Tau Exon 10 Inclusion by PrPC through Downregulating GSK3β Activity
Abstract
1. Introduction
2. Results
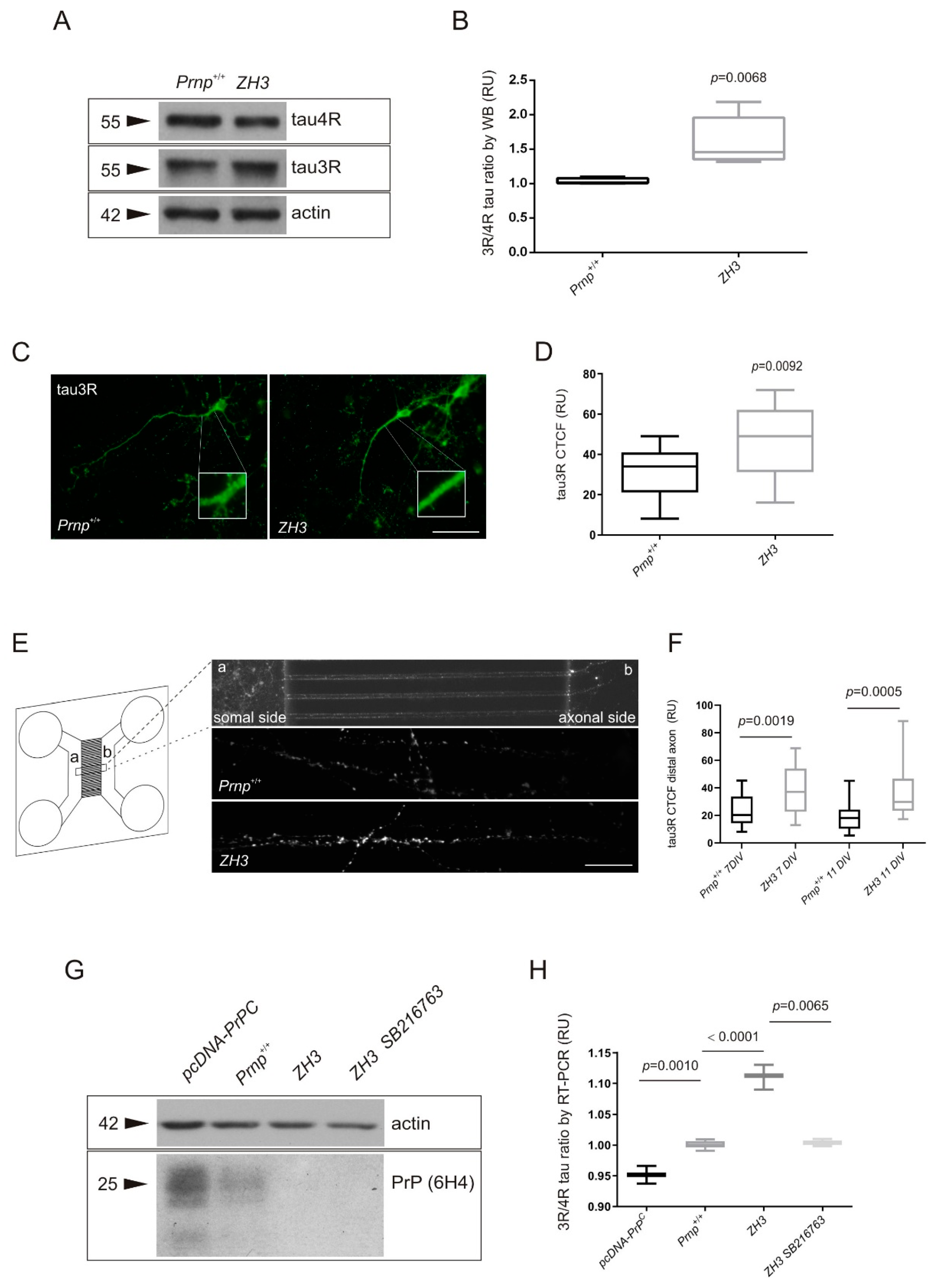
2.1. Increased 3R/4R Tau Ratio in Mice Lacking PrPC
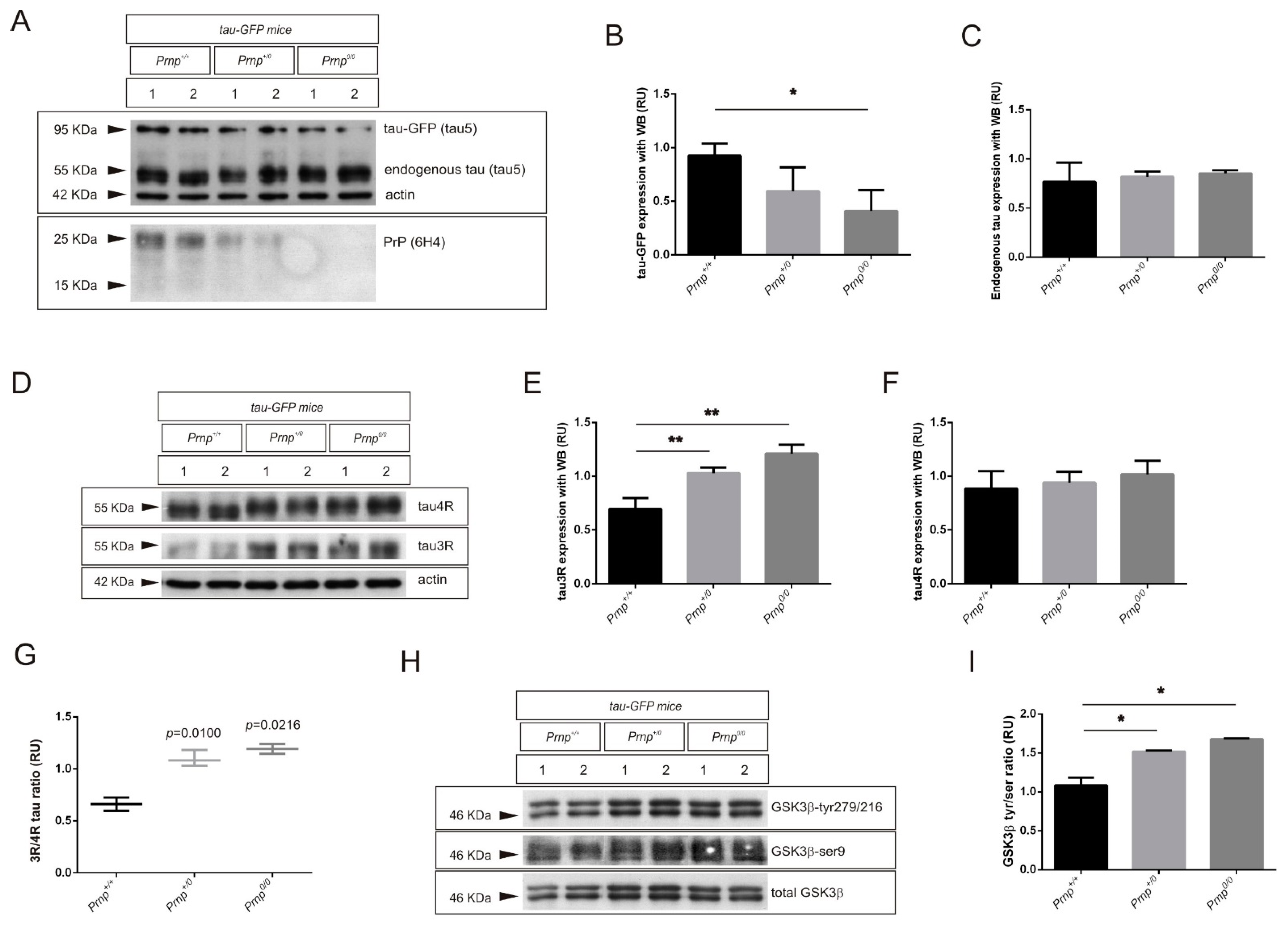
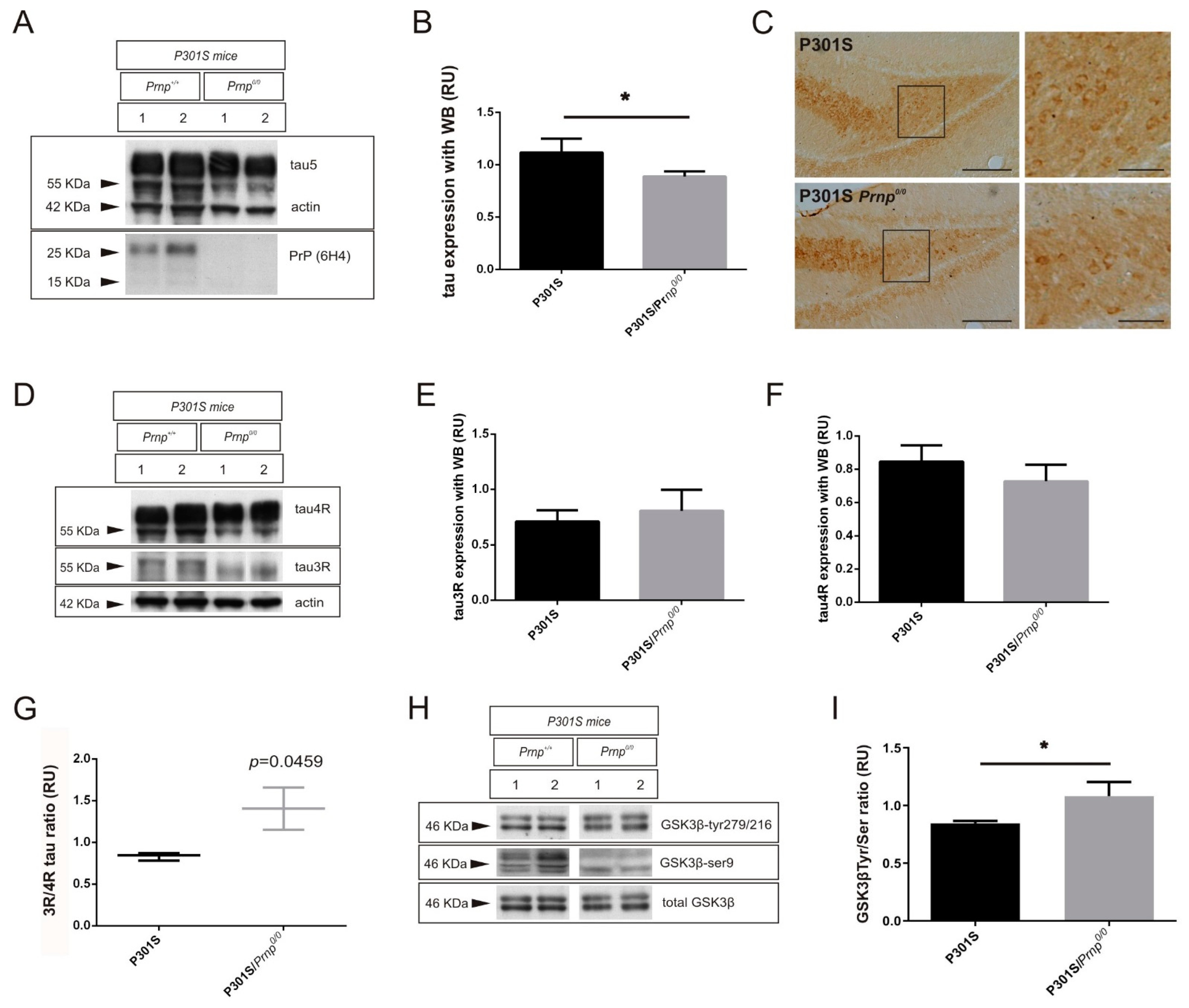
2.2. PrPC Ablation Modifies the 3R/4R Tau Ratio in Mouse Models of Tau Overexpression
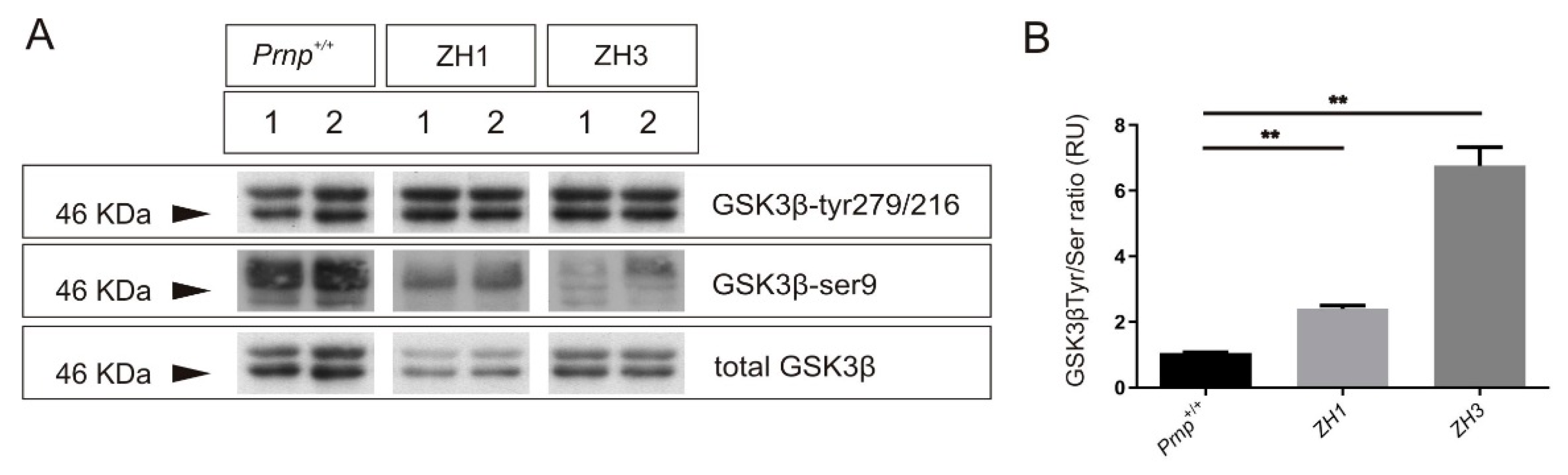
2.3. Tau Exon 10 Splicing Is Dependent on PrPC Dosage in Correlation with GSK3β
2.4. Impact of PrPC Levels on Tau Splicing in AD Brain
3. Discussion
3.1. Increase in the 3R/4R Tau Ratio Paralleled to GSK3β Activation in Mouse Models
3.2. GSK3β Activity, Correlative with PrPC Levels, Is Not Mandatory for 3R/4R Tau Ratio in AD
4. Materials and Methods
4.1. Human Hippocampal Samples
4.2. Mouse Strains and Genotyping
4.3. Primary Embryonic Cortical Cultures and Transfection
4.4. Microfluidic Devices
4.5. Western Blot Analysis
4.6. Immunohistochemical Procedures
4.7. RT-qPCR
4.8. Antibodies and Reagents
4.9. Statistical Processing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Aβ | β-amyloid |
| AD | Alzheimer’s disease |
| ADDLs | Aβ-derived diffusible ligands |
| CBD | Corticobasal degeneration |
| CNS | Central nervous system |
| CTCF | Corrected total cell fluorescence |
| DIV | Days in vitro |
| FTDP-17 | Frontotemporal dementia and parkinsonism linked to chromosome 17 |
| GFP | Green fluorescence protein |
| GR | Glutathione reductase |
| GSK3β | Glycogen Synthase Kinase 3-β |
| GSS | Gerstmann-Sträussler-Scheinker syndrome |
| iPSCs | Induced pluripotent stem cells |
| LTP | Long term potentiation |
| MAPT | Microtubule-associated protein tau |
| MT | Microtubules |
| NFT | Neurofibrillary tangles |
| PHF, | Paired helical filaments |
| PrPC | Cellular prion protein |
| PrPSC | Scrapie isoform of PrPC |
| PD | Pick’s disease |
| PSP | Progressive supranuclear palsy |
| SOD | Superoxide dismutase |
| STI-1 | Stress-inducible protein 1 |
| TSEs | Transmissible spongiform encephalopathies |
| WT | Wild type |
References
- Avila, J. Tau aggregation into fibrillar polymers: Tauopathies. FEBS Lett. 2000, 476, 89–92. [Google Scholar] [CrossRef]
- Kovacs, G.G. Invited review: Neuropathology of tauopathies: Principles and practice. Neuropathol. Appl. Neurobiol. 2015, 41, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.M.; Goedert, M.; Trojanowski, J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001, 24, 1121–1159. [Google Scholar] [CrossRef] [PubMed]
- Buee, L.; Delacourte, A. Comparative biochemistry of tau in progressive supranuclear palsy, corticobasal degeneration, FTDP-17 and Pick’s disease. Brain Pathol. 1999, 9, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.; Kalaria, R. Dementia. In Greenfield’s Neuropathology, 9th ed.; Love, S., Budka, H., Ironside, J., Perry, A., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 858–973. [Google Scholar]
- Selkoe, D.J. Alzheimer’s disease: Genes, proteins, and therapy. Physiol. Rev. 2001, 81, 741–766. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J. The relationship between amyloid and tau. J. Mol. Neurosci. 2003, 20, 203–206. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef]
- Iqbal, K.; Liu, F.; Gong, C.X.; Grundke-Iqbal, I. Tau in Alzheimer disease and related tauopathies. Curr. Alzheimer Res. 2010, 7, 656–664. [Google Scholar] [CrossRef]
- Pigino, G.; Morfini, G.; Atagi, Y.; Deshpande, A.; Yu, C.; Jungbauer, L.; LaDu, M.; Busciglio, J.; Brady, S. Disruption of fast axonal transport is a pathogenic mechanism for intraneuronal amyloid beta. Proc. Natl. Acad. Sci. USA 2009, 106, 5907–5912. [Google Scholar] [CrossRef]
