Molecular Mechanisms of PD-1 and PD-L1 Activity on a Pan-Cancer Basis: A Bioinformatic Exploratory Study
Abstract
:1. Introduction
2. Results
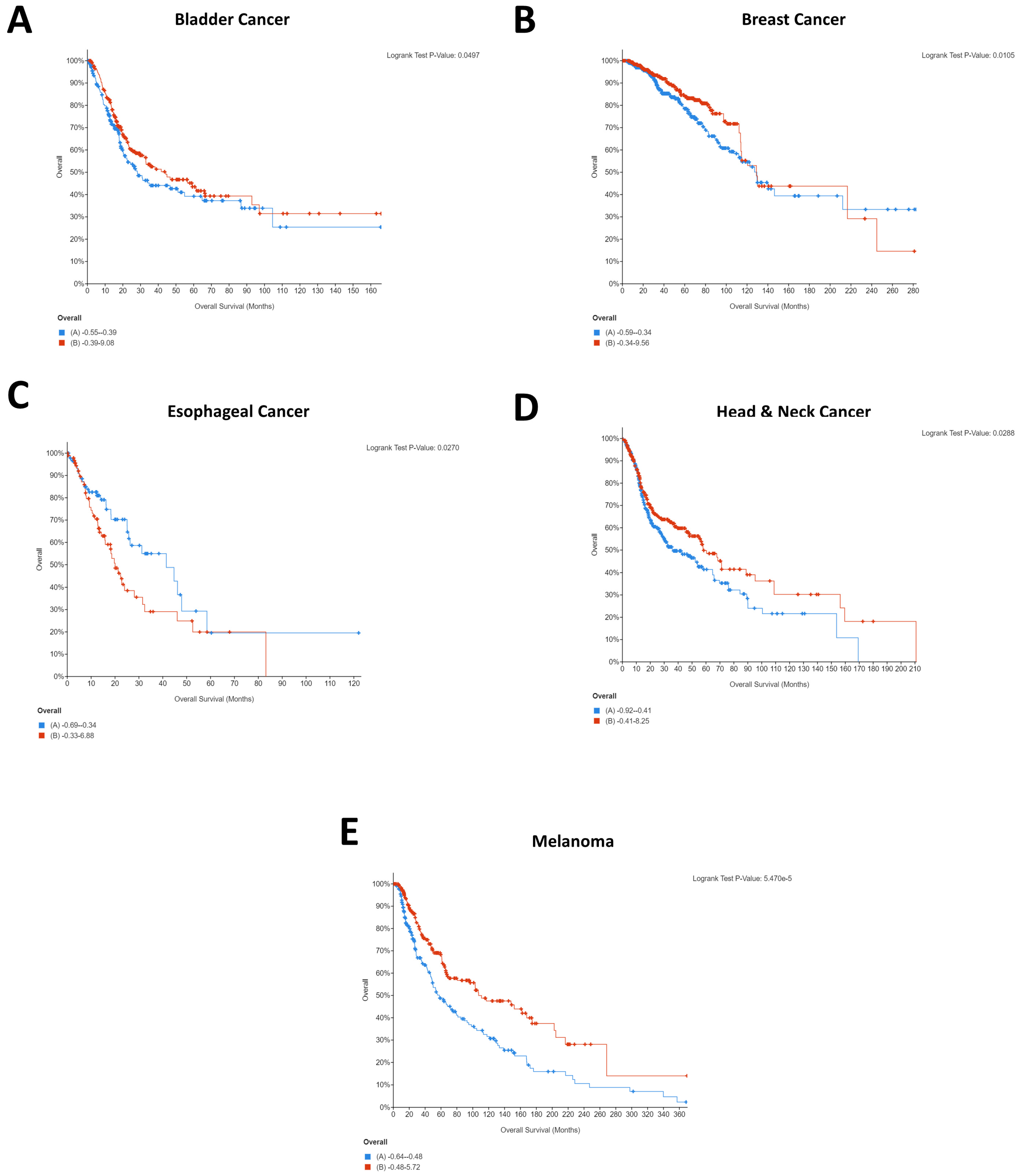
2.1. Impact of CD274 and PDCD1 Expression on Cancer Patient Survival
2.2. Identification of Genes That Correlate with PDCD1 and CD274 Pan-Cancer
2.3. Assessing Clinical Relevance of Co-Expressed Genes
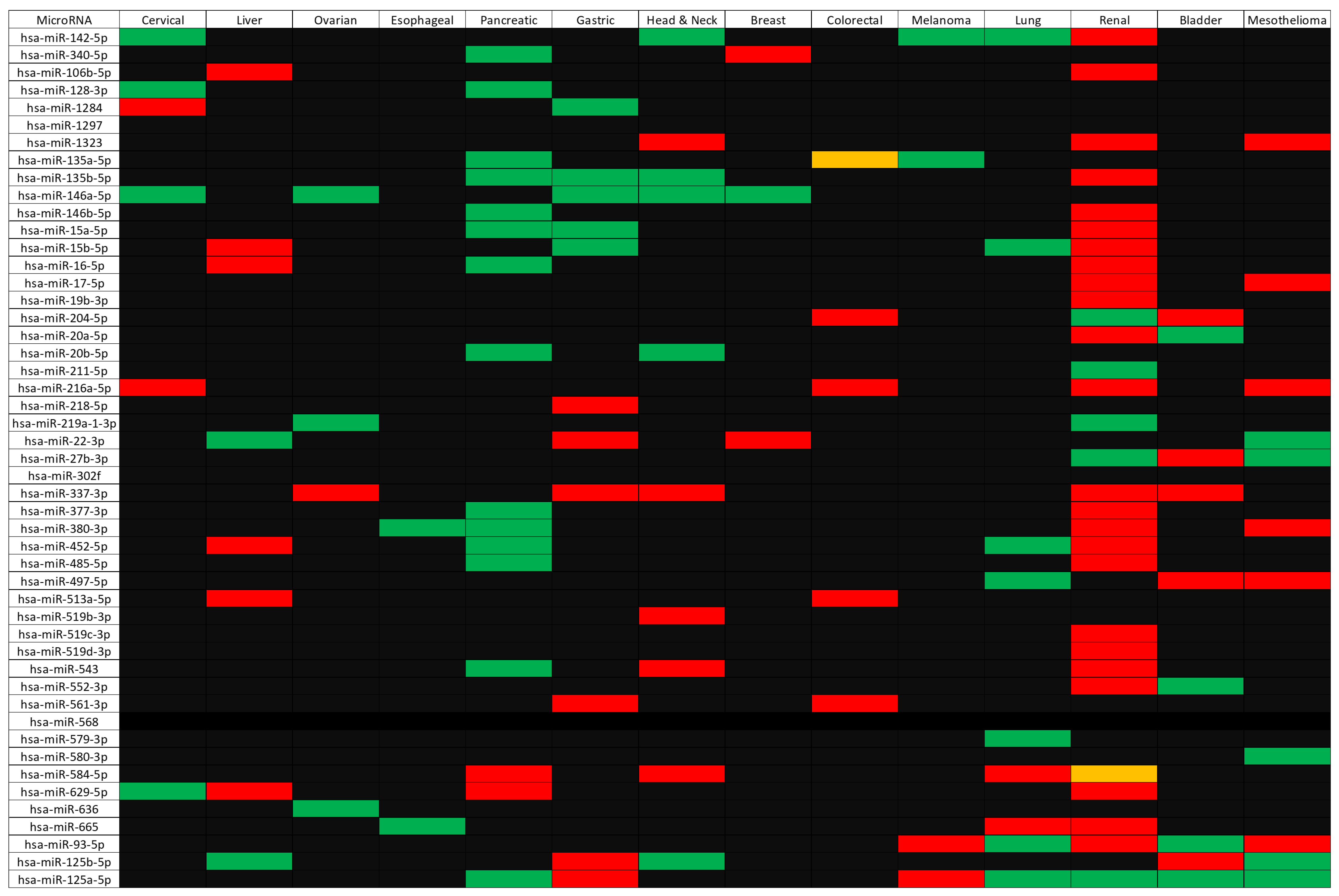
2.4. Identification of microRNAs Targeting CD274, PDCD1, and Their Correlated Genes
2.5. Assessing Clinical Relevance of the microRNAs Targeting CD274/PDCD1-Related Genes
2.6. Identification of Putative Repurposed Drugs Targeting PDCD1/CD274 Co-Expressed Genes/Proteins
3. Discussion
4. Materials and Methods
4.1. Study Selection
4.2. Assessing the Impact of Pan-Cancer PDCD1 and CD274 Expression
4.3. Identifying Co-Expressed Genes/Proteins Per Cancer Type
4.4. Identifying Common Pan-Cancer Co-Expressed Genes
4.5. Identifying Putative microRNAs Targeting Genes of Interest
4.6. Screening the Clinical Relevance of Putative microRNAs Using OncomiR
4.7. Identification of Putative Repurposed Drugs through DRUGSURV
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dobosz, P.; Dzieciątkowski, T. The Intriguing History of Cancer Immunotherapy. Front. Immunol. 2019, 10, 2965. [Google Scholar] [CrossRef] [Green Version]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene su-perfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, H.; Nose, M.; Hiai, H.; Minato, N.; Honjo, T. Development of Lupus-like Autoimmune Diseases by Disruption of the PD-1 Gene Encoding an ITIM Motif-Carrying Immunoreceptor. Immunity 1999, 11, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, H.; Okazaki, T.; Tanaka, Y.; Nakatani, K.; Hara, M.; Matsumori, A.; Sasayama, S.; Mizoguchi, A.; Hiai, H.; Minato, N.; et al. Autoimmune Dilated Cardiomyopathy in PD-1 Receptor-Deficient Mice. Science 2001, 291, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaddepally, R.K.; Kharel, P.; Pandey, R.; Garje, R.; Chandra, A.B. Review of Indications of FDA-Approved Immune Checkpoint Inhibitors per NCCN Guidelines with