Transgenic Breeding Approaches for Improving Abiotic Stress Tolerance: Recent Progress and Future Perspectives
Abstract
:1. Introduction
2. Physiological and Biochemical Mechanisms of Abiotic Stress
3. Biotechnology Applied to the Breeding of Abiotic Stress Tolerance
4. Quantitative Trait Loci (QTL)
5. miRNAs in Abiotic Stress Tolerance
6. The Development of Abiotic Stress-Tolerant Crops by CRISPR (Lustered Regularly Interspaced Short Palindromic Repeats)/Cas9
7. Transgenic Breeding to Improve Abiotic Stress Tolerance
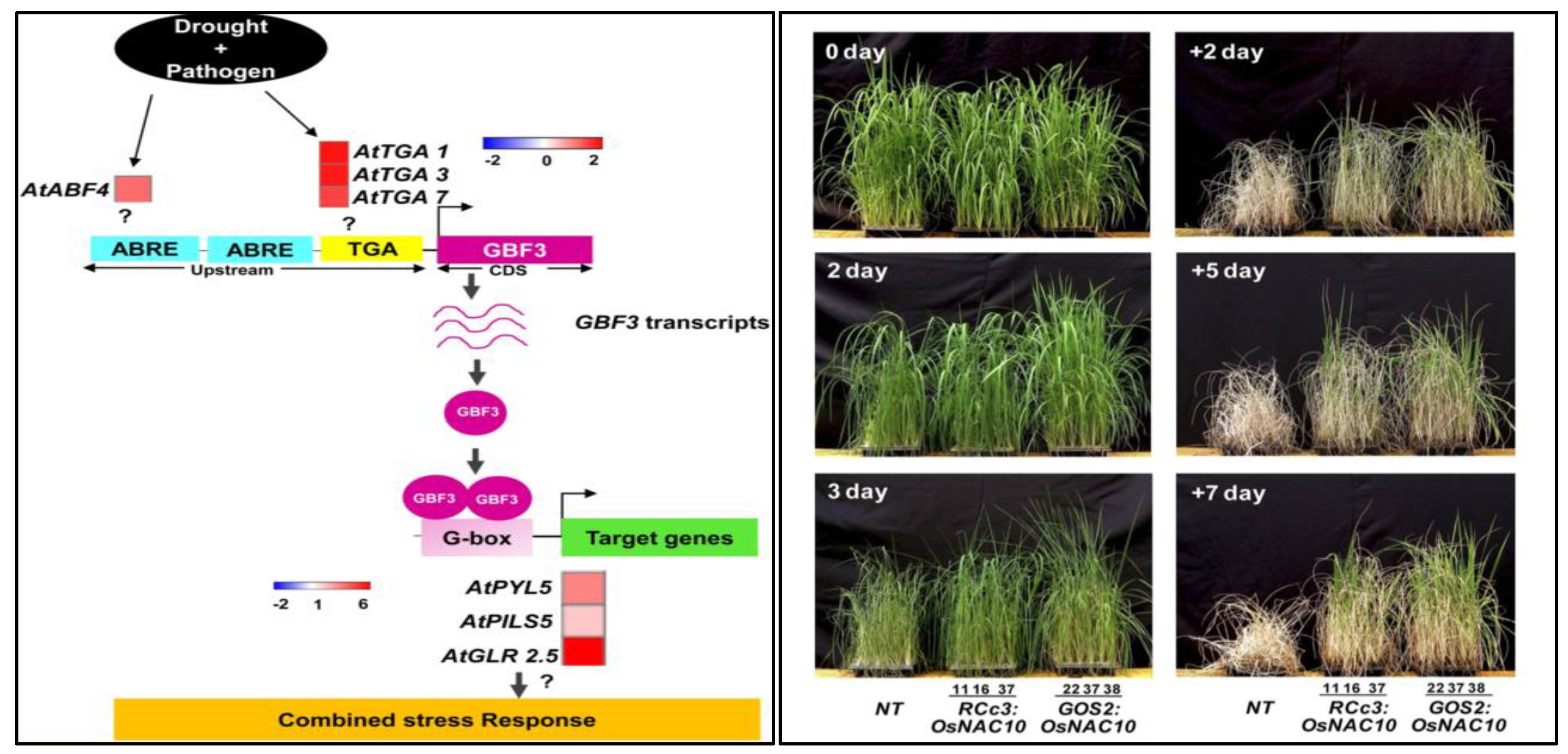
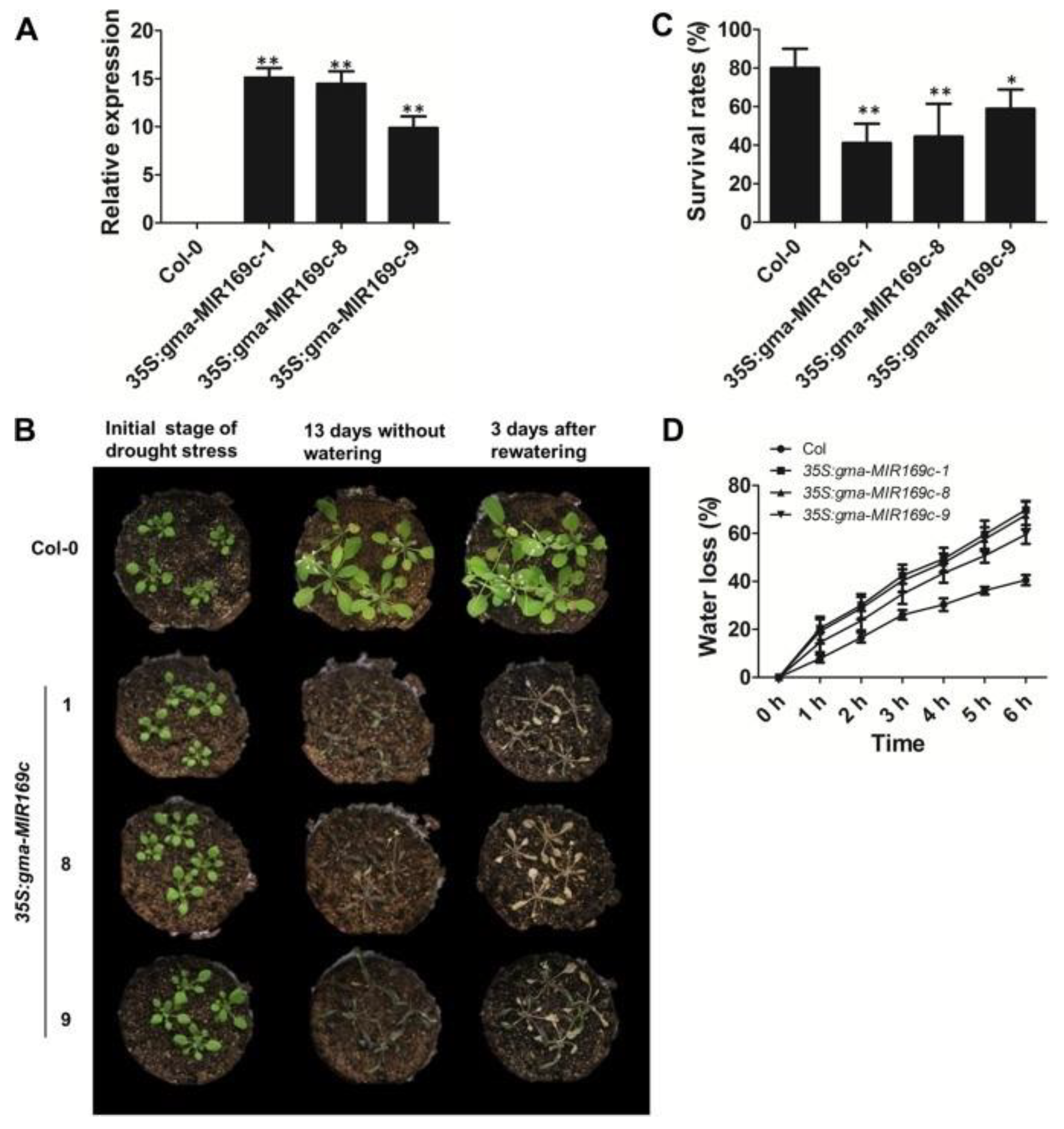
7.1. Drought Stress
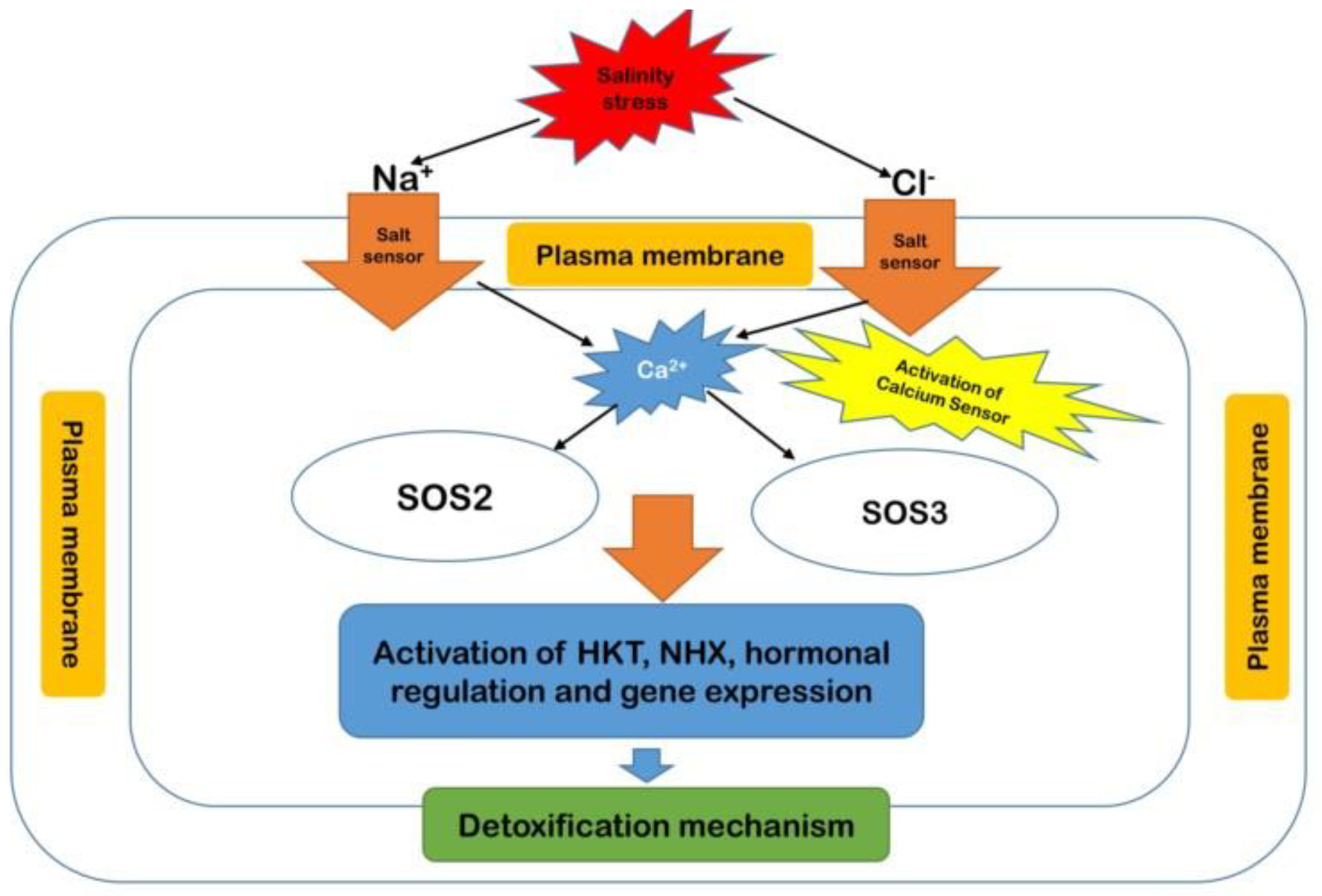
7.2. Salinity Stress
7.3. Temperature Stresses
7.4. Heavy Metal Stress
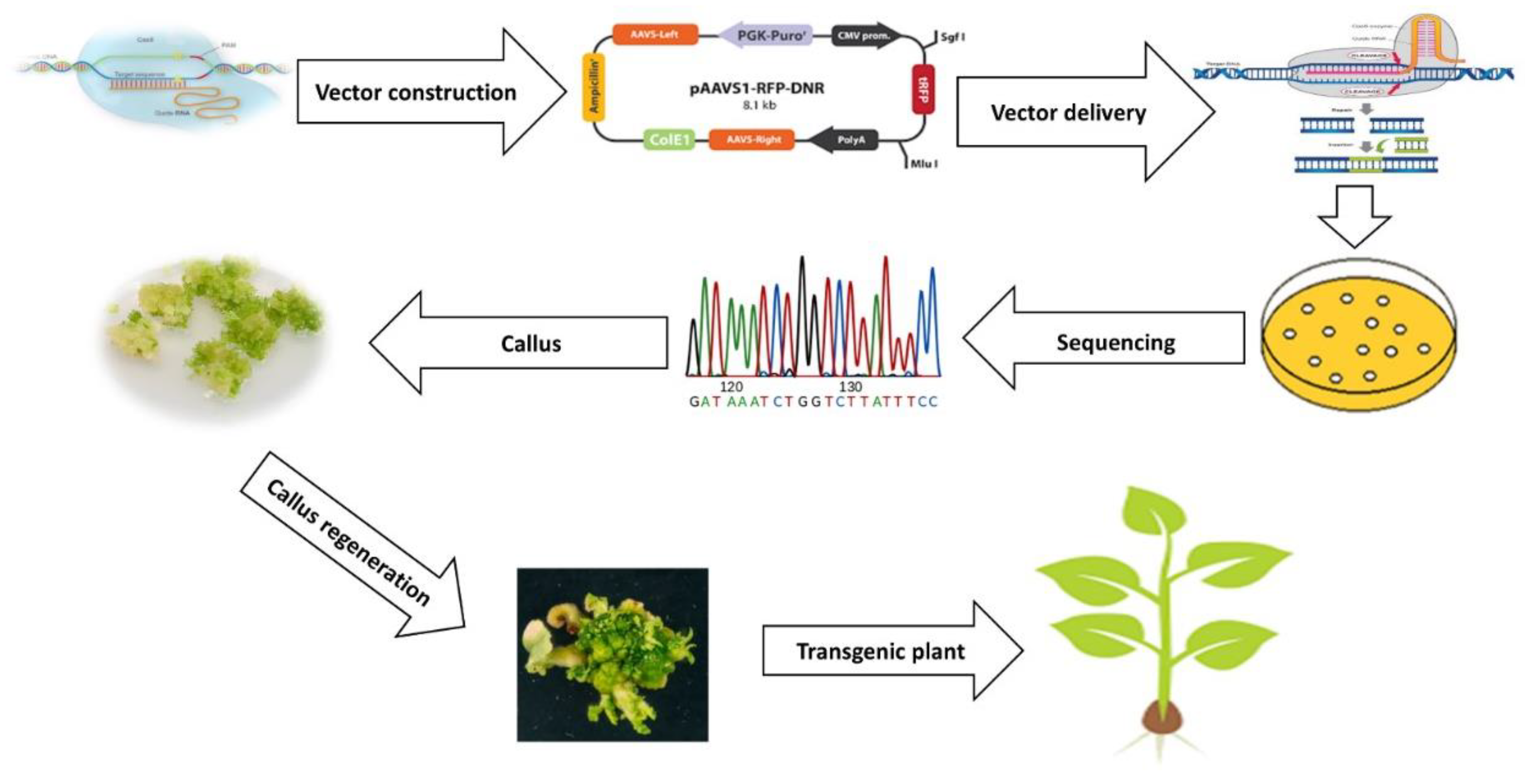
8. Recent Progresses in Genome Editing for Crop Improvement
9. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Alexandratos, N.; Bruinsma, J. World Agriculture towards 2030/2050: The 2012 Revision; FAO: Rome, Italy, 2012; ISSN 2521-1838. [Google Scholar]
- Anwar, A.; Liu, Y.; Dong, R.; Bai, L.; Yu, X.; Li, Y. The physiological and molecular mechanism of brassinosteroid in response to stress: A review. Biol. Res. 2018, 51, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, G.P.; Singh, B.; Seth, T.; Singh, A.K.; Halder, J.; Krishnan, N.; Tiwari, S.K.; Singh, P.M. Biotechnological advancements and begomovirus management in Okra (Abelmoschus esculentus L.), status and perspectives. Front. Plant Sci. 2017, 8, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X.J.; Wang, Y.J.; Zhou, Y.H.; Tao, Y.; Mao, W.H.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.Q. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwar, A.; Bai, L.; Miao, L.; Liu, Y.; Li, S.; Yu, X.; Li, Y. 24-Epibrassinolide Ameliorates endogenous hormone levels to enhance low-temperature stress tolerance in cucumber seedlings. Int. J. Mol. Sci. 2018, 19, 2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.; Fartyal, D.; Agarwal, A.; Shukla, T.; James, D.; Kaul, T.; Negi, Y.K.; Arora, S.; Reddy, M.K. Abiotic stress tolerance in plants: Myriad roles of ascorbate peroxidase. Front. Plant Sci. 2017, 8, 581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, T.; Deng, X.; Zhou, X.; Zhu, L.; Zou, L.; Li, P.; Zhang, D.; Lin, H. Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 2016, 6, 35392. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Talaat, N.B.; Shawky, B.T. Dual Application of 24-epibrassinolide and spermine confers drought stress tolerance in maize (Zea mays L.) by modulating polyamine and protein metabolism. J. Plant Growth Regul. 2016, 35, 518–533. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Deng, J.; Khalid, N.; Sanaullah, T.; Shuilin, H. Biotechnological advancements for improving floral attributes in ornamental plants. Front. Plant Sci. 2017, 8, 530. [Google Scholar] [CrossRef] [Green Version]
- Dita, M.A.; Rispail, N.; Prats, E.; Rubiales, D.; Singh, K.B. Biotechnology approaches to overcome biotic and abiotic stress constraints in legumes. Euphytica 2006, 147, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Muehlbauer, F.J.; Cho, S.; Sarker, A.; McPhee, K.E.; Coyne, C.J.; Rajesh, P.N.; Ford, R. Application of biotechnology in breeding lentil for resistance to biotic and abiotic stress. Euphytica 2006, 147, 149–165. [Google Scholar] [CrossRef]
- Choudhary, S.P.; Yu, J.Q.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012, 17, 594–605. [Google Scholar] [CrossRef]
- Chen, R.; Xu, Q.; Liu, Y.; Zhang, J.; Ren, D.; Wang, G.; Liu, Y. Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front. Plant Sci. 2018, 9, 1180. [Google Scholar] [CrossRef] [Green Version]
- Mushtaq, M.; Sakina, A.; Wani, S.H.; Shikari, A.B.; Tripathi, P.; Zaid, A.; Galla, A.; Abdelrahman, M.; Sharma, M.; Singh, A.K.; et al. Harnessing genome editing techniques to engineer disease resistance in plants. Front. Plant Sci. 2019, 10, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef]
- Li, C.; Ng, C.K.Y.; Fan, L.-M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- He, Z.; Wu, J.; Sun, X.; Dai, M. The Maize Clade A PP2C Phosphatases play critical roles in multiple abiotic stress responses. Int. J. Mol. Sci. 2019, 20, 3573. [Google Scholar] [CrossRef] [Green Version]
