Cyclin-Dependent Kinase Inhibitor Gene TaICK1 acts as a Potential Contributor to Wheat Male Sterility induced by a Chemical Hybridizing Agent
Abstract
:1. Introduction
2. Results
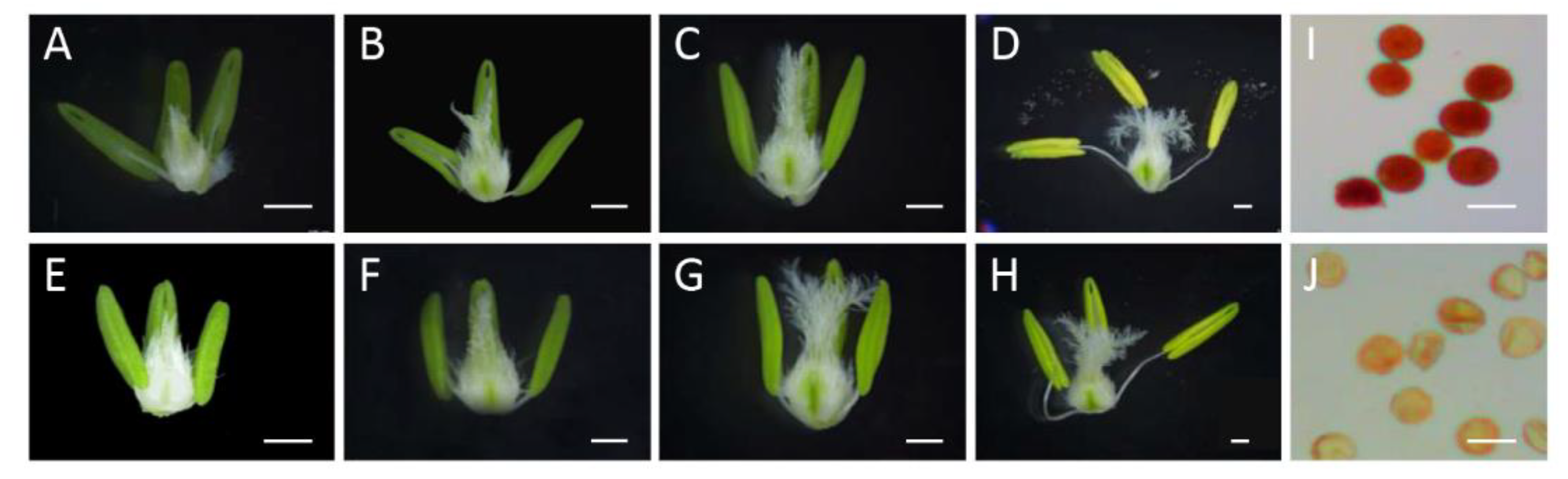
2.1. Morphological and Cytological Characteristics
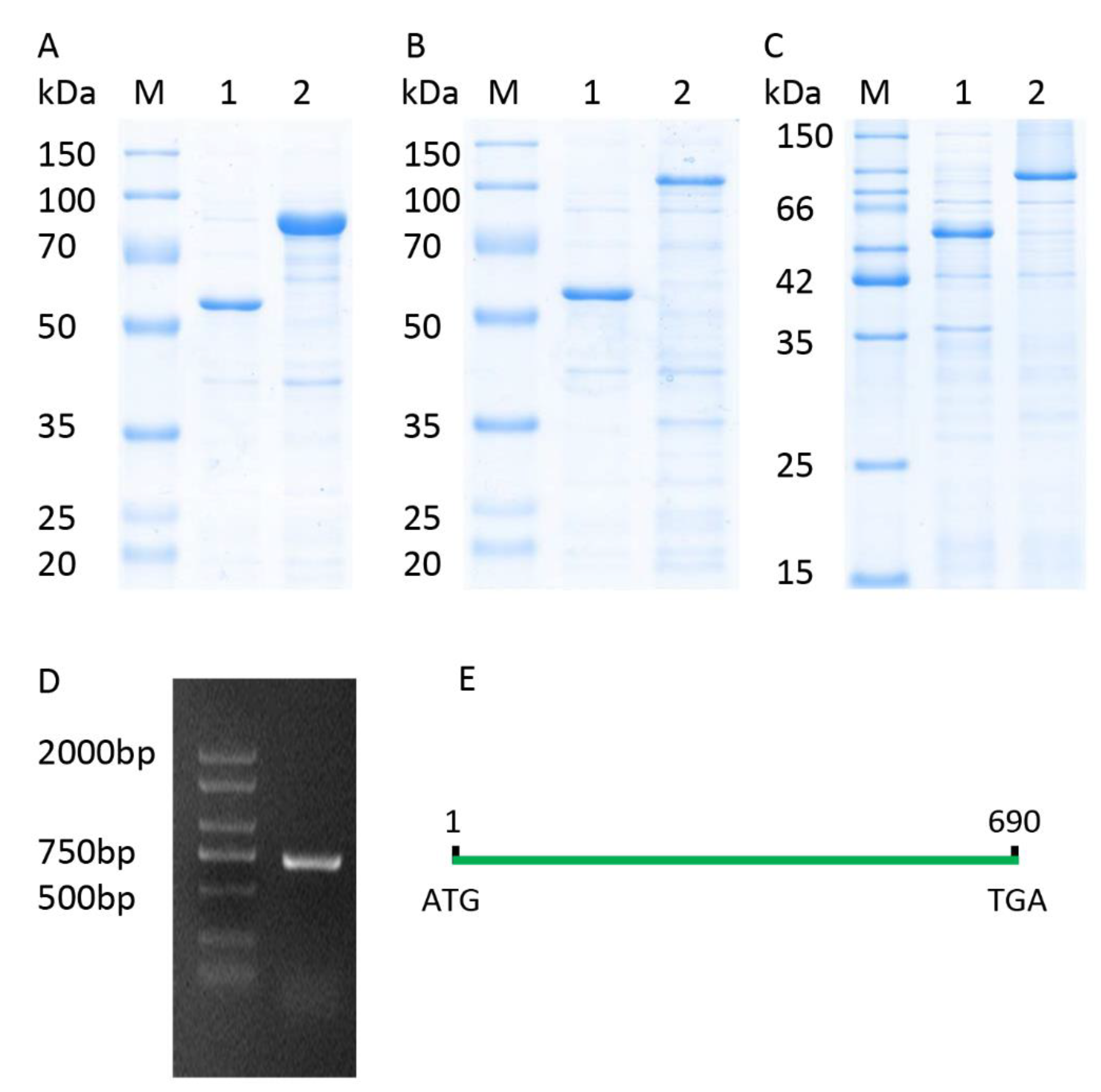
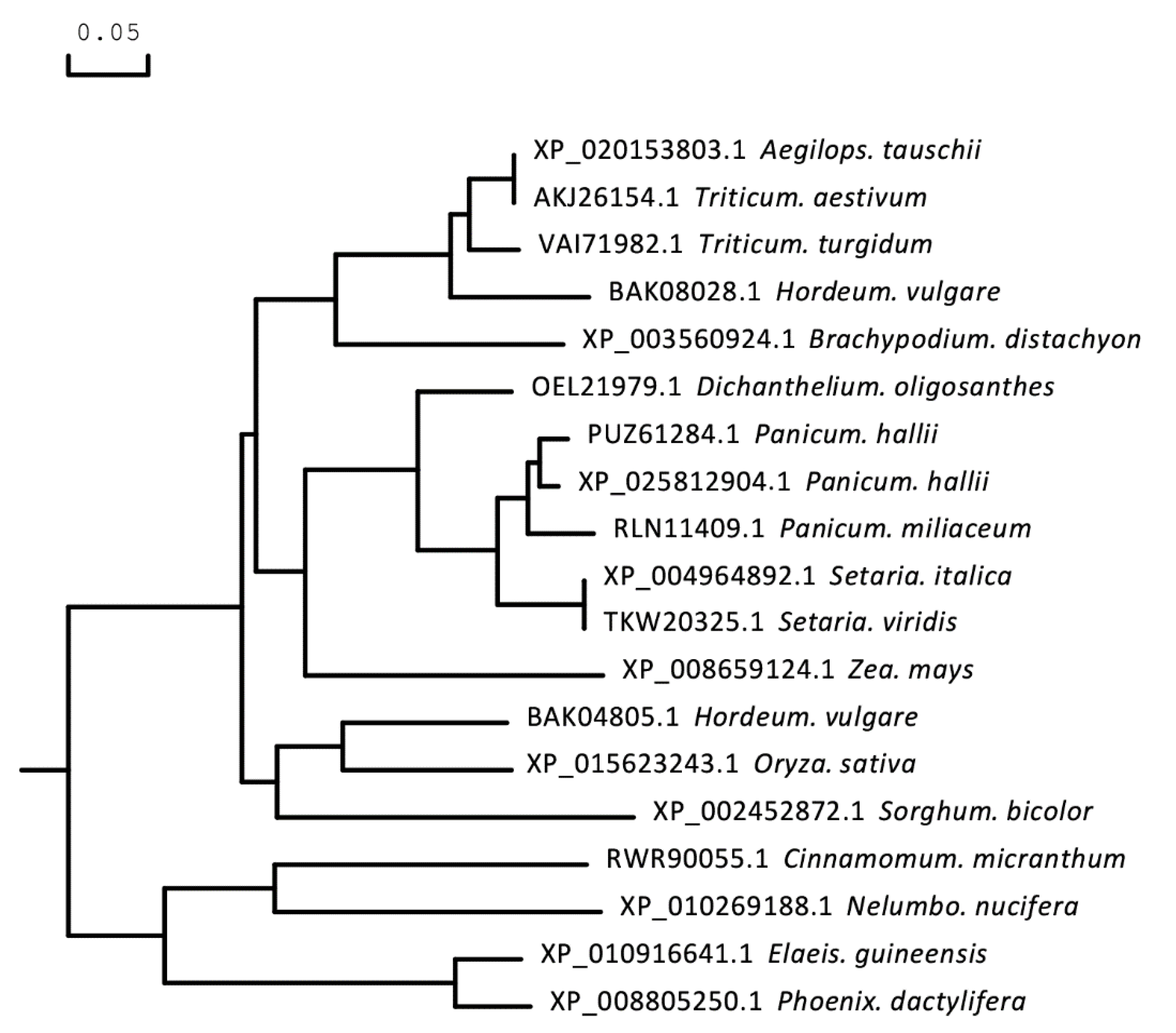
2.2. Structure of the Gene TaICK1
2.3. Expression Level of TaICK1 in XN1376-CIMS and XN1376
2.4. The Phenotype of TaICK1 Transgenic Rice Plants
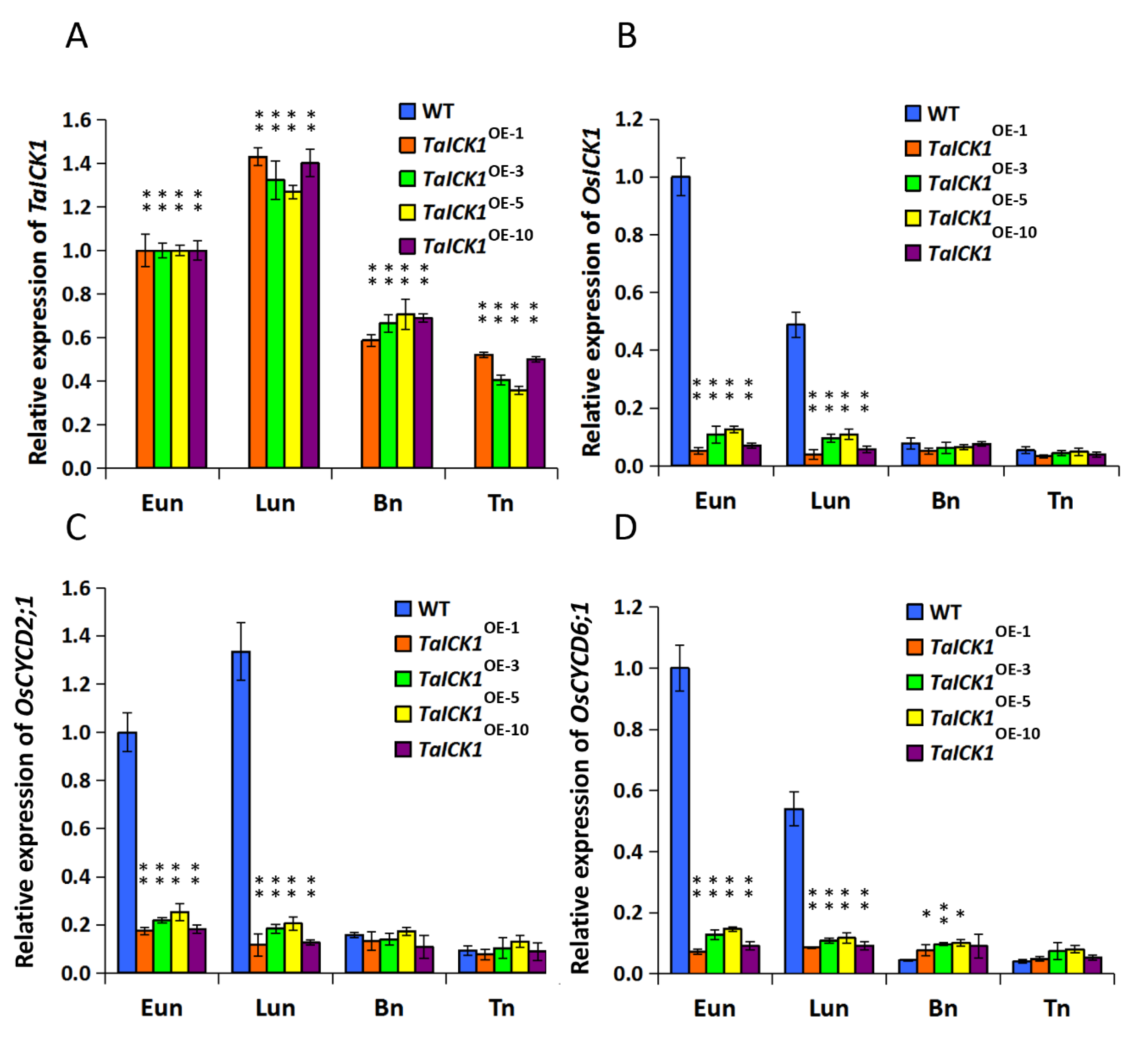
2.5. Expression Patterns of TaICK1 and OsICK1 in Transgenic Rice
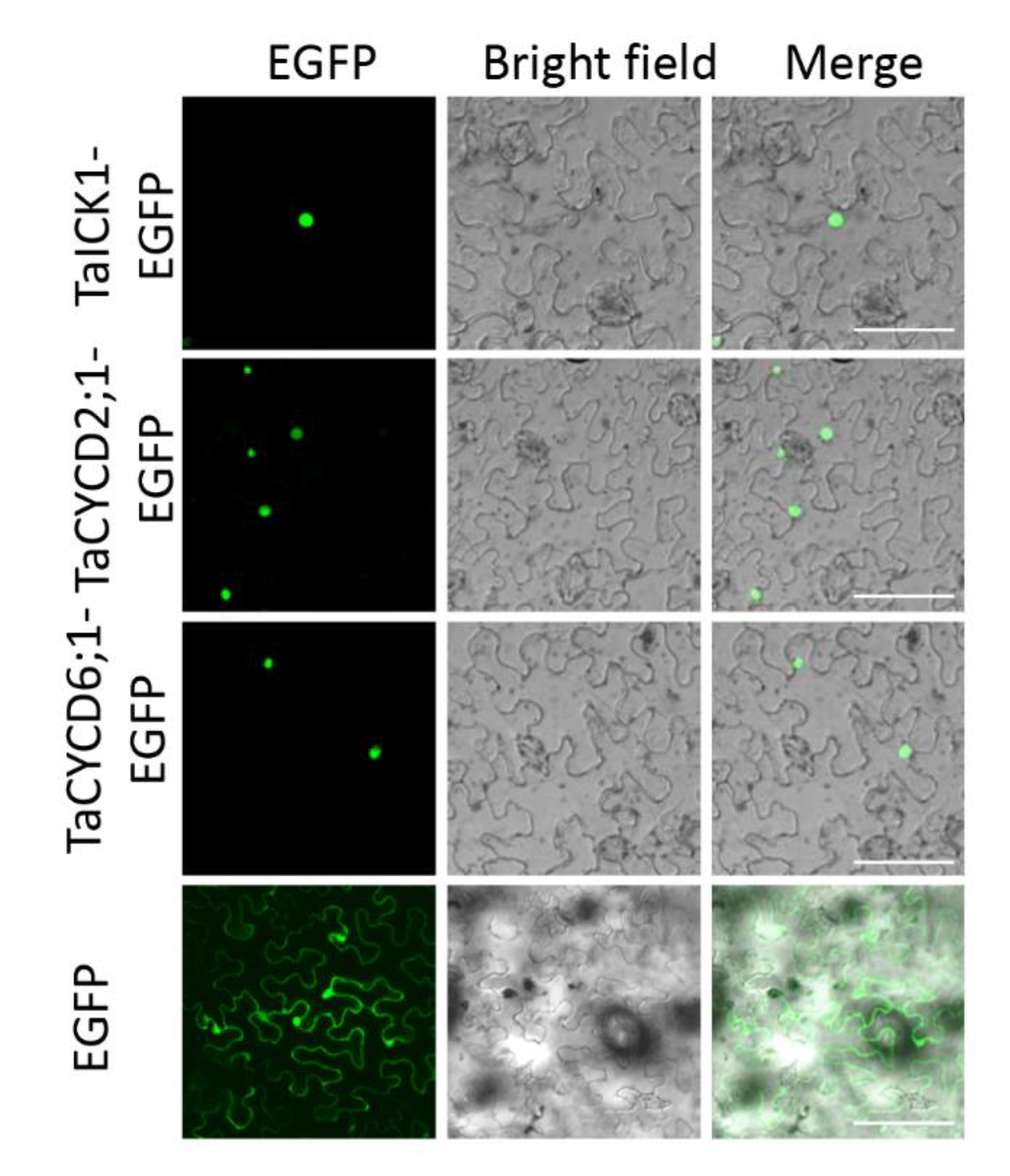
2.6. Subcellular Localization Assay of Protein TaICK1
2.7. Prokaryotic Expression of TaICK1 in E. coli
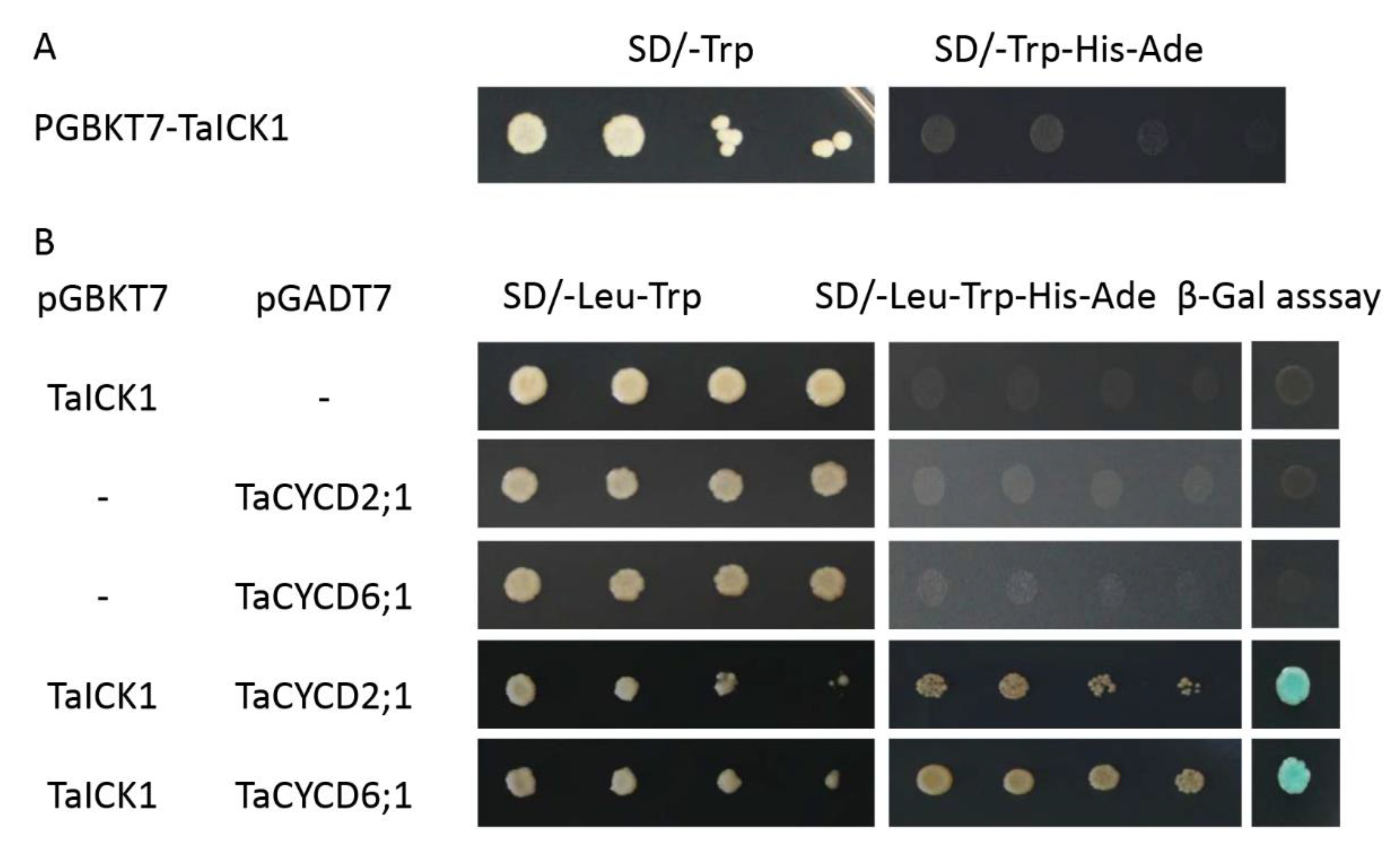
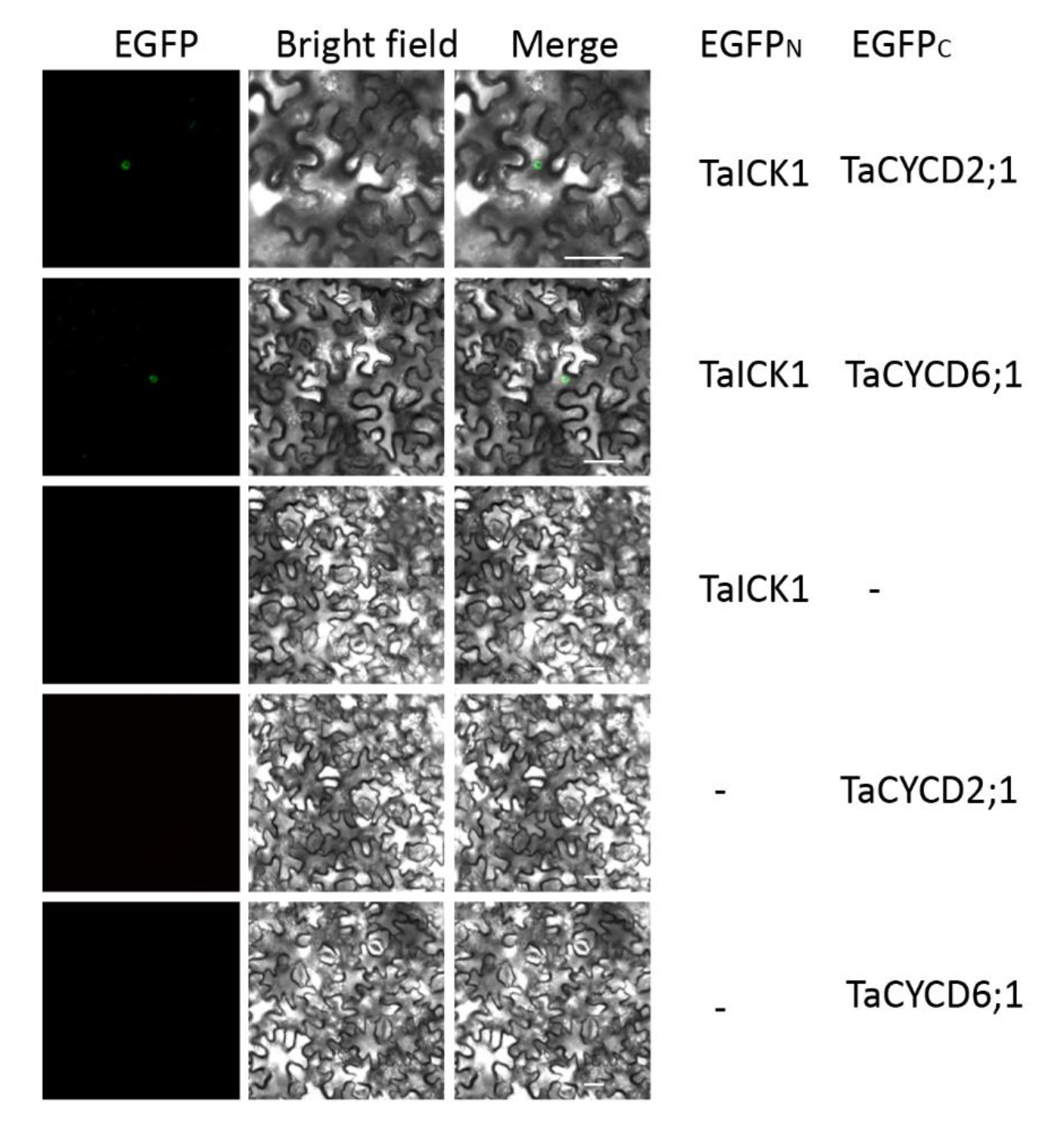
2.8. TaICK1 Interacts with TaCYCD2;1 and TaCYCD6;1
2.9. The Expression Patterns of CYCD2;1 and CYCD6;1 in Wheat and Transgenic Rice
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phenotype Characterization of Wheat Anther Development
4.3. RNA Extraction and qRT-PCR Analysis
4.4. Primer Design
4.5. Plasmid Constructs
4.6. Generation of the TaICK1 Overexpression Rice Plants
4.7. Yeast Two-Hybrid Assay
4.8. Subcellular Localization and BiFC Assay
4.9. Recombinant Protein Expression in E. coli
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gill, B.S.; Appels, R.; Botha-Oberholster, A.; Buell, C.R.; Bennetzen, J.L.; Chalhoub, B.; Chumley, F.; Dvorák, J.; Iwanaga, M.; Keller, B.; et al. A workshop report on wheat genome sequencing: International Genome Research on Wheat Consortium. Genetics 2004, 168, 1087–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.K.; Mir, R.R.; Mohan, A.; Kumar, J. Wheat genomics: Present status and future prospects. Int. J. Plant Genom. 2008, 2008, 896451. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; Del, B.A.; Dubcovsky, J.; Uauy, C. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 2016, 129, 1099–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, T.C.; Zhang, X.K.; Yin, G.H.; Wang, L.N.; Han, Y.L.; Chen, L.; Huang, F.; Tang, J.W.; Xia, X.C.; He, Z.H. Genetic gains in grain yield, net photosynthesis and stomatal conductance achieved in Henan Province of China between 1981 and 2008. Field Crop. Res. 2011, 122, 225–233. [Google Scholar] [CrossRef]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Bai, Z.; Li, X.; Wang, P.; Wu, Q.; Yang, L.; Li, L.; Li, X. SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor. Appl. Genet. 2012, 125, 1057–1068. [Google Scholar] [CrossRef]