- Lu, M.; Kosik, K.S. Competition for microtubule-binding with dual expression of tau missense and splice isoforms. Mol. Biol. Cell 2001, 12, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Sennvik, K.; Boekhoorn, K.; Lasrado, R.; Terwel, D.; Verhaeghe, S.; Korr, H.; Schmitz, C.; Tomiyama, T.; Mori, H.; Krugers, H.; et al. Tau-4R suppresses proliferation and promotes neuronal differentiation in the hippocampus of tau knockin/knockout mice. FASEB J. 2007, 21, 2149–2161. [Google Scholar] [CrossRef] [PubMed]
- Avila, J.; Lucas, J.J.; Perez, M.; Hernandez, F. Role of tau protein in both physiological and pathological conditions. Physiol. Rev. 2004, 84, 361–384. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Lewis, J.; McGowan, E.; Rockwood, J.; Melrose, H.; Nacharaju, P.; van Slegtenhorst, M.; Gwinn-Hardy, K.; Paul Murphy, M.; Baker, M.; Yu, X.; et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 2000, 25, 402–405. [Google Scholar] [CrossRef]
- Liu, F.; Gong, C.X. Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 2008, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.W.; Kouri, N.; Murray, M.E.; Josephs, K.A. Neuropathology of frontotemporal lobar degeneration-tau (FTLD-tau). J. Mol. Neurosci. 2011, 45, 384–389. [Google Scholar] [CrossRef]
- Park, S.A.; Ahn, S.I.; Gallo, J.M. Tau mis-splicing in the pathogenesis of neurodegenerative disorders. BMB Rep. 2016, 49, 405–413. [Google Scholar] [CrossRef]
- Hanger, D.P.; Byers, H.L.; Wray, S.; Leung, K.Y.; Saxton, M.J.; Seereeram, A.; Reynolds, C.H.; Ward, M.A.; Anderton, B.H. Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J. Biol. Chem. 2007, 282, 23645–23654. [Google Scholar] [CrossRef]
- Chen, K.L.; Yuan, R.Y.; Hu, C.J.; Hsu, C.Y. Amyloid-beta peptide alteration of tau exon-10 splicing via the GSK3beta-SC35 pathway. Neurobiol. Dis. 2010, 40, 378–385. [Google Scholar] [CrossRef]
- Moleres, F.J.; Velayos, J.L. Expression of PrP(C) in the rat brain and characterization of a subset of cortical neurons. Brain Res. 2005, 1056, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Ford, M.J.; Burton, L.J.; Morris, R.J.; Hall, S.M. Selective expression of prion protein in peripheral tissues of the adult mouse. Neuroscience 2002, 113, 177–192. [Google Scholar] [CrossRef]
- Moser, M.; Colello, R.J.; Pott, U.; Oesch, B. Developmental expression of the prion protein gene in glial cells. Neuron 1995, 14, 509–517. [Google Scholar] [CrossRef]
- Prusiner, S.B. Novel proteinaceous infectious particles cause scrapie. Science 1982, 216, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, L.B.; Freitas, A.R.; Zanata, S.M.; Brentani, R.R.; Martins, V.R.; Linden, R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002, 21, 3317–3326. [Google Scholar] [CrossRef]
- Zanata, S.M.; Lopes, M.H.; Mercadante, A.F.; Hajj, G.N.; Chiarini, L.B.; Nomizo, R.; Freitas, A.R.; Cabral, A.L.; Lee, K.S.; Juliano, M.A.; et al. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J. 2002, 21, 3307–3316. [Google Scholar] [CrossRef]
- Rangel, A.; Burgaya, F.; Gavin, R.; Soriano, E.; Aguzzi, A.; Del Rio, J.A. Enhanced susceptibility of Prnp-deficient mice to kainate-induced seizures, neuronal apoptosis, and death: Role of AMPA/kainate receptors. J. Neurosci. Res. 2007, 85, 2741–2755. [Google Scholar] [CrossRef]
- Brown, D.R.; Nicholas, R.S.; Canevari, L. Lack of prion protein expression results in a neuronal phenotype sensitive to stress. J. Neurosci. Res. 2002, 67, 211–224. [Google Scholar] [CrossRef]
- Sakudo, A.; Lee, D.C.; Saeki, K.; Nakamura, Y.; Inoue, K.; Matsumoto, Y.; Itohara, S.; Onodera, T. Impairment of superoxide dismutase activation by N-terminally truncated prion protein (PrP) in PrP-deficient neuronal cell line. Biochem. Biophys. Res. Commun. 2003, 308, 660–667. [Google Scholar] [CrossRef]