- Gantait, S.; Mondal, S. Transgenic approaches for genetic improvement in groundnut (Arachis hypogaea L.) against major biotic and abiotic stress factors. J. Genet. Eng. Biotechnol. 2018, 16, 537–544. [Google Scholar] [CrossRef]
- Nepolean, T.; Kaul, J.; Mukri, G.; Mittal, S. Genomics-enabled next-generation breeding approaches for developing system-specific drought tolerant hybrids in maize. Front. Plant Sci. 2018, 9, 361. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Zaidi, S.S.-E.-A.; Shakir, S.; Mansoor, S. Applications of new breeding technologies for potato improvement. Front. Plant Sci. 2018, 9, 925. [Google Scholar] [CrossRef]
- Boukar, O.; Fatokun, C.A.; Huynh, B.-L.; Roberts, P.A.; Close, T.J. Genomic tools in cowpea breeding programs: Status and perspectives. Front. Plant Sci. 2016, 7, 757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djami-Tchatchou, A.T.; Sanan-Mishra, N.; Ntushelo, K.; Dubery, I.A. Functional roles of microRNAs in agronomically important plants-potential as targets for crop improvement and protection. Front. Plant Sci. 2017, 8, 378. [Google Scholar] [CrossRef] [Green Version]
- Young, N.D. QTL mapping and quantitative disease resistance in plants. Ann. Rev. Phytopathol. 1996, 34, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wang, C.; Fu, Y.; Wang, J.; Liu, Q.; Zhang, X.; Yan, C.; Qian, Q.; Wang, K. QTL editing confers opposing yield performance in different rice varieties. J. Integr. Plant Biol. 2018, 60, 89–93. [Google Scholar] [CrossRef]
- Collins, N.C.; Tardieu, F.; Tuberosa, R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiol. 2008, 147, 469–486. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, L.; Makaju, S.O.; Missaoui, A.M. QTL mapping of flowering time and biomass yield in tetraploid alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 359. [Google Scholar] [CrossRef]
- Wen, J.; Jiang, F.; Weng, Y.; Sun, M.; Shi, X.; Zhou, Y.; Yu, L.; Wu, Z. Identification of heat-tolerance QTL and high-temperature stress-responsive genes through conventional QTL mapping, QTL-seq and RNA-seq in tomato. BMC Plant Biol. 2019, 19, 398. [Google Scholar] [CrossRef]
- Sharma, D.K.; Torp, A.M.; Rosenqvist, E.; Ottosen, C.-O.; Andersen, S.B. QTLs and potential candidate genes for heat stress tolerance identified from the mapping populations specifically segregating for Fv/Fm in wheat. Front. Plant Sci. 2017, 8, 1668. [Google Scholar] [CrossRef] [Green Version]
- Khraiwesh, B.; Zhu, J.-K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. BBA Gene Regul. Mech. 2012, 1819, 137–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abla, M.; Sun, H.; Li, Z.; Wei, C.; Gao, F.; Zhou, Y.; Feng, J. Identification of miRNAs and their response to cold stress in astragalus membranaceus. Biomolecules 2019, 9, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadarajah, K.; Kumar, I.S. Drought response in rice: The miRNA story. Int. J. Mol. Sci. 2019, 20, 3766. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Ni, Z.; Wang, Y.; Wan, H.; Hu, Z.; Jiang, Q.; Sun, X.; Zhang, H. Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Sci. 2019, 285, 68–78. [Google Scholar] [CrossRef]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Beaith, M.; Chalifoux, M.; Ying, J.; Uchacz, T.; Sarvas, C.; Griffiths, R.; Kuzma, M.; Wan, J.; Huang, Y. Shoot-specific down-regulation of protein farnesyltransferase (alpha-subunit) for yield protection against drought in canola. Mol. Plant 2009, 2, 191–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.-X.; Oono, Y.; Zhu, J.; He, X.-J.; Wu, J.-M.; Iida, K.; Lu, X.-Y.; Cui, X.; Jin, H.; Zhu, J.-K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [Green Version]
- Ni, Z.; Hu, Z.; Jiang, Q.; Zhang, H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013, 82, 113–129. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef]
- Rao, S.; Balyan, S.; Jha, S.; Mathur, S. Novel insights into expansion and functional diversification of MIR169 family in tomato. Planta 2020, 251, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-C.; Yu, Y.; Wang, C.-Y.; Li, Z.-Y.; Liu, Q.; Xu, J.; Liao, J.-Y.; Wang, X.-J.; Qu, L.-H.; Chen, F.; et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848. [Google Scholar] [CrossRef] [PubMed]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Dos Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef]