- Kindred, D.R.; Gooding, M.J. Heterosis for yield and its physiological determinants in wheat. Euphytica 2005, 142, 149–159. [Google Scholar] [CrossRef]
- Zhang, Q. Utilization of crop heterosis: A review. Euphytica 2014, 2, 161–173. [Google Scholar]
- Singh, S.P.; Srivastava, R.; Kumar, J. Male sterility systems in wheat and opportunities for hybrid wheat development. Acta Physiol. Plant. 2014, 37, 1713. [Google Scholar] [CrossRef]
- Hohn, C.E.; Lukaszewski, A.J. Engineering the 1BS chromosome arm in wheat to remove the Rf multi locus restoring male fertility in cytoplasms of Aegilops kotschyi, Ae. uniaristata and Ae. mutica. Theor. Appl. Genet. 2016, 129, 1769–1774. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Wang, Q.; Li, Z.; Cui, J.; Hu, S.; Zhao, H.; Chen, M. Cytological and comparative proteomic analyses on male sterility in Brassica napus L. induced by the chemical hybridization agent monosulphuron ester sodium. PLoS ONE 2013, 8, e80191. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.W.; Zhang, G.S.; Wang, J.W.; Wang, X.L.; Fang, Z.W. Effect of male sterility on different wheat genotype induced by SQ-1. J. Northwest Univ. Nat. Sci. Ed. 2003, 31, 15–18. [Google Scholar]
- Zhang, G.; Liu, H.; Wang, J. New variety of hybrid wheat with high resistance, quality and yield. Agric. New Technol. 2003, 4, 25–26. [Google Scholar]
- Chen, C.; Lu, Y.; Zhang, G.; Zhang, Z.; Li, P.; Zhang, B. The synthesis of genesis and the research of bioactive chemicals. Chin. J. Pestic. 2004, 43, 315–317. [Google Scholar]
- Wang, S.; Zhang, G.; Song, Q.; Zhang, Y.; Li, Y.; Guo, J.; Chen, Z.; Niu, N.; Ma, S.; Wang, J. Programmed cell death, antioxidant response and oxidative stress in wheat flag leaves induced by chemical hybridization agent SQ-1. J. Integr. Agr. 2016, 15, 76–86. [Google Scholar] [CrossRef]
- Song, Q.; Wang, S.; Zhang, G.; Li, Y.; Li, Z.; Guo, J.; Niu, N.; Wang, J.; Ma, S. Comparative proteomic analysis of a membrane-enriched fraction from flag leaves reveals responses to chemical hybridization agent SQ-1 in wheat. Front. Plant Sci. 2015, 6, 669. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Ye, J.; Zhang, G.; Wang, J.; Niu, N.; Ma, S.; Zhao, J.; Zhu, J. Differential proteomic analysis of anther proteins between cytoplasmic-nuclear male sterility line and its maintainer in wheat (Triticum aestivum L.). Prog. Biochem. Biophys. 2009, 36, 431–440. [Google Scholar]