- White, A.R.; Collins, S.J.; Maher, F.; Jobling, M.F.; Stewart, L.R.; Thyer, J.M.; Beyreuther, K.; Masters, C.L.; Cappai, R. Prion protein-deficient neurons reveal lower glutathione reductase activity and increased susceptibility to hydrogen peroxide toxicity. Am. J. Pathol. 1999, 155, 1723–1730. [Google Scholar] [CrossRef]
- Lee, Y.J.; Baskakov, I.V. The cellular form of the prion protein is involved in controlling cell cycle dynamics, self-renewal, and the fate of human embryonic stem cell differentiation. J. Neurochem. 2013, 124, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Steele, A.D.; Emsley, J.G.; Ozdinler, P.H.; Lindquist, S.; Macklis, J.D. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 3416–3421. [Google Scholar] [CrossRef]
- Loubet, D.; Dakowski, C.; Pietri, M.; Pradines, E.; Bernard, S.; Callebert, J.; Ardila-Osorio, H.; Mouillet-Richard, S.; Launay, J.M.; Kellermann, O.; et al. Neuritogenesis: The prion protein controls beta1 integrin signaling activity. FASEB J. 2012, 26, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhao, M.; Ye, W.; Huang, J.; Chu, J.; Yan, S.; Wang, C.; Zeng, R. Inhibition of glycogen synthase kinase-3 (GSK3) promotes the neural differentiation of full-term amniotic fluid-derived stem cells towards neural progenitor cells. Tissue Cell 2016, 48, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Rapp, J.; Martin-Lanneree, S.; Hirsch, T.Z.; Pradines, E.; Alleaume-Butaux, A.; Schneider, B.; Baudry, A.; Launay, J.M.; Mouillet-Richard, S. A PrP(C)-caveolin-Lyn complex negatively controls neuronal GSK3beta and serotonin 1B receptor. Sci. Rep. 2014, 4, 4881. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, I.J.; Miners, J.S.; Glennon, E.B.; Kehoe, P.G.; Love, S.; Kellett, K.A.; Hooper, N.M. Prion protein is decreased in Alzheimer’s brain and inversely correlates with BACE1 activity, amyloid-beta levels and Braak stage. PLoS ONE 2013, 8, e59554. [Google Scholar] [CrossRef]
- Lauren, J.; Gimbel, D.A.; Nygaard, H.B.; Gilbert, J.W.; Strittmatter, S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature 2009, 457, 1128–1132. [Google Scholar] [CrossRef]
- Younan, N.D.; Chen, K.F.; Rose, R.S.; Crowther, D.C.; Viles, J.H. Prion protein stabilizes amyloid-beta (Abeta) oligomers and enhances Abeta neurotoxicity in a Drosophila model of Alzheimer’s disease. J. Biol. Chem. 2018, 293, 13090–13099. [Google Scholar] [CrossRef]
- Calella, A.M.; Farinelli, M.; Nuvolone, M.; Mirante, O.; Moos, R.; Falsig, J.; Mansuy, I.M.; Aguzzi, A. Prion protein and Abeta-related synaptic toxicity impairment. EMBO Mol. Med. 2010, 2, 306–314. [Google Scholar] [CrossRef]
- Lidon, L.; Vergara, C.; Ferrer, I.; Hernandez, F.; Avila, J.; del Rio, J.A.; Gavin, R. Tau Protein as a New Regulator of Cellular Prion Protein Transcription. Mol. Neurobiol. 2020, 57, 4170–4186. [Google Scholar] [CrossRef]
- Vergara, C.; Ordonez-Gutierrez, L.; Wandosell, F.; Ferrer, I.; del Rio, J.A.; Gavin, R. Role of PrP(C) Expression in Tau Protein Levels and Phosphorylation in Alzheimer’s Disease Evolution. Mol. Neurobiol. 2015, 51, 1206–1220. [Google Scholar] [CrossRef]
- Hernandez, F.; Perez, M.; Lucas, J.J.; Mata, A.M.; Bhat, R.; Avila, J. Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35. Implications for Alzheimer’s disease. J. Biol. Chem. 2004, 279, 3801–3806. [Google Scholar] [CrossRef]
