Oxonium Ion Guided Analysis of Quantitative Proteomics Data Reveals Site-Specific O-Glycosylation of Anterior Gradient Protein 2 (AGR2)
Abstract
:1. Introduction
2. Results and Discussion
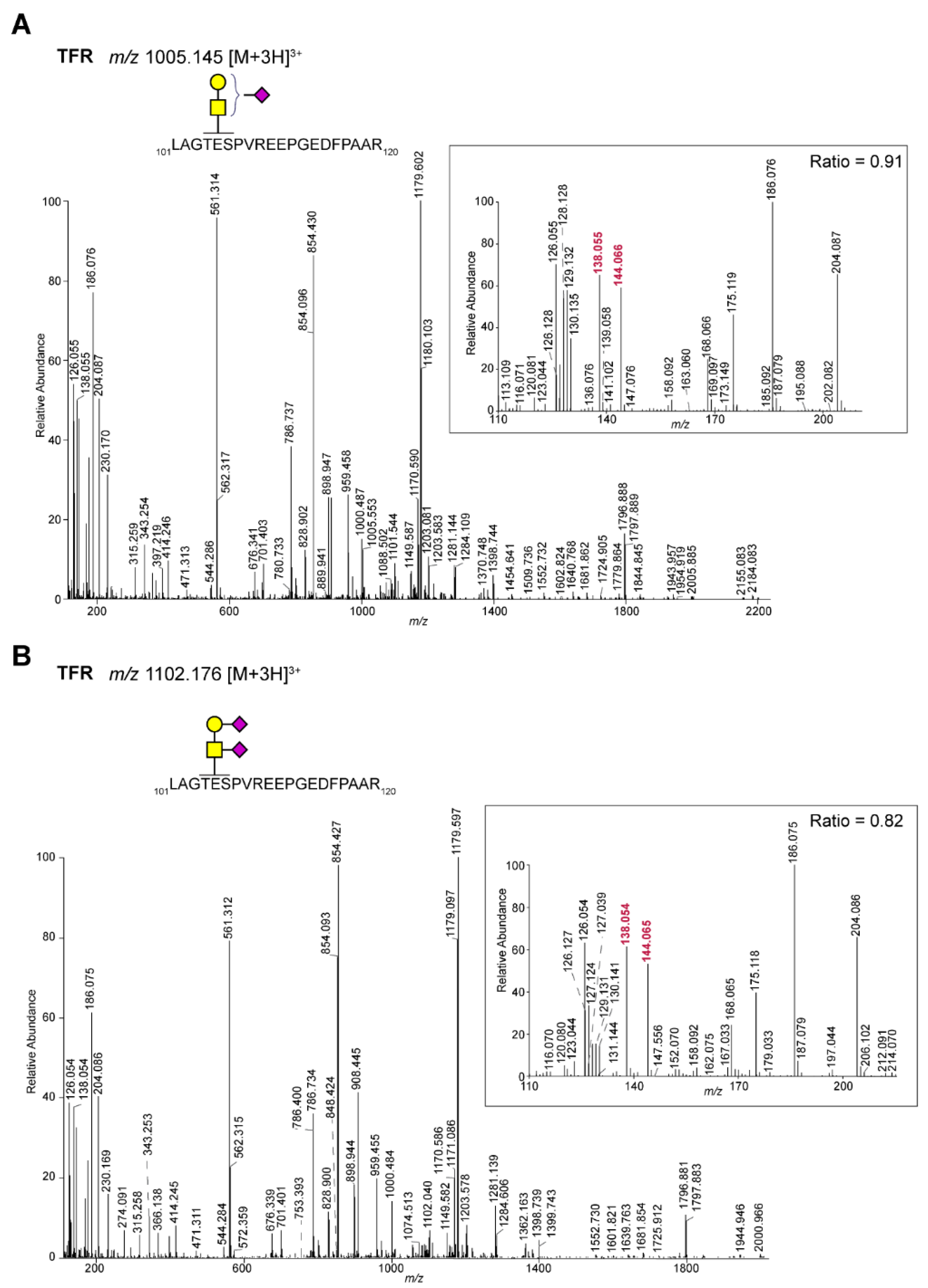
2.1. Differentiation of Glycopeptides Based on the Relative Abundance of Oxonium Ions
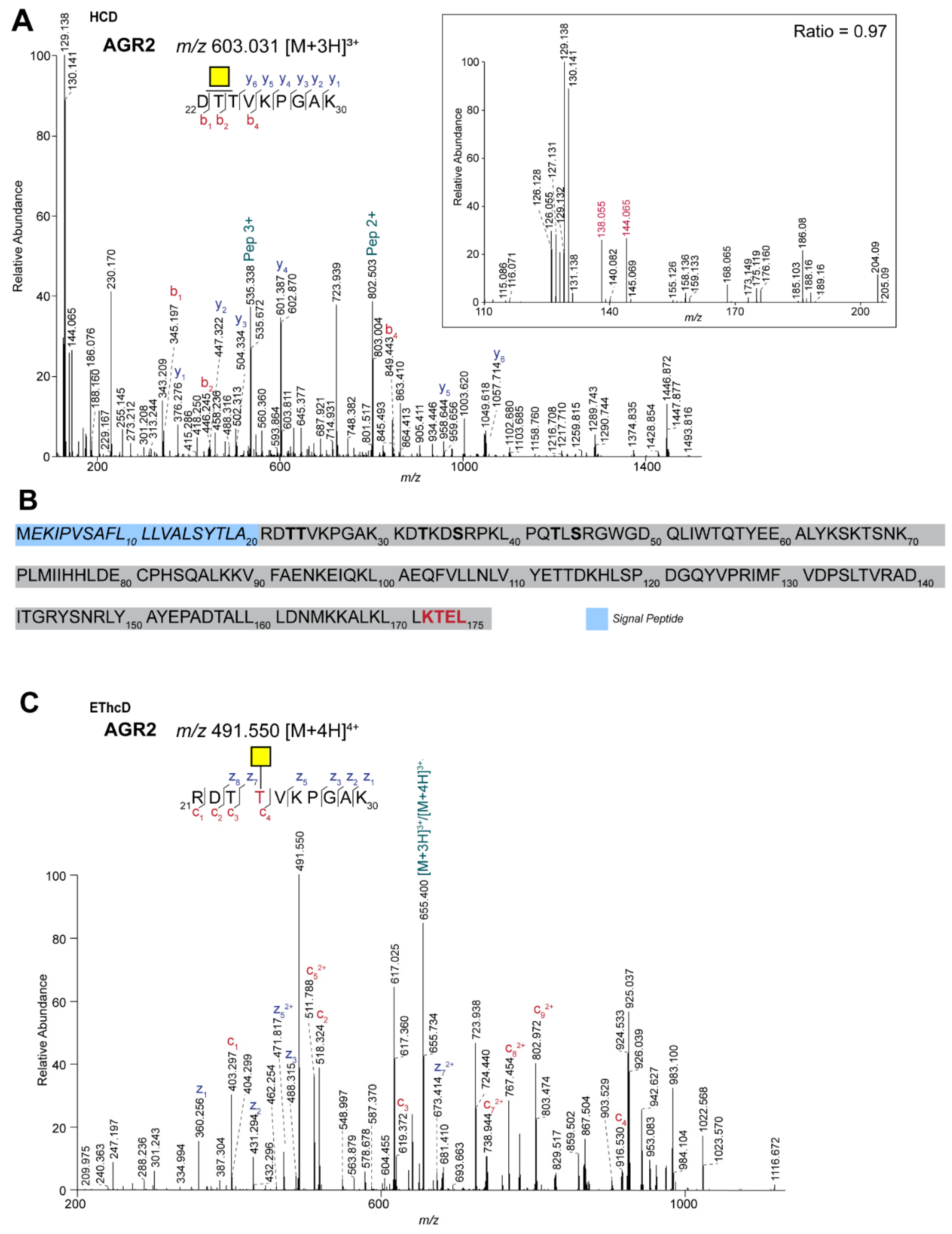
2.2. Site-Specific Identification of the O-glycosylation of Anterior Gradient Protein 2 (AGR2)
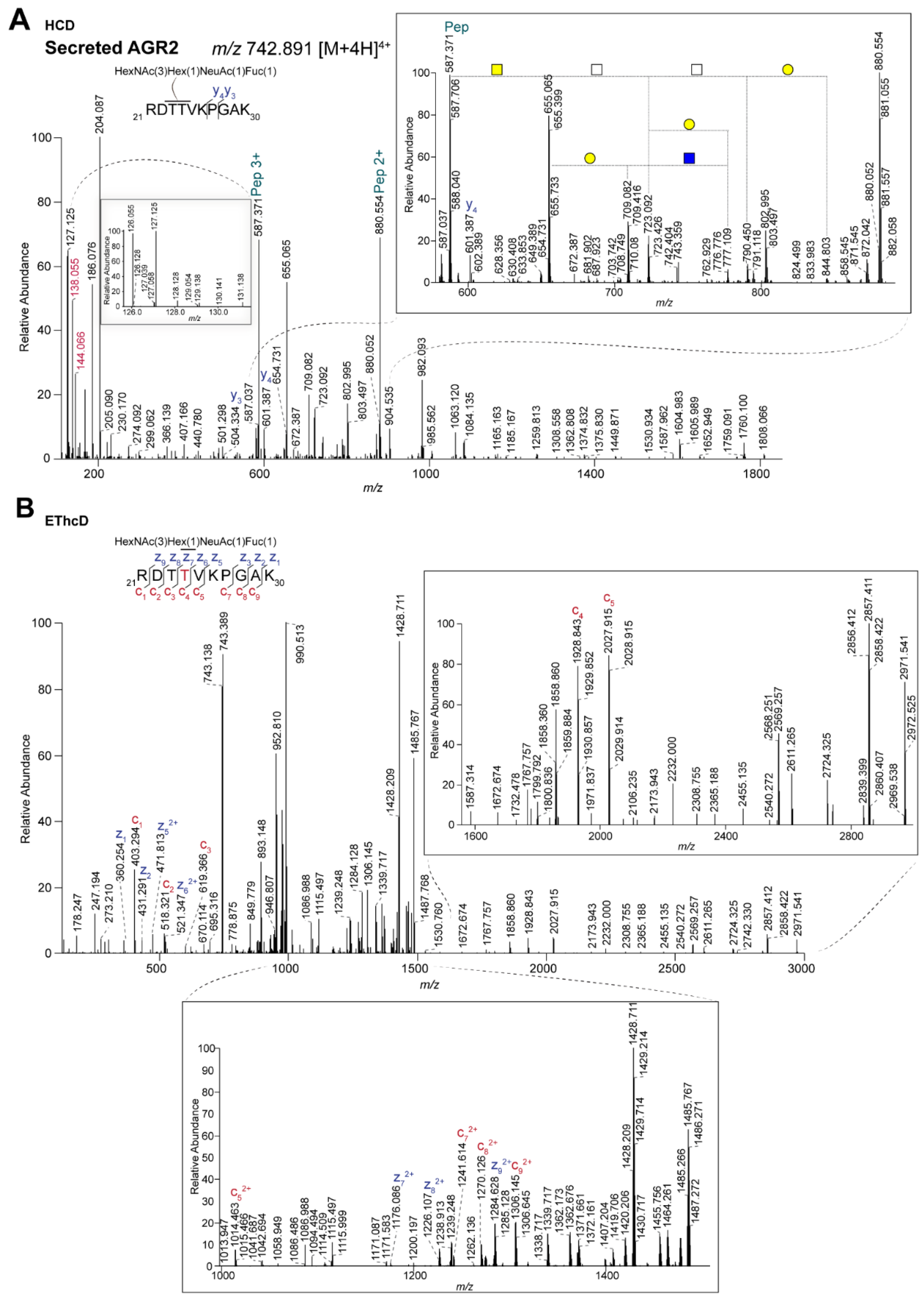
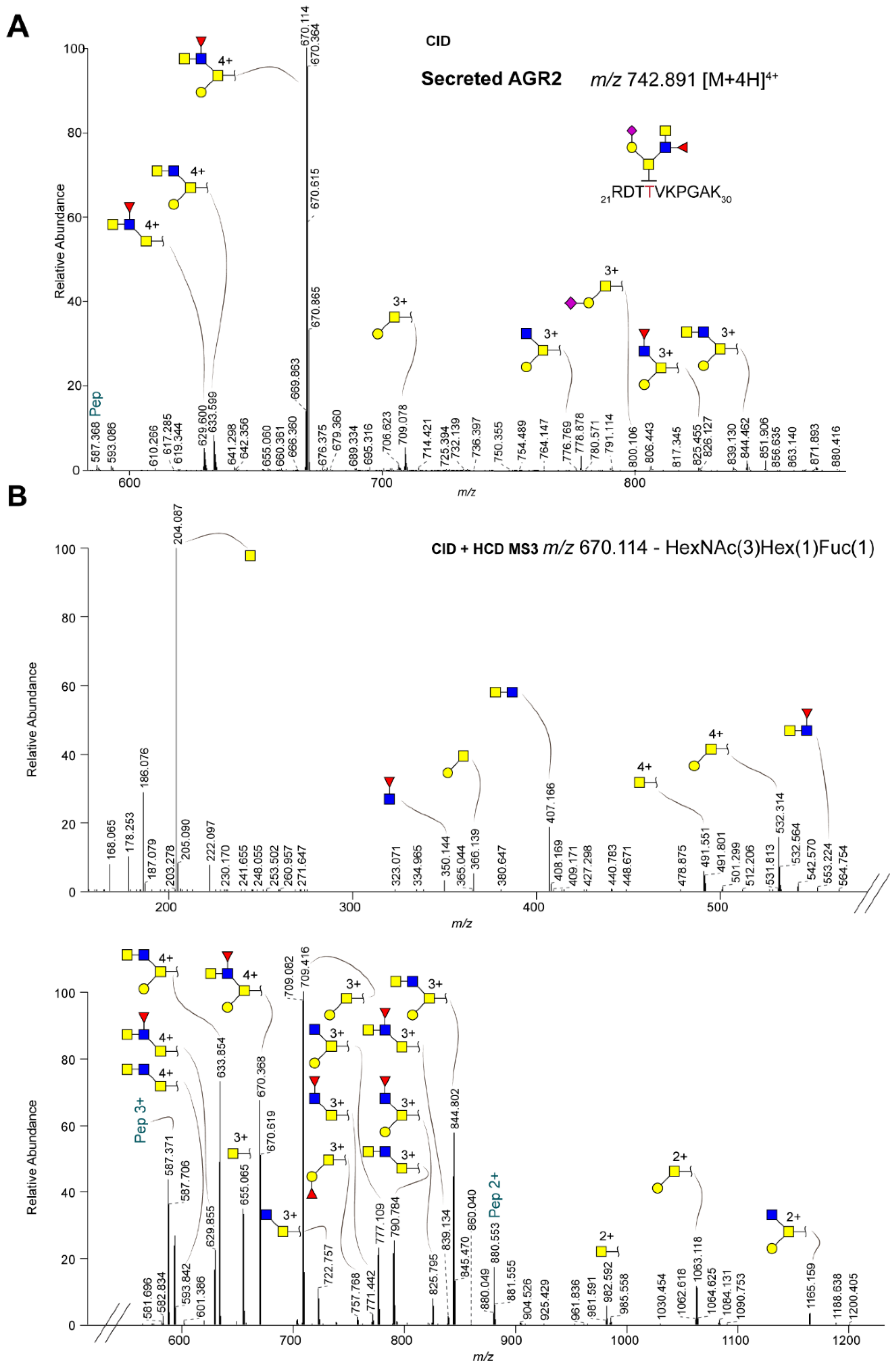
2.3. Differential Glycosylation of Secreted Versus Intracellular AGR2
3. Materials and Methods
3.1. Cell Lines Culture
3.2. Secretome Collection
3.3. TMT Labeling and LC-MS/MS Analyses
3.4. Bioinformatic Workflow for Glycopeptides Identification
3.5. Quantitative Proteomic Data Analysis of Secreted Proteins
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banazadeh, A.; Veillon, L.; Wooding, K.M.; Zabet-Moghaddam, M.; Mechref, Y. Recent advances in mass spectrometric analysis of glycoproteins. Electrophoresis 2017, 38, 162–189. [Google Scholar] [CrossRef] [Green Version]
- Wuhrer, M.; Catalina, M.I.; Deelder, A.M.; Hokke, C.H. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2007, 849, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, A.; Pipkorn, R.; Bossemeyer, D.; Lehmann, W.D. Analysis of protein phosphorylation by a combination of elastase digestion and neutral loss tandem mass spectrometry. Anal Chem. 2001, 73, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Trelle, M.B.; Jensen, O.N. Utility of immonium ions for assignment of epsilon-N-acetyllysine-containing peptides by tandem mass spectrometry. Anal. Chem. 2008, 80, 3422–3430. [Google Scholar] [CrossRef] [PubMed]