- Wang, J.; Zhang, G.; Yuan, L.; Zhang, M.; Niu, N.; Ma, S.; Ye, J. Metabolism of Reactive Oxygen Species of Physiological Male Sterile Anther Induced by Chemical Hybrid Agent in Wheat. Acta Bot. Boreali Occident. Sin. 2009, 29, 1351–1357. [Google Scholar]
- Wei, M.; Wang, J.; Zhang, G.; Zhang, L.; Yang, Z.; Sun, R.; Ye, J.; Niu, N.; Ma, S.; Li, H. Relationship between the expression of GAPDH gene and anther abortion of physiological male sterile of wheat. Mol. Plant Breed. 2009, 7, 679–684. [Google Scholar]
- Ye, J.; Zhang, G.; Wang, S.; Chen, R.; Wang, J.; Niu, N.; Ma, S.; Li, H.; Zhu, J. Differential proteomic studies on pollen grain proteins of wheat male sterile line induced by chemical hybridizing agent SQ-1. Chin. J. Biochem. Mol. Biol. 2009, 25, 949–957. [Google Scholar]
- Wang, S.; Zhang, Y.; Song, Q.; Fang, Z.; Chen, Z.; Zhang, Y.; Zhang, L.; Zhang, L.; Niu, N.; Ma, S.; et al. Mitochondrial Dysfunction Causes Oxidative Stress and Tapetal Apoptosis in Chemical Hybridization Reagent-Induced Male Sterility in Wheat. Front. Plant Sci. 2018, 8, 2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Zhang, G.; Wang, J.; Li, J.; Song, Y.; Qiao, L.; Niu, N.; Wang, J.; Ma, S.; Li, L. Chemical hybridizing agent SQ-1-induced male sterility in Triticum aestivum L.: A comparative analysis of the anther proteome. BMC Plant Biol. 2018, 18, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Zhang, Y.; Fang, Z.; Zhang, Y.; Song, Q.; Hou, Z.; Sun, K.; Song, Y.; Li, Y.; Ma, D.; et al. Cytological and Proteomic Analysis of Wheat Pollen Abortion Induced by Chemical Hybridization Agent. Int. J. Mol. Sci. 2019, 20, 1615. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yuan, L.; Yang, S.L.; Zhang, G.S.; Wang, J.S.; Song, Y.L.; Zhao, Z.J.; Niu, N.; Ma, S.C. Expression characteristic on Ta PDC- E1α gene and its regulatory enzymes gene in male sterile line of wheat(Triticum aestivum L.). Acta Agron. Sin. 2011, 37, 620–628. [Google Scholar] [CrossRef]
- Yui, R.; Iketani, S.; Mikami, T.; Kubo, T. Antisense inhibition of mitochondrial pyruvate dehydrogenase E1α subunit in anther tapetum causes male sterility. Plant J. 2003, 34, 57–66. [Google Scholar] [CrossRef] [PubMed]
- de Kok, A.; Hengeveld, A.F.; Martin, A.; Westphal, A.H. The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta BBA Protein Struct. Mol. Enzymol. 1998, 1385, 353–366. [Google Scholar] [CrossRef]