- Matamoros-Angles, A.; Gayosso, L.M.; Richaud-Patin, Y.; di Domenico, A.; Vergara, C.; Hervera, A.; Sousa, A.; Fernandez-Borges, N.; Consiglio, A.; Gavin, R.; et al. iPS Cell Cultures from a Gerstmann-Straussler-Scheinker Patient with the Y218N PRNP Mutation Recapitulate tau Pathology. Mol. Neurobiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Boutajangout, A.; Boom, A.; Leroy, K.; Brion, J.P. Expression of tau mRNA and soluble tau isoforms in affected and non-affected brain areas in Alzheimer’s disease. FEBS Lett. 2004, 576, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Chambers, C.B.; Lee, J.M.; Troncoso, J.C.; Reich, S.; Muma, N.A. Overexpression of four-repeat tau mRNA isoforms in progressive supranuclear palsy but not in Alzheimer’s disease. Ann. Neurol. 1999, 46, 325–332. [Google Scholar] [CrossRef]
- Schmitz, M.; Wulf, K.; Signore, S.C.; Schulz-Schaeffer, W.J.; Kermer, P.; Bahr, M.; Wouters, F.S.; Zafar, S.; Zerr, I. Impact of the Cellular Prion Protein on Amyloid-beta and 3PO-Tau Processing. J. Alzheimers Dis. 2014, 38, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.; Bossers, K.; Janky, R.; Salta, E.; Frigerio, C.S.; Barbash, S.; Rothman, R.; Sierksma, A.S.; Thathiah, A.; Greenberg, D.; et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 2013, 5, 1613–1634. [Google Scholar] [CrossRef]
- Pichler, S.; Gu, W.; Hartl, D.; Gasparoni, G.; Leidinger, P.; Keller, A.; Meese, E.; Mayhaus, M.; Hampel, H.; Riemenschneider, M. The miRNome of Alzheimer’s disease: Consistent downregulation of the miR-132/212 cluster. Neurobiol. Aging 2017, 50, 167.e1–167.e10. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Potier, M.C.; Ulrich, J.; Crowther, R.A. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: Differential expression of tau protein mRNAs in human brain. EMBO J. 1989, 8, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Hefti, M.M.; Farrell, K.; Kim, S.; Bowles, K.R.; Fowkes, M.E.; Raj, T.; Crary, J.F. High-resolution temporal and regional mapping of MAPT expression and splicing in human brain development. PLoS ONE 2018, 13, e0195771. [Google Scholar] [CrossRef] [PubMed]
- Sposito, T.; Preza, E.; Mahoney, C.J.; Seto-Salvia, N.; Ryan, N.S.; Morris, H.R.; Arber, C.; Devine, M.J.; Houlden, H.; Warner, T.T.; et al. Developmental regulation of tau splicing is disrupted in stem cell-derived neurons from frontotemporal dementia patients with the 10 + 16 splice-site mutation in MAPT. Hum. Mol. Genet. 2015, 24, 5260–5269. [Google Scholar] [CrossRef] [PubMed]
- Niblock, M.; Gallo, J.M. Tau alternative splicing in familial and sporadic tauopathies. Biochem. Soc. Trans. 2012, 40, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Linden, R.; Martins, V.R.; Prado, M.A.; Cammarota, M.; Izquierdo, I.; Brentani, R.R. Physiology of the prion protein. Physiol. Rev. 2008, 88, 673–728. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Yang, Y.; Wang, T.; Kouadir, M.; Zhao, D.; Hu, S. Cellular Prion Protein Promotes Neuronal Differentiation of Adipose-Derived Stem Cells by Upregulating miRNA-124. J. Mol. Neurosci. 2016, 59, 48–55. [Google Scholar] [CrossRef]
- Pantera, B.; Bini, C.; Cirri, P.; Paoli, P.; Camici, G.; Manao, G.; Caselli, A. PrPc activation induces neurite outgrowth and differentiation in PC12 cells: Role for caveolin-1 in the signal transduction pathway. J. Neurochem. 2009, 110, 194–207. [Google Scholar] [CrossRef]
- Hooper, C.; Killick, R.; Lovestone, S. The GSK3 hypothesis of Alzheimer’s disease. J. Neurochem. 2008, 104, 1433–1439. [Google Scholar] [CrossRef]
- Griebel, G.; Stemmelin, J.; Lopez-Grancha, M.; Boulay, D.; Boquet, G.; Slowinski, F.; Pichat, P.; Beeske, S.; Tanaka, S.; Mori, A.; et al. The selective GSK3 inhibitor, SAR502250, displays neuroprotective activity and attenuates behavioral impairments in models of neuropsychiatric symptoms of Alzheimer’s disease in rodents. Sci. Rep. 2019, 9, 18045. [Google Scholar] [CrossRef]
- Alvarez, G.; Munoz-Montano, J.R.; Satrustegui, J.; Avila, J.; Bogonez, E.; Diaz-Nido, J. Lithium protects cultured neurons against beta-amyloid-induced neurodegeneration. FEBS Lett. 1999, 453, 260–264. [Google Scholar] [CrossRef]