- Caval, T.; Heck, A.J.R.; Reiding, K.R. Meta-heterogeneity: Evaluating and describing the diversity in glycosylation between sites on the same glycoprotein. Mol. Cell Proteom. 2020, 20, 100010. [Google Scholar] [CrossRef] [PubMed]
- Hinneburg, H.; Stavenhagen, K.; Schweiger-Hufnagel, U.; Pengelley, S.; Jabs, W.; Seeberger, P.H.; Silva, D.V.; Wuhrer, M.; Kolarich, D. The Art of Destruction: Optimizing Collision Energies in Quadrupole-Time of Flight (Q-TOF) Instruments for Glycopeptide-Based Glycoproteomics. J. Am. Soc. Mass. Spectrom. 2016, 27, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiding, K.R.; Bondt, A.; Franc, V.; Hec, A.J.R. The benefits of hybrid fragmentation methods for glycoproteomics. Trac-Trends Anal. Chem. 2018, 108, 260–268. [Google Scholar] [CrossRef]
- Campbell, M.P. A Review of Software Applications and Databases for the Interpretation of Glycopeptide Data. Trends Glycosci. Glycotechnol. 2017, 29, E51–E62. [Google Scholar] [CrossRef]
- Lee, L.Y.; Moh, E.S.X.; Parker, B.L.; Bern, M.; Packer, N.H.; Thaysen-Anderse, M. Toward Automated N-Glycopeptide Identification in Glycoproteomics. J. Proteome Res. 2016, 15, 3904–3915. [Google Scholar] [CrossRef]
