Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics
Abstract
:1. Introduction
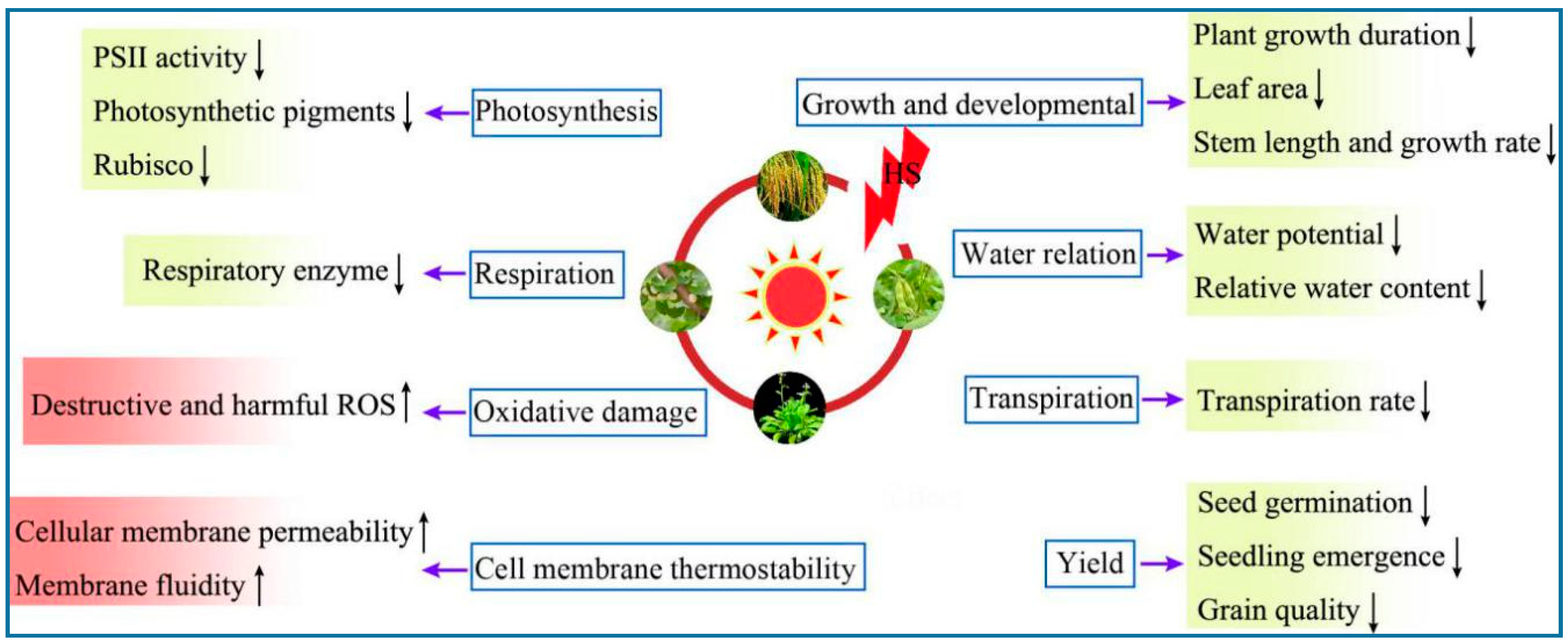
2. Physiological Responses of Plants under HS
2.1. Photosynthesis
2.2. Cell Membrane Thermostability
2.3. Oxidative Damage
2.4. Other Physiological Responses
3. Molecular Responses of Plants under HS
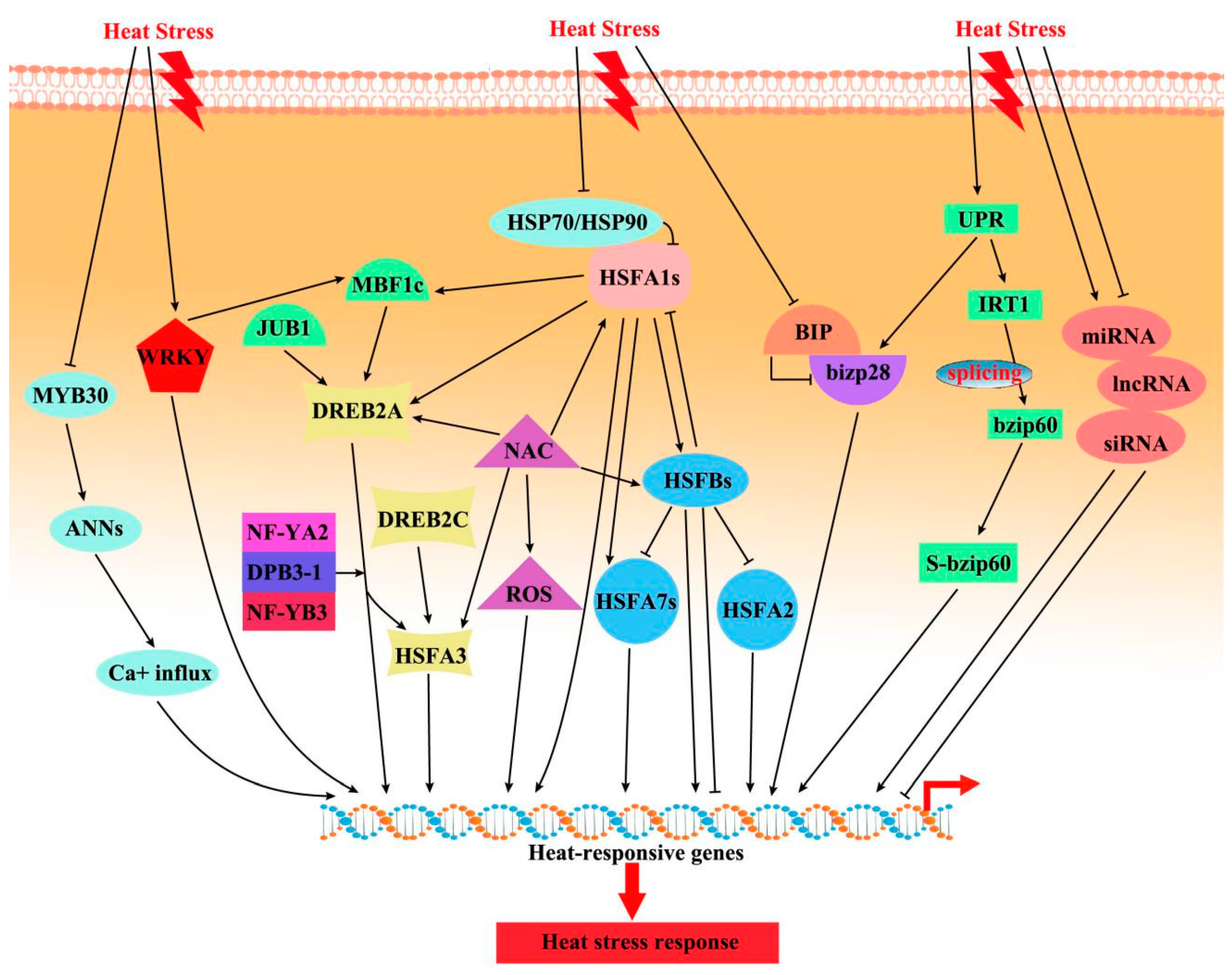
3.1. Transcriptional Regulation of HS Responses
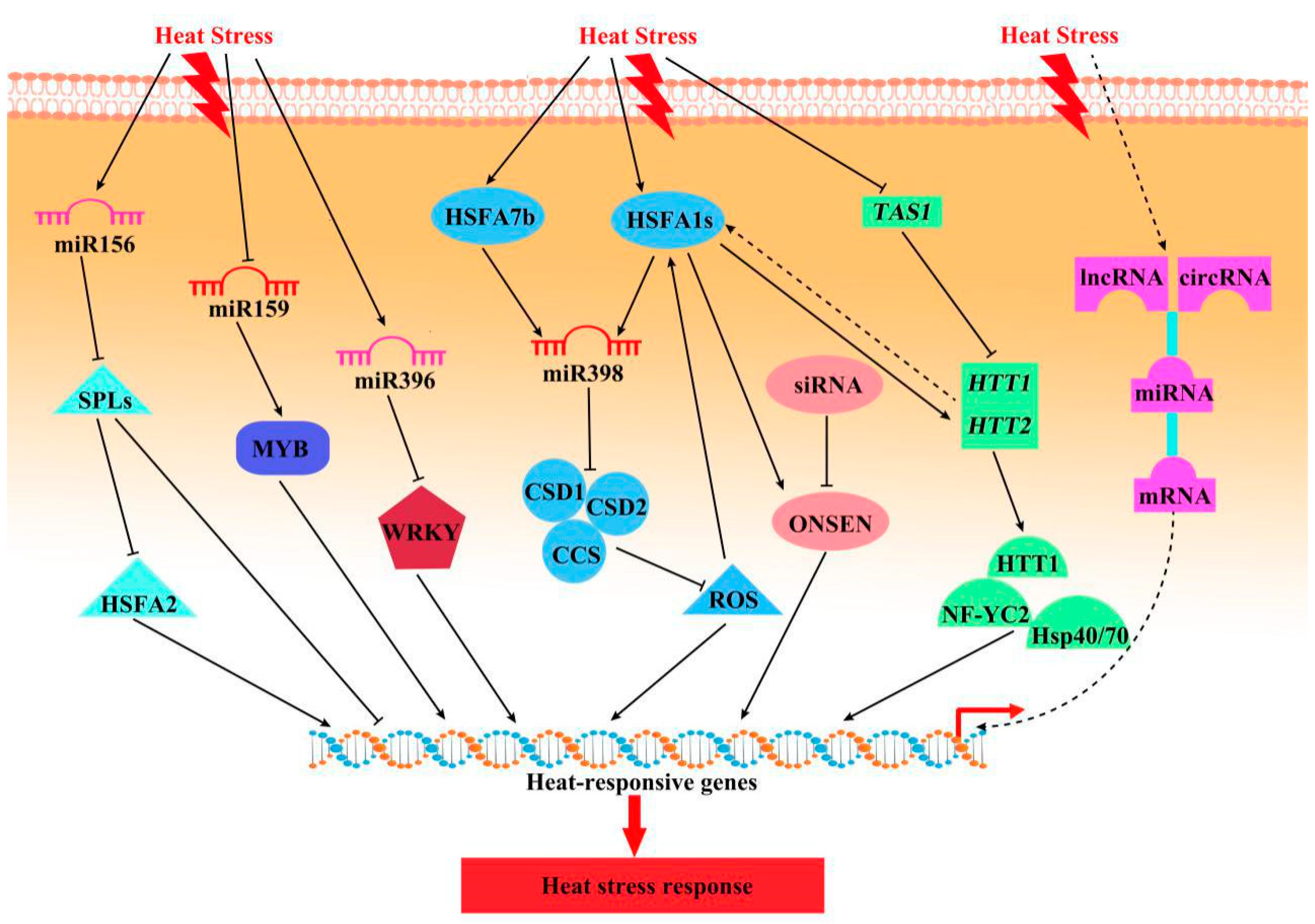
3.2. Regulation of HS Responses by Non-Coding RNAs
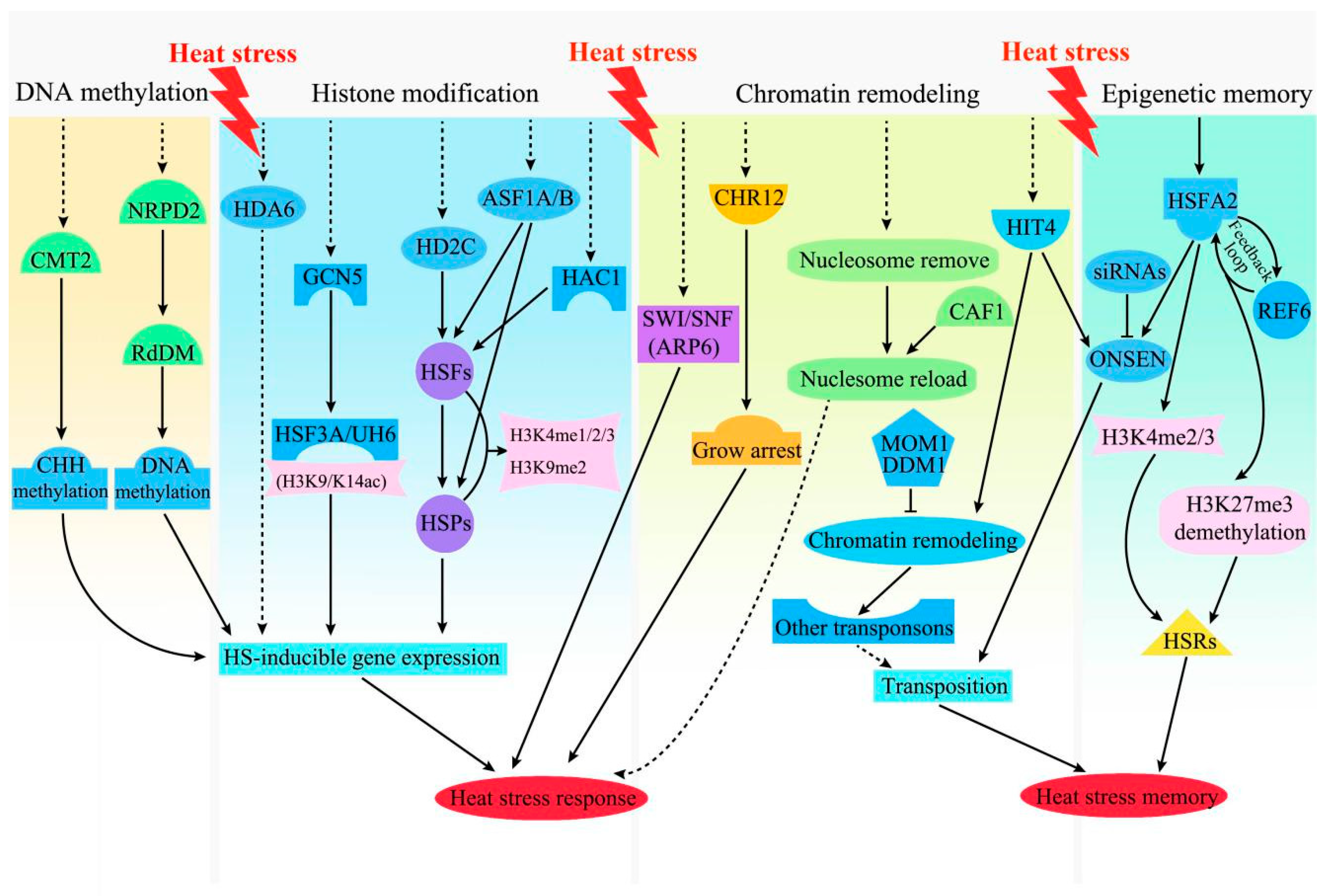
3.3. Epigenetic Regulation in the Plant HS Response and Memory
3.3.1. DNA Methylation
3.3.2. Histone Modification
3.3.3. Chromatin Remodeling
3.3.4. Epigenetic Memory
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Lippmann, R.; Babben, S.; Menger, A.; Delker, C.; Quint, M. Development of wild and cultivated plants under global warming conditions. Curr. Biol. 2019, 29, R1326–R1338. [Google Scholar] [CrossRef] [PubMed]
- Hedhly, A.; Hormaza, J.; Herrero, M. Global warming and sexual plant reproduction. Trends Plant Sci. 2009, 14, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, R.; Xu, X.; Fowler, J.C.; Miller, T.E.X.; Dong, T. Effect of summer warming on growth, photosynthesis and water status in female and male Populus cathayana: Implications for sex-specific drought and heat tolerances. Tree Physiol. 2020, 40, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Akter, N.; Islam, M.R. Heat stress effects and management in wheat. A review. Agron. Sustain. Dev. 2017, 37, 37. [Google Scholar] [CrossRef]
- Xalxo, R.; Yadu, B.; Chandra, J.; Chandrakar, V.; Keshavkant, S. Alteration in Carbohydrate Metabolism Modulates Thermotolerance of Plant under Heat Stress. In Heat Stress Tolerance in Plants: Physiological, Molecular and Genetic Perspectives, 1st ed.; Wani, S.H., Kumar, V., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2020; pp. 77–115. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ding, Y.; Zhu, C. Sensitivity and responses of chloroplasts to heat stress in plants. Front. Plant Sci. 2020, 11, 375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renata Szymańska Lesak, I.; Orzechowska, A.; Kruk, J. Physiological and biochemical responses to high light and temperature stress in plants. Environ. Exp. Bot. 2017, 139, 165–177. [Google Scholar]