the Level of Evidence. Cancers 2020, 12, 738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Zhang, L.; Yu, J.; Zhang, Y.; Pang, X.; Ma, C.; Shen, M.; Ruan, S.; Wasan, H.S.; Qiu, S. Clinical efficacy and safety of anti-PD-1/PD-L1 inhibitors for the treatment of advanced or metastatic cancer: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 2083. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Zhao, H.; Zhao, J. Efficacy of PD-1/PD-L1 blockade monotherapy in clinical trials. Ther. Adv. Med. Oncol. 2020, 12, 1758835920937612. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalin-ka-Warzocha, E.; et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. N. Engl. J. Med. 2014, 372, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Ribas, A.; Hamid, O.; Daud, A.; Wolchok, J.D.; Joshua, A.M.; Hwu, W.-J.; Weber, J.S.; Gangadhar, T.C.; Joseph, R.W.; et al. Durable Complete Response After Discontinuation of Pembrolizumab in Patients With Metastatic Melanoma. J. Clin. Oncol. 2018, 36, 1668–1674. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Zhao, H.; Zhao, J. Serious adverse events and fatal adverse events associated with nivolumab treatment in cancer patients. J. Immunother. Cancer 2018, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wan, B.; Chen, X.; Zhan, P.; Zhao, Y.; Zhang, T.; Liu, H.; Afzal, M.Z.; Dermime, S.; Hochwald, S.N.; et al. The association of PD-L1 expression with the efficacy of anti-PD-1/PD-L1 immuno-therapy and survival of non-small cell lung cancer patients: A meta-analysis of randomized controlled trials. Transl. Lung Cancer Res. 2019, 8, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Arfè, A.; Fell, G.; Alexander, B.; Awad, M.M.; Rodig, S.J.; Trippa, L.; Schoenfeld, J.D. Meta-Analysis of PD-L1 Expression as a Predictor of Survival After Checkpoint Blockade. JCO Precis. Oncol. 2020, 4, 1196–1206. [Google Scholar] [CrossRef]