- Hwang, E.-W.; Shin, S.-J.; Yu, B.-K.; Byun, M.-O.; Kwon, H.-B. miR171 family members are involved in drought response in Solanum tuberosum. J. Plant Biol. 2011, 54, 43–48. [Google Scholar] [CrossRef]
- Bustamante, A.; Marques, M.C.; Sanz-Carbonell, A.; Mulet, J.M.; Gomez, G. Alternative processing of its precursor is related to miR319 decreasing in melon plants exposed to cold. Sci. Rep. 2018, 8, 15538. [Google Scholar] [CrossRef]
- Sun, M.; Shen, Y.; Yang, J.; Cai, X.; Li, H.; Zhu, Y.; Jia, B.; Sun, X. miR535 negatively regulates cold tolerance in rice. Mol. Breed. 2020, 40, 14. [Google Scholar] [CrossRef]
- Yang, C.; Li, D.; Mao, D.; Liu, X.; Ji, C.; Li, X.; Zhao, X.; Cheng, Z.; Chen, C.; Zhu, L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013, 36, 2207–2218. [Google Scholar] [CrossRef]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [Green Version]
- Pan, W.-J.; Tao, J.-J.; Cheng, T.; Bian, X.-H.; Wei, W.; Zhang, W.-K.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Soybean miR172a Improves salt tolerance and can function as a long-distance signal. Mol. Plant 2016, 9, 1337–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Liu, X.; Yang, J.; Liu, W.; Du, Q.; Wang, H.; Fu, C.; Li, W.-X. MicroRNA528 Affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol. Plant 2018, 11, 806–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schommer, C.; Debernardi, J.M.; Bresso, E.G.; Rodriguez, R.E.; Palatnik, J.F. Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 2014, 7, 1533–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Mai, Y.-X.; Zhang, Y.-C.; Luo, Q.; Yang, H.-Q. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV Genes regulate shoot branching in arabidopsis. Mol. Plant 2010, 3, 794–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Shi, S.; Liu, X.; Zhao, S.; Yang, Z.; et al. Suppression of Jasmonic Acid-Mediated Defense by Viral-Inducible MicroRNA319 facilitates virus infection in rice. Mol. Plant 2016, 9, 1302–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, S.; Yang, Z.; Yang, R.; Huang, Y.; Guo, G.; Kong, X.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.; et al. Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol. Plant 2019, 12, 1114–1122. [Google Scholar] [CrossRef]
- Yang, R.; Li, P.; Mei, H.; Wang, D.; Sun, J.; Yang, C.; Hao, L.; Cao, S.; Chu, C.; Hu, S.; et al. Fine-Tuning of MiR528 accumulation modulates flowering time in rice. Mol. Plant 2019, 12, 1103–1113. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wu, F.; Bai, J.; He, Y. BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 2014, 12, 12–321. [Google Scholar] [CrossRef]
- Shikata, M.; Yamaguchi, H.; Sasaki, K.; Ohtsubo, N. Overexpression of Arabidopsis miR157b induces bushy architecture and delayed phase transition in Torenia fournieri. Planta 2012, 236, 1027–1035. [Google Scholar] [CrossRef]
- Alonso-Peral, M.M.; Li, J.; Li, Y.; Allen, R.S.; Schnippenkoetter, W.; Ohms, S.; White, R.G.; Millar, A.A. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010, 154, 757–771. [Google Scholar] [CrossRef] [Green Version]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.Y.; Zhang, L.; Li, W.W.; Hu, X.L.; Wang, M.-B.; Fan, Y.L.; Zhang, C.Y.; Wang, L. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Xue, X.-Y.; Zhao, B.; Chao, L.-M.; Chen, D.-Y.; Cui, W.-R.; Mao, Y.-B.; Wang, L.-J.; Chen, X.-Y. Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet. 2014, 10, e1004266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, H.; Aya, K.; Ueguchi-Tanaka, M.; Shimada, Y.; Nakazono, M.; Watanabe, R.; Nishizawa, N.K.; Gomi, K.; Shimada, A.; Kitano, H. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 2006, 47, 427–444. [Google Scholar] [CrossRef]
- Salvi, S.; Sponza, G.; Morgante, M.; Tomes, D.; Niu, X.; Fengler, K.A.; Meeley, R.; Ananiev, E.V.; Svitashev, S.; Bruggemann, E. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 2007, 104, 11376–11381. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Ahn, H.J.; Chiou, T.-J.; Ahn, J.H. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol. Cells 2011, 32, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, N.; Jalajakumari, M.B.; Blackman, L.; Shen, E.; Verma, S.; Wang, M.-B.; Millar, A.A. miR159 represses a constitutive pathogen defense response in tobacco. Plant Physiol. 2020, 182, 2182–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 3–17018. [Google Scholar]