- Song, Y.; Chen, Q.; Ci, D.; Shao, X.; Zhang, D. Effects of high temperature on photosynthesis and related gene expression in poplar. BMC Plant Biol. 2014, 14, 111. [Google Scholar] [CrossRef] [Green Version]
- Perdomo, J.A.; Capó-Bauçà, S.; Carmo-Silva, E.; Galmés, J. Rubisco and rubisco activase play an important role in the biochemical limitations of photosynthesis in rice, wheat, and maize under high temperature and water deficit. Front. Plant Sci. 2017, 8, 490. [Google Scholar] [CrossRef] [Green Version]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [Green Version]
- Dhanda, S.S.; Munjal, R. Heat tolerance in relation to acquired thermotolerance for membrane lipids in bread wheat. Field Crops Res. 2012, 135, 30–37. [Google Scholar] [CrossRef]
- Abdul-Razack, M.; Tarpley, L. Impact of high nighttime temperature on respiration, membrane stability, antioxidant capacity, and yield of rice plants. Crop. Sci. 2009, 49, 313–322. [Google Scholar]
- Nosaka, Y.; Nosaka, A.Y. Generation and detection of reactive oxygen species in photocatalysis. Chem. Rev. 2017, 117, 11302–11336. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xin, C.; Cai, j.; Zhou, Q.; Dai, T.; Cao, W.; Jiang, D. Heat priming induces trans-generational tolerance to high temperature stress in wheat. Front. Plant Sci. 2016, 7, 501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djanaguiraman, M.; Prasad, P.V.V.; Seppanen, M. Selenium protects sorghum leaves from oxidative damage under high temperature stress by enhancing antioxidant defense system. Plant Physiol. Biochem. 2010, 48, 999–1007. [Google Scholar] [CrossRef]
- Jespersen, D. Heat shock induced stress tolerance in plants: Physiological, biochemical, and molecular mechanisms of acquired tolerance. In Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants, 1st ed.; Hossain, M.A., Liu, F., David, J., Burritt, D., Eds.; Academic Press Elsevier Inc.: London, UK, 2020; pp. 161–174. [Google Scholar] [CrossRef]
- Wang, L.; Ma, K.B.; Lu, Z.G.; Ren, S.X.; Jiang, H.R.; Cui, J.W.; Chen, G.; Teng, N.J.; Lam, H.M.; Jin, B. Differential physiological, transcriptomic and metabolomic responses of Arabidopsis leaves under prolonged warming and heat shock. BMC Plant Biol. 2020, 20, 86. [Google Scholar] [CrossRef]