- Ugurel, S.; Schadendorf, D.; Horny, K.; Sucker, A.; Schramm, S.; Utikal, J.; Pföhler, C.; Herbst, R.; Schilling, B.; Blank, C.; et al. Elevated baseline serum PD-1 or PD-L1 predicts poor outcome of PD-1 inhibition therapy in metastatic melanoma. Ann. Oncol. 2020, 31, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Fan, Y.; Che, X.; Zhang, M.; Li, Z.; Li, C.; Wang, S.; Wen, T.; Hou, K.; Shao, X.; et al. Anti-PD-1 Therapy Response Predicted by the Combination of Exosomal PD-L1 and CD28. Front. Oncol. 2020, 10, 760. [Google Scholar] [CrossRef] [PubMed]
- Upadhaya, S.; Neftelino, S.T.; Hodge, J.P.; Oliva, C.; Campbell, J.R.; Yu, J.X. Combinations take centre stage in PD1/PDL1 inhibitor clinical trials. Nat. Rev. Drug Discov. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wolchok, J.D.; Kluger, H.; Callahan, M.K.; Postow, M.A.; Rizvi, N.A.; Lesokhin, A.M.; Segal, N.H.; Ariyan, C.E.; Gordon, R.-A.; Reed, K.; et al. Nivolumab plus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2013, 369, 122–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Wang, H.; Li, C.; Fang, J.-Y.; Xu, J. Cancer Cell-Intrinsic PD-1 and Implications in Combinatorial Immunotherapy. Front. Immunol. 2018, 9, 1774. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Quillet, A.; Saad, C.; Ferry, G.; Anouar, Y.; Vergne, N.; Lecroq, T.; Dubessy, C. Improving Bioinformatics Prediction of mi-croRNA Targets by Ranks Aggregation. Front. Genet. 2020, 10, 1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, N.W.; Chen, Y.; Chen, S.; Wang, X. OncomiR: An online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 2018, 34, 713–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonov, A.V. BioProfiling.de: Analytical web portal for high-throughput cell biology. Nucleic Acids Res. 2011, 39, W323–W327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonov, A.V.; Krestyaninova, M.; A Knight, R.; Rodchenkov, I.; Melino, G.; A Barlev, N. PPISURV: A novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene 2013, 33, 1621–1628. [Google Scholar] [CrossRef]
- Yang, J.; Dong, M.; Shui, Y.; Zhang, Y.; Zhang, Z.; Mi, Y.; Zuo, X.; Jiang, L.; Liu, K.; Liu, Z.; et al. A pooled analysis of the prognostic value of PD-L1 in melanoma: Evidence from 1062 patients. Cancer Cell Int. 2020, 20, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Liu, S.; Duan, Q.; Chen, L.; Wu, T.; Qian, H.; Yang, S.; Xin, D.; He, Z.; Guo, Y. MicroRNA-142-5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am. J. Transl. Res. 2017, 9, 2394–2402. [Google Scholar] [PubMed]