- Xiao, Y.; Zhou, L.; Lei, X.; Cao, H.; Wang, Y.; Dou, Y.; Tang, W.; Xia, W. Genome-wide identification of WRKY genes and their expression profiles under different abiotic stresses in Elaeis guineensis. PLoS ONE 2017, 12, e0189224. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Xu, G.; Alli, A.; Yu, L. Plant HAK/KUP/KT K(+) transporters: Function and regulation. Semin. Cell Dev. Biol. 2018, 74, 133–141. [Google Scholar] [CrossRef]
- Ou, W.; Mao, X.; Huang, C.; Tie, W.; Yan, Y.; Ding, Z.; Wu, C.; Xia, Z.; Wang, W.; Zhou, S.; et al. Genome-Wide Identification and expression analysis of the KUP family under abiotic stress in cassava (Manihot esculenta Crantz). Front. Physiol. 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Yang, H.; Shi, H.; Wei, Y.; Tie, W.; Ding, Z.; Yan, Y.; Luo, Y.; Xia, Z.; Wang, W.; et al. The MAPKKK gene family in cassava: Genome-wide identification and expression analysis against drought stress. Sci. Rep. 2017, 7, 14939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanta, T.K.; Park, Y.-H.; Bae, H. Novel genomic and evolutionary insight of WRKY transcription factors in plant lineage. Sci. Rep. 2016, 6, 37309. [Google Scholar] [CrossRef] [Green Version]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Gen. Genom. 2016, 43, 529. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [Green Version]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.; Liu, Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef]
- Chappell, J.; Watters, K.E.; Takahashi, M.K.; Lucks, J.B. A renaissance in RNA synthetic biology: New mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2015, 28, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [Green Version]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2015, 13, 578–589. [Google Scholar] [CrossRef]
- Laganà, A.; Shasha, D.; Croce, C.M. Synthetic RNAs for gene regulation: Design principles and computational tools. Front. Bioeng. Biotechnol. 2014, 2, 65. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [Green Version]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas genome-editing tool: Application in improvement of crops. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Sovero, V.; Gemenet, D. Genome-assisted breeding for drought resistance. Curr. Genom. 2016, 17, 4–330. [Google Scholar] [CrossRef] [Green Version]
- Rady, M.M. Effect of 24-epibrassinolide on growth, yield, antioxidant system and cadmium content of bean (Phaseolus vulgaris L.) plants under salinity and cadmium stress. Sci. Hortic. 2011, 129, 232–237. [Google Scholar] [CrossRef]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The antagonistic action of Abscisic Acid and Cytokinin signaling mediates drought stress response in arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Yang, L.; Liu, X.; Tang, R.; Wang, Y.; Ge, H.; Wu, M.; Zhang, J.; Zhao, F.; Luan, S.; et al. Overexpression of poplar pyrabactin resistance-like abscisic acid receptors promotes abscisic acid sensitivity and drought resistance in transgenic arabidopsis. PLoS ONE 2016, 11, e0168040. [Google Scholar] [CrossRef]
- Geiger, D.; Scherzer, S.; Mumm, P.; Marten, I.; Ache, P.; Matschi, S.; Liese, A.; Wellmann, C.; Al-Rasheid, K.A.S.; Grill, E.; et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 2010, 107, 8023–8028. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Devi, M.J.; Reddy, D.S.; Lavanya, M.; Vadez, V.; Serraj, R.; Yamaguchi-Shinozaki, K.; Sharma, K.K. Stress-inducible expression of at DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 2007, 26, 2071–2082. [Google Scholar] [CrossRef] [Green Version]
- Sharma, G.; Upadyay, A.K.; Biradar, H.; Hittalmani, S. OsNAC-like transcription factor involved in regulating seed-storage protein content at different stages of grain filling in rice under aerobic conditions. J. Genet. 2019, 98, 18. [Google Scholar] [CrossRef]