- Lu, L.; Riley, N.M.; Shortreed, M.R.; Bertozzi, C.R.; Smith, L.M. O-Pair Search with MetaMorpheus for O-glycopeptide characterization. Nat. Methods 2020, 17, 1133–1138. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.-B.G.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [Green Version]
- Ruhaak, L.R.; Xu, G.; Li, Q.; Goonatilleke, E.; Lebrilla, C.B. Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chem. Rev. 2018, 118, 7886–7930. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Zhang, W.; Shi, Z.; Tian, F.; Deng, Y.; Sun, C.; Wang, G.; Qin, W.; Qian, X. A novel strategy for global mapping of O-GlcNAc proteins and peptides using selective enzymatic deglycosylation, HILIC enrichment and mass spectrometry identification. Talanta 2017, 169, 195–202. [Google Scholar] [CrossRef]
- Zheng, J.; Xiao, H.; Wu, R. Specific Identification of Glycoproteins Bearing the Tn Antigen in Human Cells. Angew. Chem. Int. Ed. Engl. 2017, 56, 7107–7111. [Google Scholar] [CrossRef] [Green Version]
- Brockhausen, I.; Stanley, P. O-GalNAc Glycans. In Essentials of Glycobiology; Varki, A., Ed.; Cold Spring Harbor (NY): Long Island, NY, USA, 2015; pp. 113–123. [Google Scholar]
- Zachara, N.; Akimoto, Y.; Hart, G.W. The O-GlcNAc Modification. In Essentials of Glycobiology; Varki, A., Ed.; Cold Spring Harbor (NY): Long Island, NY, USA, 2015; pp. 239–251. [Google Scholar]
- Halim, A.; Westerlind, U.; Pett, C.; Schorleme, M.; Rüetschi, U.; Brinkmalm, G.; Sihlbom, C.; Lengqvist, J.; Larson, G.; Nilsson, J. Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 2014, 13, 6024–6032. [Google Scholar] [CrossRef] [PubMed]