- Tovar-Méndez, A.; Miernyk, J.A.; Randall, D.D. Regulation of pyruvate dehydrogenase complex activity in plant cells. Eur. J. Biochem. 2003, 270, 1043–1049. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Zhao, X.L.; Liu, H.Z.; Song, Y.L.; Zhang, G. Proliferating cell nuclear antigen (TaPCNA), a pyruvate dehydrogenase kinase (TaPDK)-interacting protein is involved in the regulation of microspore cell cycle in wheat (Triticum aestivum). J. Agric. Biotechnol. 2013, 21, 1045–1051. [Google Scholar]
- Yu, Y. Study on Expression Analysis of TaPCNA from Regulation of Microspore Cell Cycle and Its Interactive Protein of Physiological Male- Sterility in Wheat; Northwest A&F university: Xianyang, China, 2015. [Google Scholar]
- Wang, H.; Fowke, L.C.; Crosby, W.L. A plant cyclin-dependent kinase inhibitor gene. Nature 1997, 386, 451–452. [Google Scholar] [CrossRef]
- Lui, H.; Wang, H.; DeLong, C.; Fowke, L.C.; Crosby, W.L.; Fobert, P.R. The Arabidopsis Cdc2a-interacting protein ICK2 is structurally related to ICK1 and is a potent inhibitor of cyclin-dependent kinase activity in vitro. Plant J. 2000, 21, 379–385. [Google Scholar] [CrossRef] [Green Version]
- Gusti, A.; Baumberger, N.; Nowack, M.; Pusch, S.; Eisler, H.; Potuschak, T.; De Veylder, L.; Schnittger, A.; Genschik, P. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS ONE 2009, 4, e4780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jasinski, S.; Perennes, C.; Bergounioux, C.; Nathalie, G. Comparative Molecular and Functional Analyses of the Tobacco Cyclin-Dependent Kinase Inhibitor NtKIS1a and Its Spliced Variant NtKIs1b. Plant Physiol. 2002, 130, 1871–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, C.M.; Dante, R.A.; Sabelli, P.A.; Sun, Y.; Dilkes, B.P.; Gordon-Kamm, W.J.; Larkins, B.A. Cyclin-Dependent Kinase Inhibitors in Maize Endosperm and Their Potential Role in Endoreduplication. Plant Physiol. 2005, 138, 2323–2336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisbis, B.; Delmas, F.; Joubès, J.; Sicard, A.; Hernould, M.; Inzé, D.; Mouras, A.; Chevalier, C. Cyclin-dependent Kinase (CDK) Inhibitors Regulate the CDK-Cyclin Complex Activities in Endoreduplicating Cells of Developing Tomato Fruit. J. Biol. Chem. 2006, 281, 7374–7383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrôco, R.M.; Peres, A.; Droual, A.; De Veylder, L.; Nguyen, L.S.L.; De Wolf, J.; Mironov, V.; Peerbolte, R.; Beemster, G.T.S.; Inzé, D.; et al. The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice. Plant Physiol. 2006, 142, 1053–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Qi, Q.; Schorr, P.; Cutler, A.J.; Crosby, W.L.; Fowke, L.C. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 1998, 15, 501–510. [Google Scholar] [CrossRef]