- Koehler, D.; Shah, Z.A.; Williams, F.E. The GSK3beta inhibitor, TDZD-8, rescues cognition in a zebrafish model of okadaic acid-induced Alzheimer’s disease. Neurochem. Int. 2019, 122, 31–37. [Google Scholar] [CrossRef]
- Llorens-Martin, M.; Jurado, J.; Hernandez, F.; Avila, J. GSK-3beta, a pivotal kinase in Alzheimer disease. Front. Mol. Neurosci. 2014, 7, 46. [Google Scholar] [CrossRef]
- Saraswati, A.P.; Ali Hussaini, S.M.; Krishna, N.H.; Babu, B.N.; Kamal, A. Glycogen synthase kinase-3 and its inhibitors: Potential target for various therapeutic conditions. Eur. J. Med. Chem. 2018, 144, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Bradley, C.A.; Peineau, S.; Taghibiglou, C.; Nicolas, C.S.; Whitcomb, D.J.; Bortolotto, Z.A.; Kaang, B.K.; Cho, K.; Wang, Y.T.; Collingridge, G.L. A pivotal role of GSK-3 in synaptic plasticity. Front. Mol. Neurosci. 2012, 5, 13. [Google Scholar] [CrossRef]
- Ostapchenko, V.G.; Beraldo, F.H.; Mohammad, A.H.; Xie, Y.F.; Hirata, P.H.; Magalhaes, A.C.; Lamour, G.; Li, H.; Maciejewski, A.; Belrose, J.C.; et al. The prion protein ligand, stress-inducible phosphoprotein 1, regulates amyloid-beta oligomer toxicity. J. Neurosci. 2013, 33, 16552–16564. [Google Scholar] [CrossRef] [PubMed]
- Kosik, K.S.; Orecchio, L.D.; Bakalis, S.; Neve, R.L. Developmentally regulated expression of specific tau sequences. Neuron 1989, 2, 1389–1397. [Google Scholar] [CrossRef]
- McMillan, P.; Korvatska, E.; Poorkaj, P.; Evstafjeva, Z.; Robinson, L.; Greenup, L.; Leverenz, J.; Schellenberg, G.D.; D’Souza, I. Tau isoform regulation is region- and cell-specific in mouse brain. J. Comp. Neurol. 2008, 511, 788–803. [Google Scholar] [CrossRef]
- Nuvolone, M.; Hermann, M.; Sorce, S.; Russo, G.; Tiberi, C.; Schwarz, P.; Minikel, E.; Sanoudou, D.; Pelczar, P.; Aguzzi, A. Strictly co-isogenic C57BL/6J-Prnp−/− mice: A rigorous resource for prion science. J. Exp. Med. 2016, 213, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Gustke, N.; Trinczek, B.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. Domains of tau protein and interactions with microtubules. Biochemistry 1994, 33, 9511–9522. [Google Scholar] [CrossRef]
- Butner, K.A.; Kirschner, M.W. Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 1991, 115, 717–730. [Google Scholar] [CrossRef]
- Tarhan, M.C.; Orazov, Y.; Yokokawa, R.; Karsten, S.L.; Fujita, H. Biosensing MAPs as “roadblocks”: Kinesin-based functional analysis of tau protein isoforms and mutants using suspended microtubules (sMTs). Lab Chip 2013, 13, 3217–3224. [Google Scholar] [CrossRef]
- Vershinin, M.; Xu, J.; Razafsky, D.S.; King, S.J.; Gross, S.P. Tuning microtubule-based transport through filamentous MAPs: The problem of dynein. Traffic 2008, 9, 882–892. [Google Scholar] [CrossRef]
- Urrea, L.; Segura-Feliu, M.; Masuda-Suzukake, M.; Hervera, A.; Pedraz, L.; Aznar, J.M.; Vila, M.; Samitier, J.; Torrents, E.; Ferrer, I.; et al. Involvement of Cellular Prion Protein in alpha-Synuclein Transport in Neurons. Mol. Neurobiol. 2017, 55, 1847–1860. [Google Scholar] [CrossRef] [PubMed]
- Aulic, S.; Masperone, L.; Narkiewicz, J.; Isopi, E.; Bistaffa, E.; Ambrosetti, E.; Pastore, B.; de Cecco, E.; Scaini, D.; Zago, P.; et al. alpha-Synuclein Amyloids Hijack Prion Protein to Gain Cell Entry, Facilitate Cell-to-Cell Spreading and Block Prion Replication. Sci. Rep. 2017, 7, 10050. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Greis, C.; Ottis, P.; Silva, C.J.; Schulz-Schaeffer, W.J.; Wrede, A.; Koppe, K.; Onisko, B.; Requena, J.R.; Govindarajan, N.; et al. Loss of prion protein leads to age-dependent behavioral abnormalities and changes in cytoskeletal protein expression. Mol. Neurobiol. 2014, 50, 923–936. [Google Scholar] [CrossRef]