- Park, G.W.; Lee, J.W.; Lee, H.K.; Shin, J.H.; Kim, J.Y.; Yoo, J.S. Classification of Mucin-Type O-Glycopeptides Using Higher-Energy Collisional Dissociation in Mass Spectrometry. Anal. Chem. 2020, 92, 9772–9781. [Google Scholar] [CrossRef]
- Riley, N.M.; Malaker, S.A.; Driessen, M.D.; Bertozzi, C.R. Optimal Dissociation Methods Differ for N- and O-Glycopeptides. J. Proteome Res. 2020, 19, 3286–3301. [Google Scholar] [CrossRef] [PubMed]
- Toghi Eshghi, S.; Eshghi, S.T.; Yan, W.; Hu, Y.; Shah, P.; Sun, S.; Li, X.; Zhang, H. Classification of Tandem Mass Spectra for Identification of N- and O-linked Glycopeptides. Sci. Rep. 2016, 6, 37189. [Google Scholar] [CrossRef] [PubMed]
- Pirro, M.; Schoof, E.; van Vliet, S.; Rombouts, Y.; Stella, A.; de Ru, A.; Mohammed, Y.; Wuhrer, M.; van Veelen, P.A.; Hensbergen, P.J. Glycoproteomic Analysis of MGL-Binding Proteins on Acute T-Cell Leukemia Cells. J. Proteome Res. 2019, 18, 1125–1132. [Google Scholar] [CrossRef]
- Pirro, M.; Rombouts, Y.; Stella, A.; Neyrolles, O.; Burlet-Schiltz, O.; van Vliet, S.J.; de Ru, A.; Mohammed, Y.; Wuhrer, M.; van Veelen, P.A.; et al. Characterization of Macrophage Galactose-type Lectin (MGL) ligands in colorectal cancer cell lines. Biochim Biophys. Acta Gen. Subj 2020, 1864, 129513. [Google Scholar] [CrossRef]
- Pirro, M.; Mohammed, Y.; van Vliet, S.J.; Rombouts, Y.; Sciacca, A.; de Ru, A.H.; Janssen, G.M.C.; Tjokrodirijo, R.T.N.; Wuhrer, M.; van Veelen, P.A.; et al. N-Glycoproteins Have a Major Role in MGL Binding to Colorectal Cancer Cell Lines: Associations with Overall Proteome Diversity. Int. J. Mol. Sci. 2020, 21, 5522. [Google Scholar] [CrossRef]
