Ubiquinone Metabolism and Transcription HIF-1 Targets Pathway Are Toxicity Signature Pathways Present in Extracellular Vesicles of Paraquat-Exposed Human Brain Microvascular Endothelial Cells
Abstract
:1. Introduction
2. Results
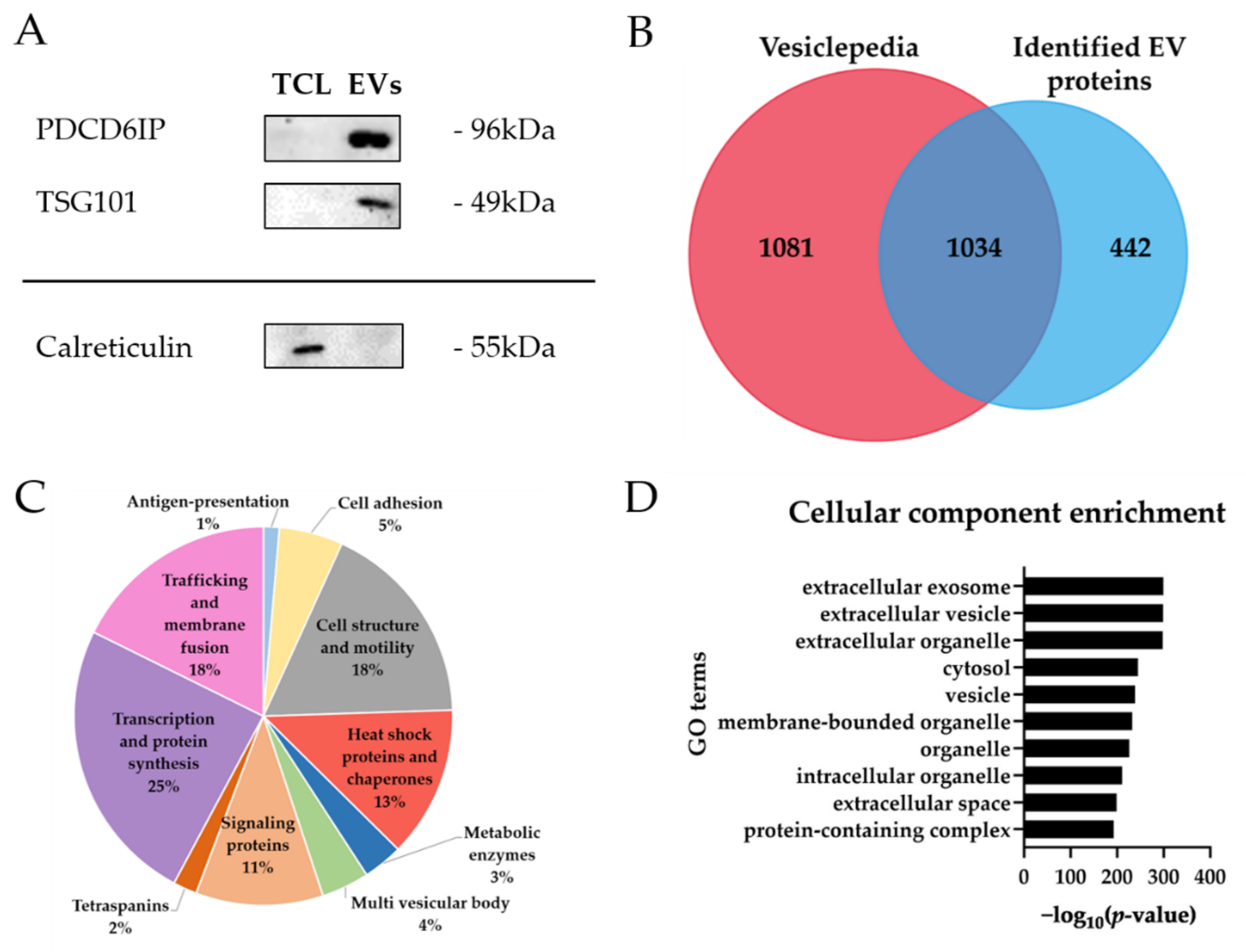
2.1. Characterization of Extracellular Vesicles from Human Brain Microvascular Endothelial Cells
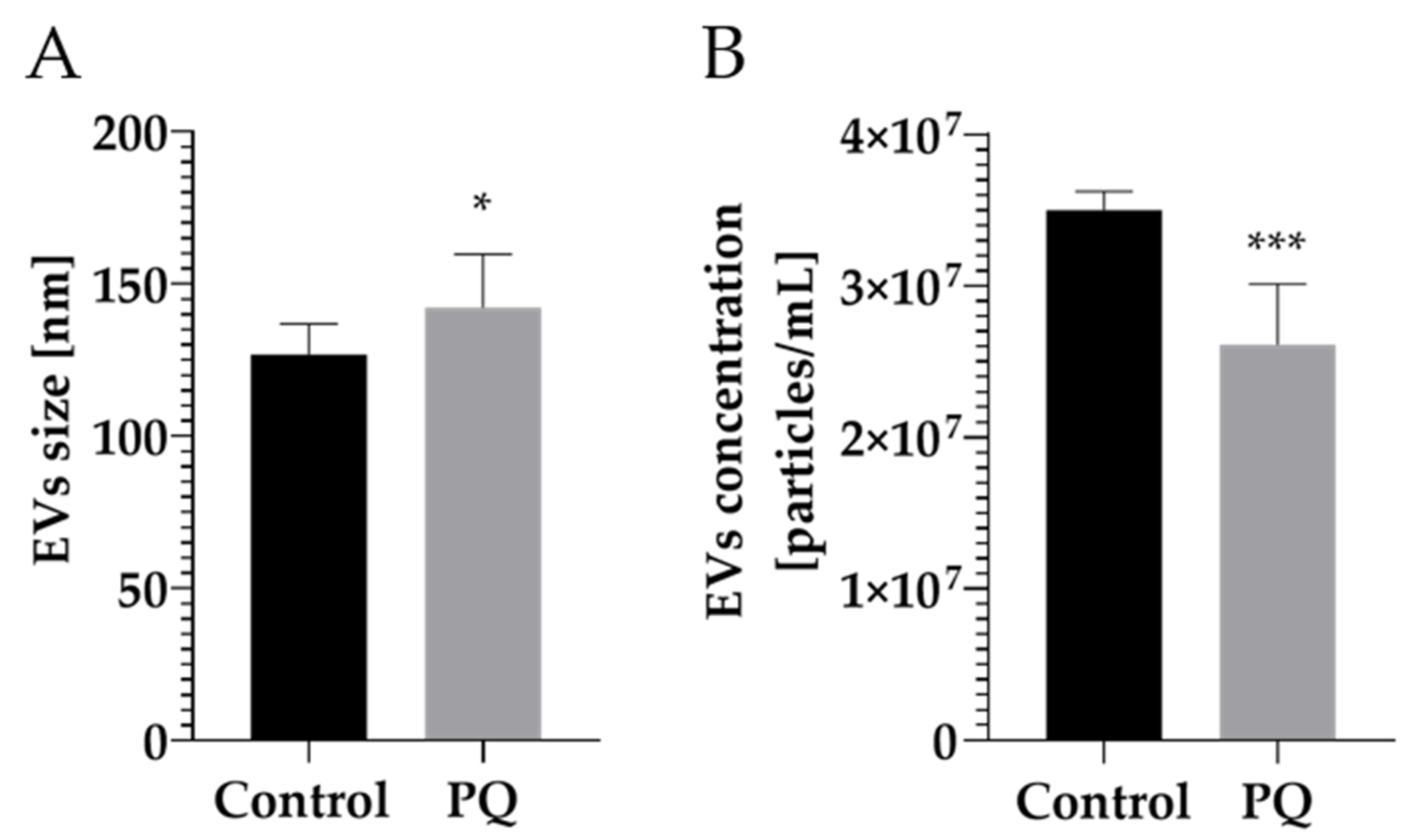
2.2. Modulation of Vesicular Protein Content after Paraquat (PQ) Exposure
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. EV Isolation
4.3. Nanoparticle Tracking Analysis (NTA)
4.4. Protein Extraction and Quantificatio
4.5. Western Blot Analysis
4.6. Sample Preparation for Mass Spectrometry-Based Proteomics
4.7. Data Independent Acquisition Mass Spectrometry (DIA-MS) and Data Analysis
4.8. Enrichment Pathway Analysis
4.9. MST Proliferation and LDH Cytotoxicity Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- De Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; van Balkom, B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef]
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiatry 2019, 9, 122. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Scalia, F.; Marino Gammazza, A.; Carlisi, D.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C. Extracellular Vesicle-Mediated Cell(-)Cell Communication in the Nervous System: Focus on Neurological Diseases. Int. J. Mol. Sci. 2019, 20, 434. [Google Scholar] [CrossRef] [Green Version]
- Serlin, Y.; Shelef, I.; Knyazer, B.; Friedman, A. Anatomy and physiology of the blood-brain barrier. Semin. Cell Dev. Biol. 2015, 38, 2–6. [Google Scholar] [CrossRef] [Green Version]
- Varatharaj, A.; Galea, I. The blood-brain barrier in systemic inflammation. Brain Behav. Immunity 2017, 60, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Poller, B.; Gutmann, H.; Krahenbuhl, S.; Weksler, B.; Romero, I.; Couraud, P.O.; Tuffin, G.; Drewe, J.; Huwyler, J. The human brain endothelial cell line hCMEC/D3 as a human blood-brain barrier model for drug transport studies. J. Neurochem. 2008, 107, 1358–1368. [Google Scholar] [CrossRef]