- Chung, P.J.; Jung, H.; Choi, Y.D.; Kim, J.K. Genome-wide analyses of direct target genes of four rice NAC-domain transcription factors involved in drought tolerance. BMC Genom. 2018, 19, 40. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Ahn, H.; Jung, I.; Shin, S.J.; Park, J.; Rhee, S.; Kim, J.K.; Jung, W.; Kwon, H.B.; Kim, S. Transcriptional network analysis reveals drought resistance mechanisms of AP2/ERF transgenic rice. Front. Plant Sci. 2017, 8, 1044. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Choi, Y.D.; Kim, J.K. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 2015, 241, 199–210. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef]
- Redillas, M.C.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 2012, 10, 792–805. [Google Scholar] [CrossRef]
- Patil, M.; Ramu, S.V.; Jathish, P.; Sreevathsa, R.; Reddy, P.C.; Prasad, T.G.; Udayakumar, M. Overexpression of AtNAC2 (ANAC092) in groundnut (Arachis hypogaea L.) improves abiotic stress tolerance. Plant Biotechnol. Rep. 2014, 8, 161–169. [Google Scholar] [CrossRef]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.-I.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- Pandurangaiah, M.; Lokanadha Rao, G.; Sudhakarbabu, O.; Nareshkumar, A.; Kiranmai, K.; Lokesh, U.; Thapa, G.; Sudhakar, C. Overexpression of Horse gram (Macrotyloma uniflorum Lam.Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Mol. Biotechnol. 2014, 56, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.K.; Gupta, A.; Fatima, U.; Senthil-Kumar, M. AtGBF3 confers tolerance to Arabidopsis thaliana against combined drought and Pseudomonas syringae stress. Environ. Exp. Bot. 2019, 168, 103881. [Google Scholar] [CrossRef]
- Lu, G.; Paul, A.L.; McCarty, D.R.; Ferl, R.J. Transcription factor veracity: Is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 1996, 8, 847–857. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, W.; Zhao, N.; Xiao, B.; Cao, P.; Wu, X.; Ye, C.; Shen, E.; Qiu, J.; Zhu, Q.-H.; et al. Regulation of nicotine biosynthesis by an endogenous target mimicry of MicroRNA in tobacco. Plant Physiol. 2015, 169, 1062–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauter, N.; Kampani, A.; Carlson, S.; Goebel, M.; Moose, S.P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 2005, 102, 9412–9417. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. The State of Food Security and Nutrition in the World 2008: Building Climate Resilience for Food Security and Nutrition; Food & Agriculture Org.: Rome, Italy, 2008. [Google Scholar]
- Kumar, M.; Choi, J.; Kumari, N.; Pareek, A.; Kim, S. Molecular breeding in Brassica for salt tolerance: Importance of microsatellite (SSR) markers for molecular breeding in Brassica. Front. Plant Sci. 2015, 6, 688. [Google Scholar] [CrossRef] [Green Version]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Niu, X.; Narasimhan, M.L.; Salzman, R.A.; Bressan, R.A.; Hasegawa, P.M. NaCl regulation of plasma membrane H(+)-ATPase gene expression in a glycophyte and a halophyte. Plant Physiol. 1993, 103, 713–718. [Google Scholar] [CrossRef]
- Hou, Q.; Bartels, D. Comparative study of the aldehyde dehydrogenase (ALDH) gene superfamily in the glycophyte Arabidopsis thaliana and Eutrema halophytes. Ann. Bot. 2015, 115, 465–479. [Google Scholar] [CrossRef] [Green Version]
- Hassidim, M.; Braun, Y.; Lerner, H.R.; Reinhold, L. Na/H and k/h antiport in root membrane vesicles isolated from the halophyte atriplex and the glycophyte cotton. Plant Physiol. 1990, 94, 1795–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, T.G.A.; Sancho, L.G.; Pintado, A.; Saco, D.; Martín, S.; Arróniz-Crespo, M.; Angel Casermeiro, M.; de la Cruz Caravaca, M.T.; Cameron, S.; Rozzi, R. Sodium chloride accumulation in glycophyte plants with cyanobacterial symbionts. AoB Plants 2017, 9, plx053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Song, G.; Shan, X.; Wei, Z.; Liu, Y.; Jiang, C.; Jiang, Y.; Jin, F.; Li, Y. Association analysis and identification of ZmHKT1;5 variation with salt-stress tolerance. Front. Plant Sci. 2018, 9, 1485. [Google Scholar] [CrossRef] [PubMed]
- Schachtman, D.P.; Schroeder, J.I. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 1994, 370, 655–658. [Google Scholar] [CrossRef]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 2010, 33, 552–565. [Google Scholar] [CrossRef]
- Horie, T.; Costa, A.; Kim, T.H.; Han, M.J.; Horie, R.; Leung, H.Y.; Miyao, A.; Hirochika, H.; An, G.; Schroeder, J.I. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007, 26, 3003–3014. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.-H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 2011, 62, 2939–2947. [Google Scholar] [CrossRef] [Green Version]