- Clarke, C.; Rudland, P.; Barraclough, R. The metastasis-inducing protein AGR2 is O-glycosylated upon secretion from mammary epithelial cells. Mol. Cell Biochem. 2015, 408, 245–252. [Google Scholar] [CrossRef] [Green Version]
- Fessart, D.; Domblides, C.; Avri, T.; Eriksson, L.A.; Begueret, H.; Pineau, R.; Malrieux, C.; Dugot-Senant, N.; Lucchesi, C.; Chevet, E.; et al. Secretion of protein disulphide isomerase AGR2 confers tumorigenic properties. Elife 2016, 5, e13887. [Google Scholar] [CrossRef] [PubMed]
- Ros, M.; Nguyen, A.T.; Chia, J.; Tran, S.L.; Guezennec, X.L.; McDowall, R.; Vakhrushev, S.; Clausen, H.; Humphries, M.J.; Saltel, F.; et al. ER-resident oxidoreductases are glycosylated and trafficked to the cell surface to promote matrix degradation by tumour cells. Nat. Cell Biol. 2020, 22, 1371–1381. [Google Scholar] [CrossRef]
- van Klinken, B.J.; Oussoren, E.; Weenink, J.J.; Strous, G.J.; Büller, H.A.; Dekker, J.; Einerhand, A.W. The human intestinal cell lines Caco-2 and LS174T as models to study cell-type specific mucin expression. Glycoconj J. 1996, 13, 757–768. [Google Scholar] [CrossRef]
- Schroeder, B.W.; Verhaeghe, C.; Park, S.-W.; Nguyenvu, L.T.; Huang, X.; Zhen, G.; Erle, D.J. AGR2 is induced in asthma and promotes allergen-induced mucin overproduction. Am. J. Respir. Cell Mol. Biol. 2012, 47, 178–185. [Google Scholar] [CrossRef]