- Haqqani, A.S.; Delaney, C.E.; Tremblay, T.L.; Sodja, C.; Sandhu, J.K.; Stanimirovic, D.B. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS 2013, 10, 4. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Zhao, Z.; Montagne, A.; Nelson, A.R.; Zlokovic, B.V. Blood-Brain Barrier: From Physiology to Disease and Back. Physiol. Rev. 2019, 99, 21–78. [Google Scholar] [CrossRef]
- Barar, J.; Rafi, M.A.; Pourseif, M.M.; Omidi, Y. Blood-brain barrier transport machineries and targeted therapy of brain diseases. BioImpacts BI 2016, 6, 225–248. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, M.D.; Sagare, A.P.; Zlokovic, B.V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018, 14, 133. [Google Scholar] [CrossRef]
- Yegambaram, M.; Manivannan, B.; Beach, T.G.; Halden, R.U. Role of environmental contaminants in the etiology of Alzheimer’s disease: A review. Curr. Alzheimer Res. 2015, 12, 116–146. [Google Scholar] [CrossRef]
- Chen, H.; Ritz, B. The Search for Environmental Causes of Parkinson’s Disease: Moving Forward. J. Parkinsons Dis. 2018, 8, S9–S17. [Google Scholar] [CrossRef] [Green Version]
- Drechsel, D.A.; Patel, M. Chapter 21 Paraquat-induced production of reactive oxygen species in brain mitochondria. Methods Enzymol. 2009, 456, 381–393. [Google Scholar] [CrossRef]
- Tawara, T.; Fukushima, T.; Hojo, N.; Isobe, A.; Shiwaku, K.; Setogawa, T.; Yamane, Y. Effects of paraquat on mitochondrial electron transport system and catecholamine contents in rat brain. Arch. Toxicol. 1996, 70, 585–589. [Google Scholar] [CrossRef]
- Fukushima, T.; Yamada, K.; Hojo, N.; Isobe, A.; Shiwaku, K.; Yamane, Y. Mechanism of cytotoxicity of paraquat: III. The effects of acute paraquat exposure on the electron transport system in rat mitochondria. Exp. Toxicol. Pathol. 1994, 46, 437–441. [Google Scholar] [CrossRef]
- Zhang, X.-f.; Thompson, M.; Xu, Y.-h. Multifactorial theory applied to the neurotoxicity of paraquat and paraquat-induced mechanisms of developing Parkinson’s disease. Lab. Investig. 2016, 96, 496. [Google Scholar] [CrossRef]