- Zhang, X.; Cai, J.; Wollenweber, B.; Liu, F.; Dai, T.; Cao, W.; Jiang, D. Multiple heat and drought events affect grain yield and accumulations of high molecular weight glutenin subunits and glutenin macropolymers in wheat. J. Cereal Sci. 2013, 57, 134–140. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Ren, S.; Ma, K.; Lu, Z.; Chen, G.; Cui, J.; Tong, P.; Wang, L.; Teng, N.; Jin, B. Transcriptomic andmetabolomic analysis of the heat-Stress response of Populus tomentosa Carr. Forests 2019, 10, 383. [Google Scholar] [CrossRef] [Green Version]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Berendzen, K.W.; Schoffl, F. Promoter specificity and interactions between early and late Arabidopsis heat hock factors. Plant Mol. Biol. 2010, 73, 559–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wang, X.; Cai, Y.; Wu, J.; Mo, B.; Yu, E. Arabidopsis heat stress transcription factors A2 (HSFA2) and A3 (HSFA3) function in the same heat regulation pathway. Acta Physiol. Plantarum. 2017, 39, 1–9. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Tanaka, H.; Maruyama, K.; Qin, F.; Osakabe, Y.; Morimoto, K.; Ohori, T.; Kusakabe, K.; Nagata, M.; et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 2014, 26, 4954–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, A.L.; Ding, Y.F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef]
- Ding, Y.L.; Shi, Y.T.; Yang, S.H. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef] [Green Version]
- Xue, G.P.; Sadat, S.; Drenth, J.; Mcintyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.Y.; Tao, P.; Li, B.Y.; Wang, W.H.; Yue, Z.C.; Lei, J.L.; Zhong, X.M. Genome-wide identification, classification, and analysis of heat shock transcription factor family in Chinese cabbage (Brassica rapa pekinensis). Genet. Mol. Res. 2015, 14, 2189–2204. [Google Scholar] [CrossRef]
- Li, P.S.; Yu, T.F.; He, G.H.; Chen, M.; Zhou, Y.B.; Chai, S.C.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Sejima, H.; Tam, R.; Schlauch, K.; Mittler, R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011, 66, 844–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, Q.; Yue, X.; Zeng, H.; Zhu, J. The protein phosphatase rcf2 and its interacting partner nac019 are critical for heat stress–responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 2014, 26, 438–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2019, 18, 1317–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Zhang, J.; Zhang, N.; Xin, M.; Peng, H.; Hu, Z.; Ni, Z.; Du, J. The Wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic Arabidopsis. PLoS ONE 2015, 10, e0135667. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef] [Green Version]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhou, X.; Chen, L.; Huang, W.; Yu, D. Functional characterization of Arabidopsis thaliana wrky39 in heat stress. Mol. Cells 2010, 29, 475. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing oswrky11 under the control of hsp101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef]
- Lai, Y.S.; Renna, L.; Yarema, J.; Ruberti, C.; He, S.Y.; Brandizzi, F. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2018, 115, E5203–E5212. [Google Scholar] [CrossRef] [Green Version]
- Kataoka, R.; Takahashi, M.; Suzuki, N. Coordination between bZIP28 and HSFA2 in the regulation of heat response signals in Arabidopsis. Plant Signal. Behav. 2017, 12, e1376159. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liang, J.; Wang, C.; Zhao, X.; Zhong, X.; Cao, X.; Li, G.; He, J.; Yi, M. Overexpression of lily HsfA3s in Arabidopsis confers increased thermotolerance and salt sensitivity via alterations in proline catabolism. J. Exp. Bot. 2018, 69, 2005–2021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, C.; Zheng, Y.; Guo, Y. Myb30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 2017, 216, 163–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Kereamy, A.; Bi, Y.M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, A.; Nguyen, A.H.; Nakaminami, K.; Seki, M. Arabidopsis non-coding RNA regulation in abiotic stress responses. Int. J. Mol. Sci. 2013, 14, 22642–22654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, K.; Chen, X. Biogenesis, turnover and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [Green Version]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef]
- Zhao, J.; He, Q.; Chen, G.; Wang, L.; Jin, B. Regulation of non-coding RNAs in heat stress responses of plants. Front. Plant Sci. 2016, 7, 1213. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Sun, F.; Cao, H.; Peng, H.R.; Ni, Z.F.; Sun, Q.X.; Yao, Y.Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2012, 7, e48445. [Google Scholar] [CrossRef] [Green Version]