- Madunic, K.; Zhang, T.; Mayboroda, O.A.; Holst, S.; Stavenhagen, K.; Jin, C.; Karlsson, N.G.; Lageveen-Kammeijer, G.S.M.; Wuhrer, M. Colorectal cancer cell lines show striking diversity of their O-glycome reflecting the cellular differentiation phenotype. Cell Mol. Life Sci. 2020, 78, 337–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westen, A.A.; Kraaijenbrink, T.; de Medina, E.A.R.; Harteveld, J.; Willemse, P.; Zuniga, S.B.; van der Gaag, K.J.; Weiler, N.E.C.; Warnaar, J.; Kayser, M.; et al. Comparing six commercial autosomal STR kits in a large Dutch population sample. Forensic Sci. Int. Genet. 2014, 10, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Vizcaino, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, 11033. [Google Scholar] [CrossRef] [Green Version]


| Uniprot ID | Protein Name | Peptide Sequence | Glycan | Byonic Score | 144/138 Ratio | Predicted * HexNAc Nature |
|---|---|---|---|---|---|---|
| Q12830 | Nucleosome-remodeling factor subunit BPTF | TVITEVTTMTSTVATESK | N1 | 457 | 0.17 | GlcNAc |
| Q96HC4 | PDZ and LIM domain protein 5 | EVVKPVPITSPAVSK | N1 | 508 | 0.18 | GlcNAc |
| P51610 | Host cell factor 1 | TIPMSAIITQAGATGVTSSPGIK | N1 | 1001 | 0.19 | GlcNAc |
| P51610 | Host cell factor 1 | SPITIITTK | N1 | 664 | 0.20 | GlcNAc |
| Q6UN15 | Pre-mRNA 3’-end-processing factor FIP1 | ETALPSTK | N1 | 362 | 0.20 | GlcNAc |
| P02545 | Prelamin-A/C | ASASGSGAQVGGPISSGSSASSVTVTR | N1 | 842 | 0.20 | GlcNAc |
| P27816 | Microtubule-associated protein 4 | ASPSKPASAPASR | N1 | 309 | 0.24 | GlcNAc |