- Torres Acosta, J.A.; Fowke, L.C.; Wang, H. Analyses of phylogeny, evolution, conserved sequences and genome-wide expression of the ICK/KRP family of plant CDK inhibitors. Ann. Bot. (Lond.) 2011, 107, 1141–1157. [Google Scholar] [CrossRef] [Green Version]
- Jasinski, S.; Riou-Khamlichi, C.; Roche, O.; Perennes, C.; Bergounioux, C.; Glab, N. The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3; 1-overexpressing plants. J. Cell Sci. 2002, 115, 973–982. [Google Scholar]
- Wang, H.; Zhou, Y.; Gilmer, S.; Whitwill, S.; Fowke, L.C. Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 2000, 24, 613–623. [Google Scholar] [CrossRef]
- De Veylder, L.; Beeckman, T.; Beemster, G.T.; Krols, L.; Terras, F.; Landrieu, I.; van der Schueren, E.; Maes, S.; Naudts, M.; Inzé, D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 2001, 13, 1653–1668. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Fowke, L.; Wang, H. Plant CDK inhibitors: Studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Rep. 2002, 20, 967–975. [Google Scholar] [CrossRef]
- Schnittger, A.; Weinl, C.; Bouyer, D.; Schöbinger, U.; Hülskamp, M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 2003, 15, 303–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Oh, S.A.; Brownfield, L.; Hong, S.H.; Ryu, H.; Hwang, I.; Twell, D.; Nam, H.G. Control of plant germline proliferation by SCFFBL17 degradation of cell cycle inhibitors. Nature 2008, 455, 1134–1137. [Google Scholar] [CrossRef] [PubMed]
- Oakenfull, E.A.; Riou-Khamlichi, C.; Murray, A.H. Plant D—Type cyclins and the control of G1 progression. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2002, 357, 749–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz, L.; Dewitte, W.; Forzani, C.; Patell, F.; Nieuwland, J.; Wen, B.; Quelhas, P.; De Jager, S.; Titmus, C.; Campilho, A.; et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 2011, 23, 641–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Wang, H.; Gilmer, S.; Whitwill, S.; Fowke, L.C. Effects of co-expressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta 2003, 216, 604–613. [Google Scholar] [CrossRef]