- Giacomelli, J.I.; Weigel, D.; Chan, R.L.; Manavella, P.A. Role of recently evolved mirna regulation of sunflower hawrky6 in response to temperature damage. New Phytol. 2012, 195, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Li, S.X.; Liu, J.X.; Liu, Z.Y.; Li, X.R.; Wu, F.J.; He, Y.K. Heatinduced tas1 target1 mediates thermotolerance via heat stress transcription factor A1a–directed pathways in Arabidopsis. Plant Cell 2014, 26, 1764–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Yang, J.; Li, X.; Liu, X.; Sun, C.; Wu, F.; He, Y.K. Global analysis of cisnatural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biol. 2013, 13, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Guo, S.; Wang, Y.; Wang, L.; Shu, S.; Sun, J. Systematic identification and analysis of heat-stress-responsive lncRNAs, circRNAs and miRNAs with associated co-expression and ceRNA networks in cucumber (Cucumis sativus L.). Physiol. Plant. 2020, 168, 736–754. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ci, D.; Tian, M.; Zhang, D.Q. Stable methylation of a noncoding RNA gene regulates gene expression in response to abiotic stress in Populus simonii. J. Exp. Bot. 2015, 67, 1477–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.; Hu, J.; Gao, C.; Chen, G.; Wang, B.; Lin, C.; Song, L.; Ding, Y.; Zhou, G. Genome-wide analysis of long non-coding RNAs unveils the regulatory roles in the heat tolerance of Chinese cabbage (Brassica rapa ssp.chinensis). Sci. Rep. 2019, 9, 5002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, T.; Seki, M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014, 55, 1859–1863. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S. Remembrance of stresses past: Heat shock factors and histone hypermethylation are key. Plant J. 2018, 95, 399–400. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Li, J.; He, Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015, 6, 267. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Seki, M. Histone Modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol. 2020, 182, 15–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, O.V.; Dinh, H.Q.; Aufsatz, W.; Jonak, C. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 2013, 6, 396–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Li, Y.; Duan, W.; Huang, F.; Hou, X. Cold acclimation alters DNA methylation patterns and confers tolerance to heat and increases growth rate in Brassica rapa. J. Exp. Bot. 2017, 68, 1213–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollunder, J.; Meins, F., Jr.; Kovalchuk, I. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Correia, B.; Valledor, L.; Meijon, M.; Rodriguez, J.L.; Dias, M.C.; Santos, C.; Cañal, M.J.; Rodriguez, R.; Pinto, G. Is the interplay between epigenetic markers related to the acclimation of cork oak plants to high temperatures? PLoS ONE 2013, 8, e53543. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Li, J.; Li, H.; Li, F.; Xu, K.; Yan, G.; Chen, B.; Qiao, J.; Wu, X. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed. Sci. 2014, 64, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Min, L.; Li, Y.; Hu, Q.; Zhu, L.; Gao, W.; Wu, Y.; Ding, Y.; Liu, S.; Yang, X.; Zhang, X. Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 2014, 164, 1293–1308. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Min, L.; Wang, M.; Wang, C.; Zhao, Y.; Li, Y.; Fang, Q.; Wu, Y.; Xie, S.; Ding, Y.; et al. Disrupted genome methylation in response to high temperature has distinct affects on microspore abortion and anther indehiscence. Plant Cell 2018, 30, 1387–1403. [Google Scholar] [CrossRef] [Green Version]
- Naydenov, M.; Baev, V.; Apostolova, E.; Gospodinova, N.; Sablok, G.; Gozmanova, M.; Yahubyan, G. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 2015, 87, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; De Jonge, J.; Forsberg, S.K.; Pettersson, M.E.; Sheng, Z.; Hennig, L.; Carlborg, Ö. Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet. 2014, 10, e1004842. [Google Scholar] [CrossRef]
- Strenkert, D.; Schmollinger, S.; Sommer, F.; Schulz-Raffelt, M.; Schroda, M. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 2011, 23, 2285–2301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecinka, A.; Dinh, H.Q.; Baubec, T.; Rosa, M.; Lettner, N.; Mittelsten Scheid, O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 2010, 22, 3118–3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folsom, J.J.; Begcy, K.; Hao, X.; Wang, D.; Walia, H. Rice fertilization-independent endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 2014, 165, 238–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Zhang, S.; Lin, S.; Guo, Y.; Deng, W.; Zhang, Y.; Xue, Y. WERAM: A database of writers, erasers and readers of histone acetylation and methylation in eukaryotes. Nucleic Acids Res. 2017, 45, D264–D270. [Google Scholar] [PubMed] [Green Version]