- Sandström, J.; Broyer, A.; Zoia, D.; Schilt, C.; Greggio, C.; Fournier, M.; Do, K.Q.; Monnet-Tschudi, F. Potential mechanisms of development-dependent adverse effects of the herbicide paraquat in 3D rat brain cell cultures. Neurotoxicology 2017, 60, 116–124. [Google Scholar] [CrossRef]
- Dinis-Oliveira, R.J.; Remião, F.; Carmo, H.; Duarte, J.A.; Navarro, A.S.; Bastos, M.L.; Carvalho, F. Paraquat exposure as an etiological factor of Parkinson’s disease. Neurotoxicology 2006, 27, 1110–1122. [Google Scholar] [CrossRef]
- Brown, T.P.; Rumsby, P.C.; Capleton, A.C.; Rushton, L.; Levy, L.S. Pesticides and Parkinson’s disease--is there a link? Environ. Health Perspect. 2006, 114, 156–164. [Google Scholar] [CrossRef]
- Castello, P.R.; Drechsel, D.A.; Patel, M. Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. J. Biol. Chem. 2007, 282, 14186–14193. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Li, L.; Iliuk, A.; Tao, W.A. Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res. 2018, 17, 3308–3316. [Google Scholar] [CrossRef]
- Iliuk, A.; Wu, X.; Li, L.; Sun, J.; Hadisurya, M.; Boris, R.S.; Tao, W.A. Plasma-Derived Extracellular Vesicle Phosphoproteomics through Chemical Affinity Purification. J. Proteome Res. 2020, 19, 2563–2574. [Google Scholar] [CrossRef]
- Men, Y.; Yelick, J.; Jin, S.; Tian, Y.; Chiang, M.S.R.; Higashimori, H.; Brown, E.; Jarvis, R.; Yang, Y. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun. 2019, 10, 4136. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [Green Version]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2020, 166747. [Google Scholar] [CrossRef]
- Hromada, C.; Mühleder, S.; Grillari, J.; Redl, H.; Holnthoner, W. Endothelial Extracellular Vesicles-Promises and Challenges. Front. Physiol. 2017, 8, 275. [Google Scholar] [CrossRef] [PubMed]
- Dozio, V.; Sanchez, J.C. Characterisation of extracellular vesicle-subsets derived from brain endothelial cells and analysis of their protein cargo modulation after TNF exposure. J. Extracell. Vesicles 2017, 6, 1302705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- András, I.E.; Toborek, M. Extracellular vesicles of the blood-brain barrier. Tissue Barriers 2016, 4, e1131804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simeone, P.; Bologna, G.; Lanuti, P.; Pierdomenico, L.; Guagnano, M.T.; Pieragostino, D.; Del Boccio, P.; Vergara, D.; Marchisio, M.; Miscia, S.; et al. Extracellular Vesicles as Signaling Mediators and Disease Biomarkers across Biological Barriers. Int. J. Mol. Sci. 2020, 21, 2514. [Google Scholar] [CrossRef] [Green Version]