- Tuzi, N.L.; Clarke, A.R.; Bradford, B.; Aitchison, L.; Thomson, V.; Manson, J.C. Cre-loxP mediated control of PrP to study transmissible spongiform encephalopathy diseases. Genesis 2004, 40, 1–6. [Google Scholar] [CrossRef]
- Llorens, F.; Ferrer, I.; Del Rio, J.A. Gene Expression Resulting from PrP Ablation and PrP Overexpression in Murine and Cellular Models. Mol. Neurobiol. 2014, 49, 413–423. [Google Scholar] [CrossRef]
- Simon, D.; Herva, M.E.; Benitez, M.J.; Garrido, J.J.; Rojo, A.I.; Cuadrado, A.; Torres, J.M.; Wandosell, F. Dysfunction of the PI3K-Akt-GSK-3 pathway is a common feature in cell culture and in vivo models of prion disease. Neuropathol. Appl. Neurobiol. 2014, 40, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Gavin, R.; Braun, N.; Nicolas, O.; Parra, B.; Urena, J.M.; Mingorance, A.; Soriano, E.; Torres, J.M.; Aguzzi, A.; del Rio, J.A. PrP(106-126) activates neuronal intracellular kinases and Egr1 synthesis through activation of NADPH-oxidase independently of PrPc. FEBS Lett. 2005, 579, 4099–4106. [Google Scholar] [CrossRef]
- Perez, M.; Rojo, A.I.; Wandosell, F.; Diaz-Nido, J.; Avila, J. Prion peptide induces neuronal cell death through a pathway involving glycogen synthase kinase 3. Biochem. J. 2003, 372, 129–136. [Google Scholar] [CrossRef]
- Bautista, M.J.; Gutierrez, J.; Salguero, F.J.; Fernandez de Marco, M.M.; Romero-Trevejo, J.L.; Gomez-Villamandos, J.C. BSE infection in bovine PrP transgenic mice leads to hyperphosphorylation of tau-protein. Vet. Microbiol. 2006, 115, 293–301. [Google Scholar] [CrossRef]
- Asuni, A.A.; Perry, V.H.; O’Connor, V. Change in tau phosphorylation associated with neurodegeneration in the ME7 model of prion disease. Biochem. Soc. Trans. 2010, 38, 545–551. [Google Scholar] [CrossRef][Green Version]
- Kapaki, E.; Kilidireas, K.; Paraskevas, G.P.; Michalopoulou, M.; Patsouris, E. Highly increased CSF tau protein and decreased beta-amyloid (1-42) in sporadic CJD: A discrimination from Alzheimer’s disease? J. Neurol. Neurosurg. Psychiatry 2001, 71, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Sarac, H.; Hajnsek, S.; Basic, S.; Henigsberg, N.; Rados, M.; Simic, G. Magnetic resonance spectroscopy and measurement of tau epitopes of autopsy proven sporadic Creutzfeldt-Jakob disease in a patient with non-specific initial EEG, MRI and negative 14-3-3 immunoblot. Coll. Antropol. 2008, 32 (Suppl. 1), 199–204. [Google Scholar]
- Ishizawa, K.; Komori, T.; Shimazu, T.; Yamamoto, T.; Kitamoto, T.; Shimazu, K.; Hirose, T. Hyperphosphorylated tau deposition parallels prion protein burden in a case of Gerstmann-Straussler-Scheinker syndrome P102L mutation complicated with dementia. Acta Neuropathol. 2002, 104, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.; Shiratsuchi, A.; Sato, S.; Omori, A.; Arioka, M.; Kobayashi, S.; Uchida, T.; Imahori, K. Glycogen synthase kinase 3 beta is identical to tau protein kinase I generating several epitopes of paired helical filaments. FEBS Lett. 1993, 325, 167–172. [Google Scholar] [CrossRef]
- Hernandez, F.; de Barreda, E.G.; Fuster-Matanzo, A.; Lucas, J.J.; Avila, J. GSK3: A possible link between beta amyloid peptide and tau protein. Exp. Neurol. 2010, 223, 322–325. [Google Scholar] [CrossRef]
- Leroy, K.; Yilmaz, Z.; Brion, J.P. Increased level of active GSK-3beta in Alzheimer’s disease and accumulation in argyrophilic grains and in neurones at different stages of neurofibrillary degeneration. Neuropathol. Appl. Neurobiol. 2007, 33, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Blalock, E.M.; Geddes, J.W.; Chen, K.C.; Porter, N.M.; Markesbery, W.R.; Landfield, P.W. Incipient Alzheimer’s disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. USA 2004, 101, 2173–2178. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 2004, 256, 183–194. [Google Scholar] [CrossRef]