- James, R.A.; Blake, C.; Zwart, A.B.; Hare, R.A.; Rathjen, A.J.; Munns, R. Impact of ancestral wheat sodium exclusion genes and on grain yield of durum wheat on saline soils. Funct. Plant Biol. 2012, 39, 609–618. [Google Scholar] [CrossRef]
- Han, Y.; Yin, S.; Huang, L.; Wu, X.; Zeng, J.; Liu, X.; Qiu, L.; Munns, R.; Chen, Z.H.; Zhang, G. A Sodium Transporter HvHKT1;1 Confers salt tolerance in barley via regulating tissue and cell ion homeostasis. Plant Cell Physiol. 2018, 59, 1976–1989. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-T.; Ren, Z.-J.; Liu, Z.-Q.; Feng, X.; Guo, R.-Q.; Li, B.-G.; Li, L.-G.; Jing, H.-C. SbHKT1;4, a member of the high-affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na+/K+ balance under Na+ stress. J. Integr. Plant Biol. 2014, 56, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabnoune, M.; Espeout, S.; Mieulet, D.; Fizames, C.; Verdeil, J.-L.; Conéjéro, G.; Rodríguez-Navarro, A.; Sentenac, H.; Guiderdoni, E.; Abdelly, C. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009, 150, 1955–1971. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.-K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, M.; Li, J.; Yahya, M.; Wang, M.; Ali, A.; Cheng, A.; Wang, X.; Ma, C. Grain legumes and fear of salt stress: Focus on mechanisms and management strategies. Int. J. Mol. Sci. 2019, 20, 799. [Google Scholar] [CrossRef] [Green Version]
- Park, H.J.; Kim, W.-Y.; Yun, D.-J. A new insight of salt stress signaling in plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Yan, S.; Yang, T.; Zhang, S.; Chen, Y.-Q.; Liu, B. Small RNAs in regulating temperature stress response in plants. J. Integr. Plant Biol. 2017, 59, 774–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Li, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Characterization of 19 genes encoding membrane-bound fatty acid desaturases and their expression profiles in gossypium raimondii under low temperature. PLoS ONE 2015, 10, e0123281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khodakovskaya, M.; McAvoy, R.; Peters, J.; Wu, H.; Li, Y. Enhanced cold tolerance in transgenic tobacco expressing a chloroplast omega-3 fatty acid desaturase gene under the control of a cold-inducible promoter. Planta 2006, 223, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.C.; Yamaji, N.; Horie, T.; Che, J.; Li, J.; An, G.; Ma, J.F. A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice. Plant Physiol. 2017, 174, 1837–1849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Liu, J.; Guo, H.; He, X.; Wu, W.; Du, J.; Zhang, Z.; An, X. Characterization of two highly similar CBF/DREB1-like genes, PhCBF4a and PhCBF4b, in Populus hopeiensis. Plant Physiol. Biochem. 2014, 83, 107–116. [Google Scholar] [CrossRef]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [Green Version]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef] [Green Version]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haake, V.; Cook, D.; Riechmann, J.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Li, Z.; Li, J.; Wang, A. Ectopic overexpression of SsCBF1, a CRT/DRE-binding factor from the nightshade plant Solanum lycopersicoides, confers freezing and salt tolerance in transgenic Arabidopsis. PLoS ONE 2013, 8, e61810. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, W.; Wang, M.; Zhang, H.-Y.; Liu, J.-H. The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene–polyamine homeostasis. Plant Cell Physiol. 2016, 57, 1865–1878. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Feng, L.; Li, J.; He, Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015, 6, 267. [Google Scholar] [CrossRef]
- Proveniers, M.C.; van Zanten, M. High temperature acclimation through PIF4 signaling. Trends Plant Sci. 2013, 18, 59–64. [Google Scholar] [CrossRef]
- Stavang, J.A.; Gallego-Bartolomé, J.; Gómez, M.D.; Yoshida, S.; Asami, T.; Olsen, J.E.; García-Martínez, J.L.; Alabadí, D.; Blázquez, M.A. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009, 60, 589–601. [Google Scholar] [CrossRef]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Qi, L.; Li, Y.; Chu, J.; Li, C. PIF4–mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012, 8, e1002594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Anil, K.; Bhardwaj, R. Plant steroidal hormone epibrassinolide regulate—Heavy metal stress tolerance in Oryza sativa L. by modulating antioxidant defense expression. Environ. Exp. Bot. 2016, 122, 1–9. [Google Scholar] [CrossRef]