| Q8NBJ4 | Golgi membrane protein 1 | LQAAGLPHTEVPQGK | H1N1S2 | 619 | 0.44 | GalNAc |
| Q92753 | Nuclear receptor ROR-beta | QQQSGEAEALAR | H1N1S2 | 273 | 0.68 | GlcNAc |
| P14314 | Glucosidase 2 subunit beta | SEALPTDLPAPSAPDLTEPK | H1N1S2 | 775 | 0.68 | GalNAc |
| P30533 | Alpha-2-macroglobulin receptor-associated protein | QVTSNSLSGTQEDGLDDPRLEK | N1 | 575 | 0.70 | GalNAc |
| Q8IYS2 | Uncharacterized protein KIAA2013 | VAALQTVGPTAGPAPK | H1N1S2 | 695 | 0.73 | GalNAc |
| Q8NBS9 | Thioredoxin domain-containing protein 5 | TETGATETVTPSEAPVLAAEPEADK | N1 | 609 | 0.77 | GalNAc |
| P02786 | Transferrin receptor protein 1 | LAGTESPVREEPGEDFPAAR | H1N1S2 | 364 | 0.82 | GalNAc |
| P02786 | Transferrin receptor protein 1 | LAGTESPVREEPGEDFPAAR | H1N1F1 | 406 | 0.85 | GalNAc |
| Q10469 | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase | GGDHPSVAVGIR | H1N1S2 | 456 | 0.90 | GalNAc |
| P02786 | Transferrin receptor protein 1 | LAGTESPVREEPGEDFPAAR | H1N1S1 | 596 | 0.91 | GalNAc |
| P26572 | Alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase | GRVPTAAPPAQPR | H1N1S2 | 260 | 0.91 | GalNAc |
| Q92520 | Protein FAM3C | STKPPR | H1N1S2 | 285 | 0.97 | GalNAc |
| O95994 | Anterior gradient protein 2 homolog | DTTVKPGAK | N1 | 376 | 0.97 | GalNAc |
| P14314 | Glucosidase 2 subunit beta | SEALPTDLPAPSAPDLTEPK | N1 | 674 | 1.06 | GalNAc |