- Hane, F.T.; Robinson, M.; Lee, B.Y.; Bai, O.; Leonenko, Z.; Albert, M.S. Recent Progress in Alzheimer’s Disease Research, Part 3: Diagnosis and Treatment. J. Alzheimer’s Dis. 2017, 57, 645–665. [Google Scholar] [CrossRef]
- Irwin, D.J. Tauopathies as clinicopathological entities. Parkinsonism Relat. Disord. 2016, 22 (Suppl. 1), S29–S33. [Google Scholar] [CrossRef]
- Hara, M.; Hirokawa, K.; Kamei, S.; Uchihara, T. Isoform transition from four-repeat to three-repeat tau underlies dendrosomatic and regional progression of neurofibrillary pathology. Acta Neuropathol. 2013, 125, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.S.; Memmott, J.; Lafyatis, R.; Stamm, S.; Screaton, G.; Andreadis, A. Complex regulation of tau exon 10, whose missplicing causes frontotemporal dementia. J. Neurochem. 2000, 74, 490–500. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Q.S.; Wang, Y.; Lafyatis, R.; Stamm, S.; Andreadis, A. Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. J. Neurochem. 2004, 88, 1078–1090. [Google Scholar] [CrossRef] [PubMed]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.Y.; Delay, C.; Girard, J.; Papon, M.A.; Planel, E.; Sergeant, N.; Buee, L.; Hebert, S.S. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet. 2011, 20, 4016–4024. [Google Scholar] [CrossRef]
- Braak, H.; Braak, E.; Bohl, J.; Bratzke, H. Evolution of Alzheimer’s disease related cortical lesions. J. Neural Transm. Suppl. 1998, 54, 97–106. [Google Scholar]
- Braak, H.; Braak, E. Evolution of the neuropathology of Alzheimer’s disease. Acta Neurol Scand. Suppl. 1996, 165, 3–12. [Google Scholar] [CrossRef]
- Bueler, H.; Fischer, M.; Lang, Y.; Bluethmann, H.; Lipp, H.P.; DeArmond, S.J.; Prusiner, S.B.; Aguet, M.; Weissmann, C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 1992, 356, 577–582. [Google Scholar] [CrossRef]
- Pratt, T.; Sharp, L.; Nichols, J.; Price, D.J.; Mason, J.O. Embryonic stem cells and transgenic mice ubiquitously expressing a tau-tagged green fluorescent protein. Dev. Biol. 2000, 228, 19–28. [Google Scholar] [CrossRef]
- Yoshiyama, Y.; Higuchi, M.; Zhang, B.; Huang, S.M.; Iwata, N.; Saido, T.C.; Maeda, J.; Suhara, T.; Trojanowski, J.Q.; Lee, V.M. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 2007, 53, 337–351. [Google Scholar] [CrossRef]
- Taylor, A.M.; Blurton-Jones, M.; Rhee, S.W.; Cribbs, D.H.; Cotman, C.W.; Jeon, N.L. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2005, 2, 599–605. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Bribian, A.; Fontana, X.; Llorens, F.; Gavin, R.; Reina, M.; Garcia-Verdugo, J.M.; Torres, J.M.; de Castro, F.; del Rio, J.A. Role of the cellular prion protein in oligodendrocyte precursor cell proliferation and differentiation in the developing and adult mouse CNS. PLoS ONE 2012, 7, e33872. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Carulla, P.; Bribian, A.; Rangel, A.; Gavin, R.; Ferrer, I.; Caelles, C.; Del Rio, J.A.; Llorens, F. Neuroprotective role of PrPC against kainate-induced epileptic seizures and cell death depends on the modulation of JNK3 activation by GluR6/7-PSD-95 binding. Mol. Biol. Cell 2011, 22, 3041–3054. [Google Scholar] [CrossRef]


| Case Number | Braak and Braak Stage | Gender | Age | Post-Mortem Delay | Analysis |
|---|---|---|---|---|---|
| nAD1 | - | M | 39 | 9 h 15 min | WB |
| nAD2 | - | F | 46 | 14 h 15 min | WB |
| nAD3 | - | M | 53 | 7 h 25 min | WB |
| nAD4 | - | M | 46 | 15 h | WB |
| nAD5 | - | M | 43 | 4 h 35 min | WB/RT-PCR |
| nAD6 | - | M | 52 | 3 h | WB |
| nAD7 | - | M | 51 | 3 h 30 min | WB |
| nAD8 | - | F | 86 | 4 h | WB |