- Ali, B.; Hasan, S.A.; Hayat, S.; Hayat, Q.; Yadav, S.; Fariduddin, Q.; Ahmad, A. A role for brassinosteroids in the amelioration of aluminium stress through antioxidant system in mung bean (Vigna radiata L. Wilczek). Environ. Exp. Bot. 2008, 62, 153–159. [Google Scholar] [CrossRef]
- Jalmi, S.K.; Bhagat, P.K.; Verma, D.; Noryang, S.; Tayyeba, S.; Singh, K.; Sharma, D.; Sinha, A.K. Traversing the links between heavy metal stress and plant signaling. Front. Plant Sci. 2018, 9, 12. [Google Scholar] [CrossRef]
- Lešková, A.; Giehl, R.F.H.; Hartmann, A.; Fargašová, A.; von Wirén, N. Heavy metals induce iron deficiency responses at different hierarchic and regulatory levels. Plant Physiol. 2017, 174, 1648–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, R.-L.; Zhou, Q.-X.; Sun, F.-H.; Jin, C.-X. Antioxidative defense and proline/phytochelatin accumulation in a newly discovered Cd-hyperaccumulator, Solanum nigrum L. Environ. Exp. Bot. 2007, 60, 468–476. [Google Scholar] [CrossRef]
- Fusconi, A.; Repetto, O.; Bona, E.; Massa, N.; Gallo, C.; Dumas-Gaudot, E.; Berta, G. Effects of cadmium on meristem activity and nucleus ploidy in roots of Pisum sativum L. cv. Frisson seedlings. Environ. Exp. Bot. 2006, 58, 253–260. [Google Scholar] [CrossRef]
- Opdenakker, K.; Remans, T.; Vangronsveld, J.; Cuypers, A. Mitogen-Activated Protein (MAP) kinases in plant metal stress: Regulation and responses in comparison to other biotic and abiotic stresses. Int. J. Mol. Sci. 2012, 13, 7828–7853. [Google Scholar]
- Jonak, C.; Nakagami, H.; Hirt, H. Heavy Metal Stress. Activation of distinct mitogen-activated protein kinase pathways by Copper and Cadmium. Plant Physiol. 2004, 136, 3276–3283. [Google Scholar] [CrossRef] [Green Version]
- Ricachenevsky, F.K.; Menguer, P.K.; Sperotto, R.A.; Williams, L.E.; Fett, J.P. Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 2013, 4, 144. [Google Scholar] [CrossRef] [Green Version]
- Gustin, J.L.; Zanis, M.J.; Salt, D.E. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 2011, 11, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delhaize, E.; Gruber, B.D.; Pittman, J.K.; White, R.G.; Leung, H.; Miao, Y.; Jiang, L.; Ryan, P.R.; Richardson, A.E. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007, 51, 198–210. [Google Scholar] [CrossRef]
- Van der Zaal, B.J.; Neuteboom, L.W.; Pinas, J.E.; Chardonnens, A.N.; Schat, H.; Verkleij, J.A.; Hooykaas, P.J. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999, 119, 1047–1055. [Google Scholar] [CrossRef] [Green Version]
- Desbrosses-Fonrouge, A.G.; Voigt, K.; Schroder, A.; Arrivault, S.; Thomine, S.; Kramer, U. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005, 579, 4165–4174. [Google Scholar] [CrossRef]
- Shahzad, Z.; Gosti, F.; Frerot, H.; Lacombe, E.; Roosens, N.; Saumitou-Laprade, P.; Berthomieu, P. The five AhMTP1 zinc transporters undergo different evolutionary fates towards adaptive evolution to zinc tolerance in Arabidopsis halleri. PLoS Genet. 2010, 6, e1000911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrivault, S.; Senger, T.; Kramer, U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006, 46, 861–879. [Google Scholar] [CrossRef] [PubMed]
- Peiter, E.; Montanini, B.; Gobert, A.; Pedas, P.; Husted, S.; Maathuis, F.J.; Blaudez, D.; Chalot, M.; Sanders, D. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 8532–8537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, H.X.; Wang, Z.F.; Wang, Q.H.; Wang, M.M.; Bao, Y.M.; Huang, J.; Zhang, H.S. Characterization of a vacuolar zinc transporter OZT1 in rice (Oryza sativa L.). Mol. Biol. Rep. 2013, 40, 1201–1210. [Google Scholar] [CrossRef]
- Yuan, L.; Yang, S.; Liu, B.; Zhang, M.; Wu, K. Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 2012, 31, 67–79. [Google Scholar] [CrossRef]
- Lequeux, H.; Hermans, C.; Lutts, S.; Verbruggen, N. Response to copper excess in Arabidopsis thaliana: Impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 2010, 48, 673–682. [Google Scholar] [CrossRef]