- Song Ba, Q.; Sheng Zhang, G.; Sheng Wang, J.; Xue Che, H.; Liu, H.; Niu, N.; Cai Ma, S.; Wei Wang, J. Relationship between metabolism of reactive oxygen species and chemically induced male sterility in wheat (Triticum aestivum L.). Can. J. Plant Sci. 2013, 93, 675–681. [Google Scholar]
- Ba, Q.; Zhang, G.; Che, H.; Liu, H.; Ng, T.; Zhang, L.; Wang, J.; Sheng, Y.; Niu, N.; Ma, S.; et al. Aliphatic Metabolism during Anther Development Interfered by Chemical Hybridizing Agent in Wheat. Crop Sci. 2014, 54, 1458–1467. [Google Scholar] [CrossRef]
- Zhang, M.; Yuan, L.; Wang, J.; Zhang, G.; Guo, W.; Zhang, L. Expressing Analysis of cACO Gene in Male Sterile Wheat Induced by Chemical Hybridizing Agent SQ-1. Chin. J. Biochem. Mol. Biol. 2010, 26, 740–748. [Google Scholar]
- Wang, S.; Zhang, G.; Zhang, Y.; Song, Q.; Chen, Z.; Wang, J.; Guo, J.; Niu, N.; Wang, J.; Ma, S. Comparative studies of mitochondrial proteomics reveal an intimate protein network of male sterility in wheat (Triticum aestivum L.). J. Exp. Bot. 2015, 66, 6190–6203. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yuan, L.; Zhang, M.; Zhang, G.; Niu, N.; Ma, S.; Li, H. Expression of Ubiquitin-26S Proteasome Pathway in Physiological Male Sterility of Wheat Induced by Chemical Hybrid Agents SQ-1. J. Agric. Biotechnol. 2010, 18, 695–701. [Google Scholar]
- Sang, Q.; Liu, H.; Zhang, G.; Zhu, Q.; Zhu, W.; Zhang, L.; Che, H. Study on the Relationship between the Ubiquitination of Histones on Chromosomes and the Physiological Male Sterility Induced by Chemical Hybridizing Agent SQ-1 in Wheat (Triticum aestivum L.). J. Agric. Biotechnol. 2013, 21, 396–406. [Google Scholar]
- Ba, Q.; Zhang, G.; Wang, J.; Niu, N.; Ma, S.; Wang, J. Gene expression and DNA methylation alterations in chemically induced male sterility anthers in wheat (Triticum aestivum L.). Acta Physiol. Plant. 2014, 36, 503–512. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, G.; Zhu, W.; Wu, W.K.K.; Ba, Q.; Zhang, L.; Zhang, L.; Niu, N.; Ma, S.; Wang, J. Differential proteomic analysis of polyubiquitin-related proteins in chemical hybridization agent-induced wheat (Triticum aestivum L.) male sterility. Acta Physiol. Plant. 2014, 36, 1473–1489. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Y.; Fowke, L.C. The emerging importance of cyclin-dependent kinase inhibitors in the regulation of the plant cell cycle and related processesThis review is one of a selection of papers published in the Special Issue on Plant Cell Biology. Can. J. Bot. 2006, 84, 640–650. [Google Scholar] [CrossRef]