| O60502 | Protein O-GlcNAcase | QVAHSGAK | N1 | 374 | 1.13 | GlcNAc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pirro, M.; Mohammed, Y.; de Ru, A.H.; Janssen, G.M.C.; Tjokrodirijo, R.T.N.; Madunić, K.; Wuhrer, M.; van Veelen, P.A.; Hensbergen, P.J. Oxonium Ion Guided Analysis of Quantitative Proteomics Data Reveals Site-Specific O-Glycosylation of Anterior Gradient Protein 2 (AGR2). Int. J. Mol. Sci. 2021, 22, 5369. https://doi.org/10.3390/ijms22105369
Pirro M, Mohammed Y, de Ru AH, Janssen GMC, Tjokrodirijo RTN, Madunić K, Wuhrer M, van Veelen PA, Hensbergen PJ. Oxonium Ion Guided Analysis of Quantitative Proteomics Data Reveals Site-Specific O-Glycosylation of Anterior Gradient Protein 2 (AGR2). International Journal of Molecular Sciences. 2021; 22(10):5369. https://doi.org/10.3390/ijms22105369
Chicago/Turabian StylePirro, Martina, Yassene Mohammed, Arnoud H. de Ru, George M. C. Janssen, Rayman T. N. Tjokrodirijo, Katarina Madunić, Manfred Wuhrer, Peter A. van Veelen, and Paul J. Hensbergen. 2021. "Oxonium Ion Guided Analysis of Quantitative Proteomics Data Reveals Site-Specific O-Glycosylation of Anterior Gradient Protein 2 (AGR2)" International Journal of Molecular Sciences 22, no. 10: 5369. https://doi.org/10.3390/ijms22105369
APA StylePirro, M., Mohammed, Y., de Ru, A. H., Janssen, G. M. C., Tjokrodirijo, R. T. N., Madunić, K., Wuhrer, M., van Veelen, P. A., & Hensbergen, P. J. (2021). Oxonium Ion Guided Analysis of Quantitative Proteomics Data Reveals Site-Specific O-Glycosylation of Anterior Gradient Protein 2 (AGR2). International Journal of Molecular Sciences, 22(10), 5369. https://doi.org/10.3390/ijms22105369