- Deng, X.; Qiu, Q.; He, K.; Cao, X. The seekers: How epigenetic modifying enzymes find their hidden genomic targets in Arabidopsis. Curr. Opin. Plant Biol. 2018, 45, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Song, N.; Zheng, M.; Liu, X.; Liu, Z.; Xing, J.; Ma, J.; Guo, W.; Yao, Y.; Peng, H.; et al. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 2015, 84, 1178–1191. [Google Scholar] [CrossRef]
- Earley, K.W.; Pontvianne, F.; Wierzbicki, A.T.; Blevins, T.; Tucker, S.; Costa-Nunes, P.; Pontes, O.; Pikaard, C.S. Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 2010, 24, 1119–1132. [Google Scholar] [CrossRef] [Green Version]
- Buszewicz, D.; Archacki, R.; Palusiński, A.; Kotliński, M.; Fogtman, A.; Iwanicka-Nowicka, R.; Sosnowska, K.; Kuciński, J.; Pupel, P.; Olędzki, J.; et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016, 39, 2108–2122. [Google Scholar] [CrossRef]
- Park, J.; Lim, C.J.; Shen, M.; Park, H.J.; Cha, J.Y.; Iniesto, E.; Rubio, V.; Mengiste, T.; Zhu, J.K.; Bressan, R.A.; et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 2018, 115, E5400–E5409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.J.; Baek, D.; Cha, J.Y.; Liao, X.; Kang, S.H.; McClung, C.R.; Lee, S.Y.; Yun, D.J.; Kim, W.Y. HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 2019, 31, 37–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharti, K.; Von Koskull-Döring, P.; Bharti, S.; Kumar, P.; Tintschl-Körbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, M.; Yang, Y.; Feng, H.; Pan, Z.; Shen, W.H.; Zhu, Y.; Dong, A. Histone chaperone ASF1 is involved in gene transcription activation in response to heat stress in Arabidopsis thaliana. Plant Cell Environ. 2014, 37, 2128–2138. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Wigge, P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alos, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Mlynarova, L.; Nap, J.P.; Bisseling, T. The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 2007, 51, 874–885. [Google Scholar] [CrossRef]
- Iwasaki, M.; Paszkowski, J. Identification of genes preventing transgenerational transmission of stress induced epigenetic states. Proc. Natl. Acad. Sci. USA 2014, 111, 8547–8552. [Google Scholar] [CrossRef] [Green Version]