- Mir, B.; Goettsch, C. Extracellular Vesicles as Delivery Vehicles of Specific Cellular Cargo. Cells 2020, 9, 1601. [Google Scholar] [CrossRef]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W.E. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar] [CrossRef]
- Fan, Z.; Zheng, W.; Li, H.; Wu, W.; Liu, X.; Sun, Z.; Hu, H.; Du, L.; Jia, Q.; Liu, Q. LOXL2 upregulates hypoxia-inducible factor-1α signaling through Snail-FBP1 axis in hepatocellular carcinoma cells. Oncol. Rep. 2020, 43, 1641–1649. [Google Scholar] [CrossRef]
- Ben-Yosef, Y.; Miller, A.; Shapiro, S.; Lahat, N. Hypoxia of endothelial cells leads to MMP-2-dependent survival and death. Am J. Physiol. Cell Physiol. 2005, 289, C1321–C1331. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Li, B.; He, Q.-Y. Significance of prohibitin domain family in tumorigenesis and its implication in cancer diagnosis and treatment. Cell Death Dis. 2018, 9, 580. [Google Scholar] [CrossRef]
- Zhu, Y.; Tan, J.; Xie, H.; Wang, J.; Meng, X.; Wang, R. HIF-1α regulates EMT via the Snail and β-catenin pathways in paraquat poisoning-induced early pulmonary fibrosis. J. Cell. Mol. Med. 2016, 20, 688–697. [Google Scholar] [CrossRef]
- Bus, J.S.; Gibson, J.E. Paraquat: Model for oxidant-initiated toxicity. Environ. Health Perspect. 1984, 55, 37–46. [Google Scholar] [CrossRef]
- McCarthy, S.; Somayajulu, M.; Sikorska, M.; Borowy-Borowski, H.; Pandey, S. Paraquat induces oxidative stress and neuronal cell death; neuroprotection by water-soluble Coenzyme Q10. Toxicol. Appl. Pharmacol. 2004, 201, 21–31. [Google Scholar] [CrossRef]
- Vujić, T.; Schvartz, D.; Sanchez, J.-C. Quantitative Data-Independent Acquisition (DIA) analysis unraveled paraquat-induced cholesterol biosynthesis dysregulation in human brain microvascular endothelial cells. Unpublished, manuscript in preparation.