- Pettkó-Szandtner, A.; Mészáros, T.; Horváth, G.V.; Bakó, L.; Csordás-Tóth, É.; Blastyák, A.; Zhiponova, M.; Miskolczi, P.; Dudits, D. Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J. 2006, 46, 111–123. [Google Scholar] [CrossRef]
- Yang, R.; Tang, Q.; Wang, H.; Zhang, X.; Pan, G.; Wang, H.; Tu, J. Analyses of two rice (Oryza sativa) cyclin-dependent kinase inhibitors and effects of transgenic expression of OsiICK6 on plant growth and development. Ann. Bot. 2011, 107, 1087–1101. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wang, H.; Gilmer, S.; Whitwill, S.; Keller, W.; Fowke, L.C. Control of petal and pollen development by the plant cyclin-dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta 2002, 215, 248–257. [Google Scholar] [CrossRef]
- Sherr, C.; Roberts, J. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.; Zhuo, X.; Li, L.; Cheng, T.; Zhang, Q. Genome-Wide Analysis of the D-type Cyclin Gene Family Reveals Differential Expression Patterns and Stem Development in the Woody Plant Prunus mume. Forests 2019, 10, 147. [Google Scholar] [CrossRef] [Green Version]
- Menges, M.; Samland, A.K.; Planchais, S.; Murray, J.A.H. The D-Type Cyclin CYCD3;1 Is Limiting for the G1-to-S-Phase Transition inArabidopsis. Plant Cell 2006, 18, 893–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, R.; John, P.C.L. Expression of genomic AtCYCD2;1 in Arabidopsis induces cell division at smaller cell sizes: Implications for the control of plant growth. Plant Physiol. 2007, 144, 1587–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockcroft, C.E.; den Boer, B.G.W.; Healy, J.M.S.; Murray, J.A.H. Cyclin D control of growth rate in plants. Nature 2000, 405, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Gong, L.; Yuan, W.; Li, X.; Chen, G.; Li, X.; Zhang, Q.; Wu, C. Replication Protein A (RPA1a) Is Required for Meiotic and Somatic DNA Repair But Is Dispensable for DNA Replication and Homologous Recombination in Rice. Plant Physiol. 2009, 151, 2162–2173. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Li, H.; Su, Y.; Li, W.; Shi, C. G1/ELE Functions in the Development of Rice Lemmas in Addition to Determining Identities of Empty Glumes. Front. Plant Sci. 2016, 7, 1006. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wu, Y.; Williams, M.; Bernard, S.; Driouich, A.; Showalter, A.M.; Faik, A. Functional Identification of Two Nonredundant Arabidopsis α(1,2) Fucosyltransferases Specific to Arabinogalactan Proteins. J. Biol. Chem. 2010, 285, 13638–13645. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Bai, X.; Wang, X.; Chu, C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007, 164, 969–979. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, X.; Wang, X.; Peng, L.; Gu, M. A Rapid Simple Method of Assaying Hygromycin Resistance in Transgenic Rice Plants. J. Agric. Biotechnol. 2001, 9, 264. [Google Scholar]
- Waadt, R.; Schmidt, L.K.; Lohse, M.; Hashimoto, K.; Bock, R.; Kudla, J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant. J. 2008, 56, 505–516. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef] [PubMed]
- Schatz, P.J.; Beckwith, J. Genetic Analysis of Protein Export in Escherichia Coli. Annu. Rev. Genet. 1990, 24, 215–248. [Google Scholar] [CrossRef] [PubMed]
- Beliveau, C.; Potvin, C.; Trudel, J.; Asselin, A.; Bellemare, G. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J. Bacteriol. 1991, 173, 5619–5623. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Plant | OE-1 | OE-3 | OE-5 | OE-7 | OE-8 | OE-10 | OE-15 | WT |
|---|---|---|---|---|---|---|---|---|
| T1 | 19.7% | 58.6% | 58.7% | 63.5% | 38.1% | 57.4% | 29.6% | 94.1% |
| T2 | 33.4% | 58.6% | 66.2% | 46.7% | 47.3% | 30.2% | 68.9% | 95.1% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wang, C.; Yu, Y.; Zhang, Y.; Song, Y.; Li, Z.; Wang, S.; Zhang, Y.; Guo, X.; Liu, D.; et al. Cyclin-Dependent Kinase Inhibitor Gene TaICK1 acts as a Potential Contributor to Wheat Male Sterility induced by a Chemical Hybridizing Agent. Int. J. Mol. Sci. 2020, 21, 2468. https://doi.org/10.3390/ijms21072468
Zhang L, Wang C, Yu Y, Zhang Y, Song Y, Li Z, Wang S, Zhang Y, Guo X, Liu D, et al. Cyclin-Dependent Kinase Inhibitor Gene TaICK1 acts as a Potential Contributor to Wheat Male Sterility induced by a Chemical Hybridizing Agent. International Journal of Molecular Sciences. 2020; 21(7):2468. https://doi.org/10.3390/ijms21072468
Chicago/Turabian StyleZhang, Lili, Chaojie Wang, Yongang Yu, Yamin Zhang, Yulong Song, Zheng Li, Shuping Wang, Yanfang Zhang, Xiaofeng Guo, Dan Liu, and et al. 2020. "Cyclin-Dependent Kinase Inhibitor Gene TaICK1 acts as a Potential Contributor to Wheat Male Sterility induced by a Chemical Hybridizing Agent" International Journal of Molecular Sciences 21, no. 7: 2468. https://doi.org/10.3390/ijms21072468