- Crisp, P.A.; Ganguly, D.; Eichten, S.R.; Borevitz, J.O.; Pogson, B.J. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2016, 2, e1501340. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Li, Z. Epigenetic Environmental memories in plants: Establishment, maintenance, and reprogramming. Trends Genet. 2018, 34, 856–866. [Google Scholar] [CrossRef]
- Lämke, J.; Brzezinka, K.; Altmann, S.; Bäurle, I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016, 35, 162–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Feng, L.; Gu, X.; Deng, X.; Qiu, Q.; Li, Q.; Zhang, Y.; Wang, M.; Deng, Y.; Wang, E.; et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019, 29, 379–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suter, L.; Widmer, A. Phenotypic effects of salt and heat stress over three generations in Arabidopsis thaliana. PLoS ONE 2013, 8, e80819. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.H.; Liu, J.Z.; Jin, H.; Lin, L.; Li, Q.; Chen, Y.; Yuan, Y.X.; Wang, Z.Y.; Huang, H.; Qi, Y.J.; et al. Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 9171–9176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, H.; Yoshida, T.; Tsukahara, S.; Kawabe, A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene 2013, 518, 256–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, L.D.; Gevaert, K.; De Smet, I. Feeling the heat: Searching for plant thermosensors. Trends Plant Sci. 2019, 24, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef]
- Yoo, H.; Triandafillou, C.; Drummond, D.A. Cellular sensing by phase separation: Using the process, not just the products. J. Biol. Chem. 2019, 294, 7151–7159. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Lu, Z.; Wang, L.; Jin, B. Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. Int. J. Mol. Sci. 2021, 22, 117. https://doi.org/10.3390/ijms22010117
Zhao J, Lu Z, Wang L, Jin B. Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. International Journal of Molecular Sciences. 2021; 22(1):117. https://doi.org/10.3390/ijms22010117
Chicago/Turabian StyleZhao, Jianguo, Zhaogeng Lu, Li Wang, and Biao Jin. 2021. "Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics" International Journal of Molecular Sciences 22, no. 1: 117. https://doi.org/10.3390/ijms22010117
APA StyleZhao, J., Lu, Z., Wang, L., & Jin, B. (2021). Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. International Journal of Molecular Sciences, 22(1), 117. https://doi.org/10.3390/ijms22010117




