | |
| gene | 1,737,462 | 6.02% | 52,551 | 3.35% | |
| repeat | 4,173,497 | 14.46% | 56,926 | 3.63% | |
| known_miRNA | 238,353 | 0.83% | 2462 | 0.16% | |
| unannotation | 22,670,169 | 78.57% | 1,454,477 | 92.63% |
| Novel ID | MiRDeep2 Score | Consensus Mature Sequence | Consensus Precursor Sequence | Precursor Coordinate |
|---|---|---|---|---|
| novel1 | 6.2 | cggcucgauuuguuugaaauac | guuucacucaucaucgagcuguuggacggcucgauuuguuugaaauac | TRINITY_DN19462_c0_g1_i2:301…349:+ |
| novel2 | 4.4 | ccggcggcggcgaucaugaagugc | ccggcggcggcgaucaugaagugcuucggcauggaauguuuucgucgucgcuaguucgg | TRINITY_DN23739_c0_g5_i2:245…304:+ |
| novel3 | 4.2 | uaacggaaacaacgaucagcc | cugauggaggugaagguaaugguggauauaacggaaacaacgaucagcc | TRINITY_DN14496_c0_g2_i1:253…302:+ |
| novel4 | 2.1 | cggcgcgccgggcccggc | cagcccggcgugcagggcccccagggcccgccgggauacccuggcgacaugggccccgugggccgcaccggcgcgccgggcccggc | TRINITY_DN14556_c0_g1_i1:557…643:+ |
| novel5 | 1.1 | uuuauuucgguaugucugc | aagcauauugaacuuuacaauuaaauucuggguguccucuguggugaggauuucugaguaaauaccucuuuauuucgguaugucugc | TRINITY_DN17506_c0_g1_i1:476…563:+ |
| novel6 | 0.9 | aaggcugaaacuuaaagga | cuuuaaguuaggcuuugcuaauaaaggcugaaacuuaaagga | TRINITY_DN24369_c0_g1_i1:2878…2920:- |
| novel7 | 0.9 | uggacggcgggguccuugcggac | uggacggcgggguccuugcggacuuccccagcuucggucggggaagucggaugagacuucggcggucuuacg | TRINITY_DN23739_c0_g3_i2:205…277:+ |
| novel8 | 0.5 | ucgguaucaacgaacuccuuga | cacgcggggaguugcaaugaauccaaucgaucauccacacggaggacgaacgaaagcgguucgguaucaacgaacuccuuga | TRINITY_DN24930_c0_g1_i1:681…763:+ |
| No. | Target Gene Symbol | Gene Description | Biological Processes | Up Down | Related Identified miRNAs |
|---|---|---|---|---|---|
| 1 | GPX | glutathione peroxidase | glutathione peroxidase activity | up | miR-23b-3p↓ |
| 2 | asps | aspartyl-tRNA synthetase | tRNA aminoacylation for protein translation | up | miR-193b↓ |
| 3 | TARS | threonyl-tRNA synthetase | tRNA aminoacylation for protein translation | up | miR-370-3p↓ |
| 4 | glyS | glycyl-tRNA synthetase alpha chain | arginyl-tRNA aminoacylation | up | miR-370-3p↓ |
| 5 | glnA | glutamine synthetase | putrescine binding | up | miR-370-3p↓, miR-145↓ |
| 6 | UQCRFS1 | ubiquinol-cytochrome c reductase iron-sulfur subunit | ubiquinol-cytochrome-c reductase activity | up | miR-744-5p↓ |
| 7 | ACOX1 | acyl-CoA oxidase | very long-chain fatty acid metabolic process | up | miR-28↓ |
| 8 | UBE1 | ubiquitin-activating enzyme E1 | ubiquitin activating enzyme activity | up | novel2_mature↓ |
| 9 | CPT1 | carnitine O-palmitoyltransferase 1 | long-chain fatty acid metabolic process | up | novel2_mature↓ |
| 10 | F-Actin | actin beta/gamma 1 | structural constituent of cytoskeleton | up | novel2_mature↓ |
| 11 | HtpG | molecular chaperone HtpG | protein folding | up | novel2_mature↓ |
| 12 | ACSL | long-chain acyl-CoA synthetase | long-chain fatty acid-CoA ligase activity | up | miR-10178-5p↓ |
| 13 | SGK | serum/glucocorticoid-regulated kinase | neuron projection morphogenesis | down | miR-143↑ |
| 14 | RP-L2 | large subunit ribosomal protein L2 | structural constituent of ribosome; translation | down | miR-1307↑, miR-1180↑, miR-339↑ |
| 15 | rub | Rubredoxin-NAD+ reductase | rubredoxin-NADP reductase activity | down | miR-1307↑ |
| 16 | COX5B | cytochrome c oxidase subunit 5b | cytochrome-c oxidase activity | down | miR-106b-3p↑ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, N.; Bhatti, M.Z.; Zhang, H.; Ni, B.; Fan, X.; Chen, J. The Encystment-Related MicroRNAs and Its Regulation Molecular Mechanism in Pseudourostyla cristata Revealed by High Throughput Small RNA Sequencing. Int. J. Mol. Sci. 2020, 21, 2309. https://doi.org/10.3390/ijms21072309
Pan N, Bhatti MZ, Zhang H, Ni B, Fan X, Chen J. The Encystment-Related MicroRNAs and Its Regulation Molecular Mechanism in Pseudourostyla cristata Revealed by High Throughput Small RNA Sequencing. International Journal of Molecular Sciences. 2020; 21(7):2309. https://doi.org/10.3390/ijms21072309
Chicago/Turabian StylePan, Nan, Muhammad Zeeshan Bhatti, Haiyang Zhang, Bing Ni, Xinpeng Fan, and Jiwu Chen. 2020. "The Encystment-Related MicroRNAs and Its Regulation Molecular Mechanism in Pseudourostyla cristata Revealed by High Throughput Small RNA Sequencing" International Journal of Molecular Sciences 21, no. 7: 2309. https://doi.org/10.3390/ijms21072309