- Rufino-Ramos, D.; Albuquerque, P.R.; Carmona, V.; Perfeito, R.; Nobre, R.J.; Pereira de Almeida, L. Extracellular vesicles: Novel promising delivery systems for therapy of brain diseases. J. Control Release 2017, 262, 247–258. [Google Scholar] [CrossRef]
- Upadhya, R.; Zingg, W.; Shetty, S.; Shetty, A.K. Astrocyte-derived extracellular vesicles: Neuroreparative properties and role in the pathogenesis of neurodegenerative disorders. J. Control Release 2020, 323, 225–239. [Google Scholar] [CrossRef]
- Leitolis, A.; Suss, P.H.; Roderjan, J.G.; Angulski, A.B.B.; da Costa, F.D.A.; Stimamiglio, M.A.; Correa, A. Human Heart Explant-Derived Extracellular Vesicles: Characterization and Effects on the In Vitro Recellularization of Decellularized Heart Valves. Int. J. Mol. Sci. 2019, 20, 1279. [Google Scholar] [CrossRef] [Green Version]
- Vella, L.J.; Scicluna, B.J.; Cheng, L.; Bawden, E.G.; Masters, C.L.; Ang, C.-S.; Willamson, N.; McLean, C.; Barnham, K.J.; Hill, A.F. A rigorous method to enrich for exosomes from brain tissue. J. Extracell. Vesicles 2017, 6, 1348885. [Google Scholar] [CrossRef]
- Rathinam, M.L.; Watts, L.T.; Narasimhan, M.; Riar, A.K.; Mahimainathan, L.; Henderson, G.I. Astrocyte mediated protection of fetal cerebral cortical neurons from rotenone and paraquat. Environ. Toxicol. Pharmacol. 2012, 33, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Bagley, A.C.; Krall, J.; Lynch, R.E. Superoxide mediates the toxicity of paraquat for Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3189–3193. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Sun, A.Y. Paraquat-induced free radical reaction in mouse brain microsomes. Neurochem. Res. 1998, 23, 47–53. [Google Scholar] [CrossRef]
- Wang, X.; Luo, F.; Zhao, H. Paraquat-Induced Reactive Oxygen Species Inhibit Neutrophil Apoptosis via a p38 MAPK/NF-κB–IL-6/TNF-α Positive-Feedback Circuit. PLoS ONE 2014, 9, e93837. [Google Scholar] [CrossRef]
- Yang, Y.; Boza-Serrano, A.; Dunning, C.J.R.; Clausen, B.H.; Lambertsen, K.L.; Deierborg, T. Inflammation leads to distinct populations of extracellular vesicles from microglia. J. Neuroinflamm. 2018, 15, 168. [Google Scholar] [CrossRef]
- Chan, J.C.; Morgan, C.P.; Adrian Leu, N.; Shetty, A.; Cisse, Y.M.; Nugent, B.M.; Morrison, K.E.; Jašarević, E.; Huang, W.; Kanyuch, N.; et al. Reproductive tract extracellular vesicles are sufficient to transmit intergenerational stress and program neurodevelopment. Nat. Commun. 2020, 11, 1499. [Google Scholar] [CrossRef] [Green Version]
- Yuana, Y.; Sturk, A.; Nieuwland, R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013, 27, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Xi, X.J.; Zeng, J.J.; Lu, Y.; Chen, S.H.; Jiang, Z.W.; He, P.J.; Mi, H. Extracellular vesicles enhance oxidative stress through P38/NF-kB pathway in ketamine-induced ulcerative cystitis. J. Cell Mol. Med. 2020, 24, 7609–7624. [Google Scholar] [CrossRef] [PubMed]
- D’Acunzo, P.; Pérez-González, R.; Kim, Y.; Hargash, T.; Miller, C.; Alldred, M.J.; Erdjument-Bromage, H.; Penikalapati, S.C.; Pawlik, M.; Saito, M.; et al. Mitovesicles are a novel population of extracellular vesicles of mitochondrial origin altered in Down syndrome. Sci. Adv. 2021, 7, eabe5085. [Google Scholar] [CrossRef] [PubMed]