| nAD9 | - | F | 46 | 9 h 35 min | RT-PCR |
| nAD10 | - | M | 70 | 13 h | RT-PCR |
| nAD11 | - | F | 82 | 11 h | RT-PCR |
| nAD12 | - | F | 46 | 20 h | RT-PCR |
| nAD13 | - | M | 61 | 2 h 45 min | RT-PCR |
| AD1 | I | M | 61 | 3 h 40 min | WB |
| AD2 | I | M | 53 | 6 h 15 min | WB |
| AD3 | I | M | 74 | 4 h | WB |
| AD4 | I | M | 71 | 11 h 30 min | WB |
| AD5 | I | M | 64 | 2 h 15 min | WB |
| AD6 | I | F | 79 | 3 h 35 min | WB/RT-PCR |
| AD7 | I | M | 65 | 5 h 15 min | WB |
| AD8 | I | F | 75 | 4 h 55 min | WB |
| AD9 | I | M | 63 | 6 h | WB |
| AD10 | I | M | 68 | 10 h 55 min | WB |
| AD11 | I | M | 64 | 8 h 35 min | RT-PCR |
| AD12 | I | M | 61 | 5 h 35 min | RT-PCR |
| AD13 | I | M | 67 | 14 h 40 min | RT-PCR |
| AD14 | I | F | 73 | 15 h 45 min | RT-PCR |
| AD15 | I | M | 70 | 5 h | RT-PCR |
| AD16 | II | M | 65 | 5 h | RT-PCR |
| AD17 | II | F | 77 | 11 h | WB |
| AD18 | II | M | 65 | 5 h | WB |
| AD19 | II | M | 66 | 4 h 55 min | WB |
| AD20 | II | M | 72 | 8 h 45 min | WB |
| AD21 | II | M | 71 | 5 h 15 min | WB |
| AD22 | II | M | 66 | 5 h | WB |
| AD23 | II | F | 60 | 9 h 40 min | WB |
| AD24 | II | F | 80 | 3 h 30 min | WB |
| AD25 | II | F | 75 | 4 h 55 min | RT-PCR |
| AD26 | II | F | 86 | 4 h 15 min | RT-PCR |
| AD27 | II | M | 55 | 9 h 45 min | RT-PCR |
| AD28 | II | F | 57 | 4 h 30 min | RT-PCR |
| AD29 | II | M | 69 | 3 h 45 min | WB |
| AD30 | II | M | 74 | 5 h 30 min | WB/RT-PCR |
| AD31 | II | M | 86 | 5 h 35 min | WB |
| AD32 | III | F | 81 | 1 h 30 min | WB |
| AD33 | III | F | 71 | 7 h 15 min | WB/RT-PCR |
| AD34 | III | F | 77 | 11 h 30 min | WB/RT-PCR |
| AD35 | III | F | 67 | 6 h 10 min | WB |
| AD36 | III | M | 69 | 13 h 10 min | WB |
| AD37 | III | F | 83 | 2 h 30 min | RT-PCR |
| AD38 | III | M | 87 | 3 h 30 min | RT-PCR |
| AD39 | III | F | 82 | 4 h 50 min | RT-PCR |
| AD40 | III | M | 64 | 6 h | WB/RT-PCR |
| AD41 | IV | F | 80 | 2 h 45 min | WB |
| AD42 | IV | F | 81 | 12 h | WB |
| AD43 | IV | M | 84 | 12 h 45 min | WB |
| AD44 | IV | M | 79 | 50 min | WB |
| AD45 | IV | M | 83 | 7 h 25 min | WB/RT-PCR |
| AD46 | IV | F | 90 | 10 h | WB/RT-PCR |
| AD47 | IV | F | 81 | 5 h | RT-PCR |
| AD48 | V | M | 87 | 7 h 5 min | WB/RT-PCR |
| AD49 | V | M | 75 | 11 h 30 min | WB/RT-PCR |
| AD50 | V | M | 82 | 3 h 45 min | WB/RT-PCR |
| AD51 | V | M | 77 | 16 h | WB |
| AD52 | V | F | 82 | 1 h 45 min | WB/RT-PCR |
| AD53 | V | F | 75 | 4 h 15 min | WB/RT-PCR |
| AD54 | V | M | 93 | 3 h | RT-PCR |
| AD55 | VI | M | 86 | 20 h 35 min | WB |
| AD56 | VI | M | 67 | 8 h | WB/RT-PCR |
| AD57 | VI | F | 56 | 7 h | WB/RT-PCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lidón, L.; Llaó-Hierro, L.; Nuvolone, M.; Aguzzi, A.; Ávila, J.; Ferrer, I.; del Río, J.A.; Gavín, R. Tau Exon 10 Inclusion by PrPC through Downregulating GSK3β Activity. Int. J. Mol. Sci. 2021, 22, 5370. https://doi.org/10.3390/ijms22105370
Lidón L, Llaó-Hierro L, Nuvolone M, Aguzzi A, Ávila J, Ferrer I, del Río JA, Gavín R. Tau Exon 10 Inclusion by PrPC through Downregulating GSK3β Activity. International Journal of Molecular Sciences. 2021; 22(10):5370. https://doi.org/10.3390/ijms22105370
Chicago/Turabian StyleLidón, Laia, Laura Llaó-Hierro, Mario Nuvolone, Adriano Aguzzi, Jesús Ávila, Isidro Ferrer, José Antonio del Río, and Rosalina Gavín. 2021. "Tau Exon 10 Inclusion by PrPC through Downregulating GSK3β Activity" International Journal of Molecular Sciences 22, no. 10: 5370. https://doi.org/10.3390/ijms22105370
APA StyleLidón, L., Llaó-Hierro, L., Nuvolone, M., Aguzzi, A., Ávila, J., Ferrer, I., del Río, J. A., & Gavín, R. (2021). Tau Exon 10 Inclusion by PrPC through Downregulating GSK3β Activity. International Journal of Molecular Sciences, 22(10), 5370. https://doi.org/10.3390/ijms22105370