- Li, G.; Song, H.; Li, B.; Kronzucker, H.J.; Shi, W. Auxin Resistant1 and PIN-FORMED2 Protect Lateral Root Formation in Arabidopsis under Iron Stress. Plant Physiol. 2015, 169, 2608–2623. [Google Scholar] [PubMed] [Green Version]
- Hu, Y.F.; Zhou, G.; Na, X.F.; Yang, L.; Nan, W.B.; Liu, X.; Zhang, Y.Q.; Li, J.L.; Bi, Y.R. Cadmium interferes with maintenance of auxin homeostasis in Arabidopsis seedlings. J. Plant Physiol. 2013, 170, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Vitti, A.; Nuzzaci, M.; Scopa, A.; Tataranni, G.; Remans, T.; Vangronsveld, J.; Sofo, A. Auxin and cytokinin metabolism and root morphological modifications in Arabidopsis thaliana seedlings infected with Cucumber mosaic virus (CMV) or exposed to cadmium. Int. J. Mol. Sci. 2013, 14, 6889–6902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.M.; Huang, X. Inhibition of root meristem growth by cadmium involves nitric oxide-mediated repression of auxin accumulation and signalling in Arabidopsis. Plant Cell Environ. 2016, 39, 120–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamurthy, A.; Rathinasabapathi, B. Auxin and its transport play a role in plant tolerance to arsenite-induced oxidative stress in Arabidopsis. Plant Cell Environ. 2013, 36, 1838–1849. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soyabean. Plant Biotechnol. J. 2018, 16, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287. [Google Scholar] [CrossRef]
- Anwar, A.; Li, Y.; He, C.; Yu, X. 24-Epibrassinolide promotes NO3− and NH4+ ion flux rate and NRT1 gene expression in cucumber under suboptimal root zone temperature. BMC Plant Biol. 2019, 19, 225. [Google Scholar]
- Anwar, A.; Di, Q.; Yan, Y.; He, C.; Li, Y.; Yu, X. Exogenous 24-epibrassinolide alleviates the detrimental effects of suboptimal root zone temperature in cucumber seedlings. Arch, Agron. Soil Sci. 2019, 65, 1927–1940. [Google Scholar] [CrossRef]



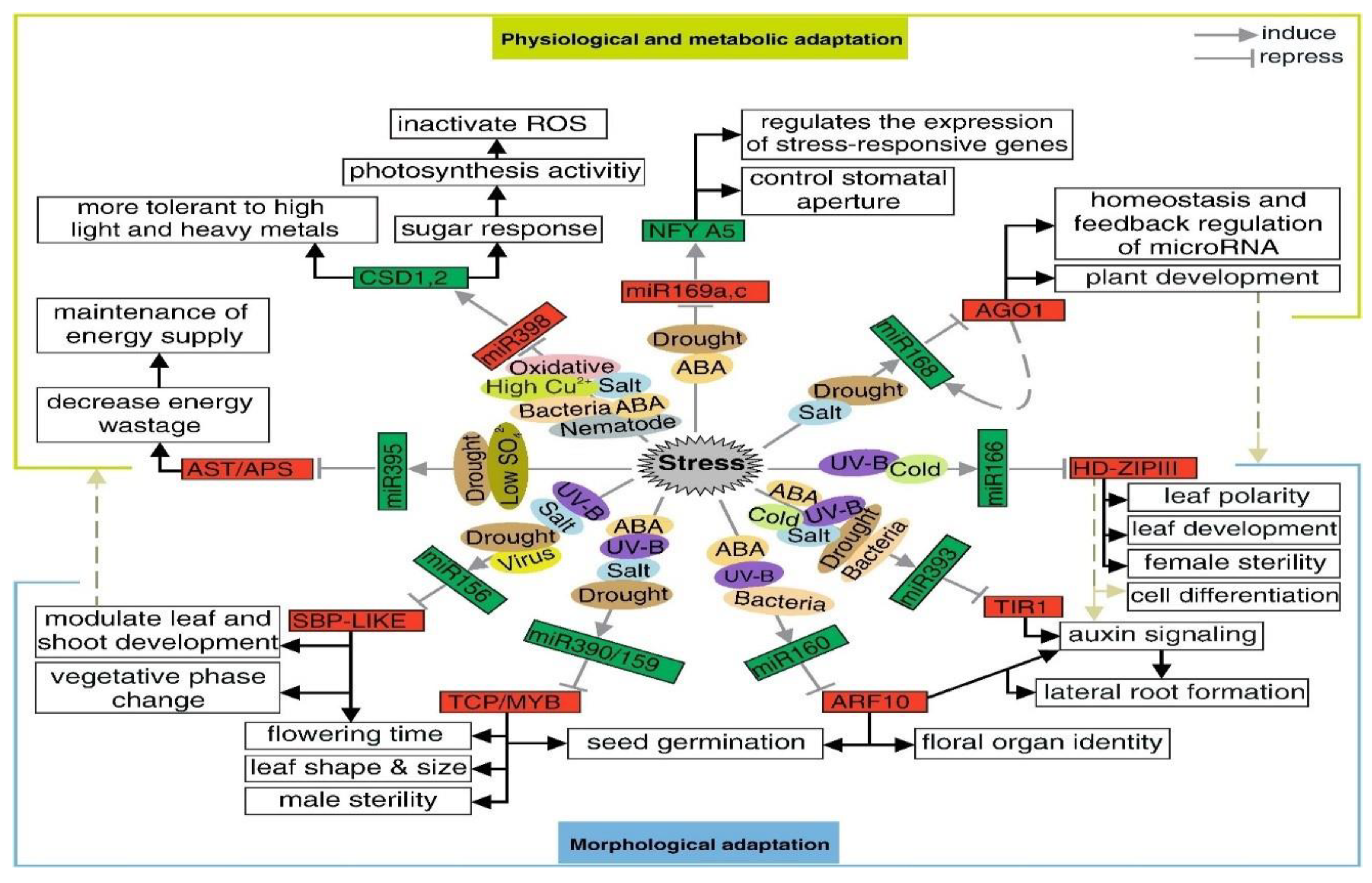

| miRNA | Species | Targeted Genes | Function | References |
|---|---|---|---|---|
| miR172a | Soybean | SSAC1 | Salt tolerance | [52] |
| miR528 | Maize | ZmLAC3, ZmLAC5 | Lignin biosynthesis | [53] |
| miR319 | Arabidopsis | TCP4 | Leaf development and hormone biosynthesis and signaling | [54] |
| miR171c | Arabidopsis | SCL6-II, SCL6-III, SCL6-IV | Shoot and branching | [55] |
| miR319 | Rice | TCP21, JA biosynthesis and signaling-related genes (PLDα1, LOX5, LOX11, CORI1b and CORI2) | Rice ragged stunt virus resistance | [56] |
| miR528 | Rice | OsSPL9 | Rice stripe virus resistance | [57] |
| miR528 | Rice | OsRFI2 | Flowering time | [58] |
| miR156 | Brassica | BrpSPL9-2 | Control heading time | [59] |
| miR157b | SPL | Control of various developmental process | [60] | |
| miR159 | Arabidopsis | AtMYB | Involved in GA signaling pathway and control programmed cell death and flowering | [61] |
| miR393 | Rice | OsAFB2, OsTIR1 | Early flowering, salt and drought stress tolerance | [62] |
| miR169 | Arabidopsis | NY-YA2 | Promote flowering and stress tolerance | [63] |
| miR171 | Arabidopsis | SPL | Control plant growth and flowering time | [64] |
| miR159 | Rice | OsGAMYBLs | Regulate heading | [65] |
| miR172 | Maize | ZmTOE1 | Regulate flowering time | [66] |
| miR169 | Arabidopsis | PH2 | Abiotic stress response | [67] |
| miR159 | Tobacco | GAMYB | Pathogen defense response | [68] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anwar, A.; Kim, J.-K. Transgenic Breeding Approaches for Improving Abiotic Stress Tolerance: Recent Progress and Future Perspectives. Int. J. Mol. Sci. 2020, 21, 2695. https://doi.org/10.3390/ijms21082695
Anwar A, Kim J-K. Transgenic Breeding Approaches for Improving Abiotic Stress Tolerance: Recent Progress and Future Perspectives. International Journal of Molecular Sciences. 2020; 21(8):2695. https://doi.org/10.3390/ijms21082695
Chicago/Turabian StyleAnwar, Ali, and Ju-Kon Kim. 2020. "Transgenic Breeding Approaches for Improving Abiotic Stress Tolerance: Recent Progress and Future Perspectives" International Journal of Molecular Sciences 21, no. 8: 2695. https://doi.org/10.3390/ijms21082695
APA StyleAnwar, A., & Kim, J.-K. (2020). Transgenic Breeding Approaches for Improving Abiotic Stress Tolerance: Recent Progress and Future Perspectives. International Journal of Molecular Sciences, 21(8), 2695. https://doi.org/10.3390/ijms21082695