- Van Dommelen, S.M.; van der Meel, R.; van Solinge, W.W.; Coimbra, M.; Vader, P.; Schiffelers, R.M. Cetuximab treatment alters the content of extracellular vesicles released from tumor cells. Nanomedicine 2016, 11, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Runz, S.; Keller, S.; Rupp, C.; Stoeck, A.; Issa, Y.; Koensgen, D.; Mustea, A.; Sehouli, J.; Kristiansen, G.; Altevogt, P. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007, 107, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef] [Green Version]
- Puhm, F.; Afonyushkin, T.; Resch, U.; Obermayer, G.; Rohde, M.; Penz, T.; Schuster, M.; Wagner, G.; Rendeiro, A.F.; Melki, I.; et al. Mitochondria Are a Subset of Extracellular Vesicles Released by Activated Monocytes and Induce Type I IFN and TNF Responses in Endothelial Cells. Circ. Res. 2019, 125, 43–52. [Google Scholar] [CrossRef]
- Jang, S.C.; Crescitelli, R.; Cvjetkovic, A.; Belgrano, V.; Olofsson Bagge, R.; Sundfeldt, K.; Ochiya, T.; Kalluri, R.; Lötvall, J. Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. J. Extracell. Vesicles 2019, 8, 1635420. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Tan, J.-t.; Wang, R.-l.; Meng, X.-X.; Tang, X.; Gao, S. Expression and significance of HIF-1α in pulmonary fibrosis induced by paraquat. Exp. Biol. Med. 2013, 238, 1062–1068. [Google Scholar] [CrossRef]
- Alural, B.; Ozerdem, A.; Allmer, J.; Genc, K.; Genc, S. Lithium protects against paraquat neurotoxicity by NRF2 activation and miR-34a inhibition in SH-SY5Y cells. Front. Cell. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhou, Y.; Gao, Q.; Ping, D.; Wang, Y.; Wu, W.; Lin, X.; Fang, Y.; Zhang, J.; Shao, A. The Role of Exosomal microRNAs and Oxidative Stress in Neurodegenerative Diseases. Oxid. Med. Cell. Longev. 2020, 2020, 3232869. [Google Scholar] [CrossRef]
- Dozio, V.; Sanchez, J.C. Profiling the proteomic inflammatory state of human astrocytes using DIA mass spectrometry. J. Neuroinflamm. 2018, 15, 331. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vujić, T.; Schvartz, D.; Iliuk, A.; Sanchez, J.-C. Ubiquinone Metabolism and Transcription HIF-1 Targets Pathway Are Toxicity Signature Pathways Present in Extracellular Vesicles of Paraquat-Exposed Human Brain Microvascular Endothelial Cells. Int. J. Mol. Sci. 2021, 22, 5065. https://doi.org/10.3390/ijms22105065
Vujić T, Schvartz D, Iliuk A, Sanchez J-C. Ubiquinone Metabolism and Transcription HIF-1 Targets Pathway Are Toxicity Signature Pathways Present in Extracellular Vesicles of Paraquat-Exposed Human Brain Microvascular Endothelial Cells. International Journal of Molecular Sciences. 2021; 22(10):5065. https://doi.org/10.3390/ijms22105065
Chicago/Turabian StyleVujić, Tatjana, Domitille Schvartz, Anton Iliuk, and Jean-Charles Sanchez. 2021. "Ubiquinone Metabolism and Transcription HIF-1 Targets Pathway Are Toxicity Signature Pathways Present in Extracellular Vesicles of Paraquat-Exposed Human Brain Microvascular Endothelial Cells" International Journal of Molecular Sciences 22, no. 10: 5065. https://doi.org/10.3390/ijms22105065