- Zuo, X.-L.; Chen, Z.-Q.; Wang, J.-F.; Wang, J.-G.; Liang, L.-H.; Cai, J. miR-337-3p suppresses the proliferation and invasion of hepatocellular carcinoma cells through targeting JAK2. Am. J. Cancer Res. 2018, 8, 662–674. [Google Scholar]
- Li, J.; Wang, T.; Jiang, X. Inhibition of miR-337-3p involved in the protection of CoCl 2 -induced injury in PC12 cells via activating JAK2/STAT3 signaling pathway. J. Cell. Biochem. 2019, 120, 19076–19086. [Google Scholar] [CrossRef]
- Iacona, J.R.; Lutz, C.S. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip. Rev. RNA 2019, 10, e1533. [Google Scholar] [CrossRef]
- Cui, Y.; She, K.; Tian, D.; Zhang, P.; Xin, X. miR-146a Inhibits Proliferation and Enhances Chemosensitivity in Epithelial Ovarian Cancer via Reduction of SOD2. Oncol. Res. 2016, 23, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Zhu, W.; Mei, L.; Lu, Z. Prognostic and clinicopathologic significance of MicroRNA-125a-5p in cancers: A meta-analysis. Medicine 2019, 98, e16685. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.-Y.; Luo, B.; An, J.-Y.; He, J.-Y.; Chen, N.-D.; Xu, L.-Y.; Huang, Y.-Y.; Liu, X.-G.; Le, H.-B.; Zhang, Y.-K. Differential Expression of miR-125a-5p and let-7e Predicts the Progression and Prognosis of Non-Small Cell Lung Cancer. Cancer Investig. 2014, 32, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Huang, Q.; Chang, J.; Wang, E.; Qiu, X. MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Exp. Lung Res. 2011, 37, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Liang, Y.; Liu, Y.; Bai, Y.; Yang, C.; Xu, S. The upregulation of TMPRSS4, partly ascribed to the downregulation of miR-125a-5p, promotes the growth of human lung adenocarcinoma via the NF-κB signaling pathway. Int. J. Oncol. 2018, 53, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Mao, W.; Zheng, S.; Ye, J. Epidermal growth factor receptor-regulated miR-125a-5p—A metastatic inhibitor of lung cancer. FEBS J. 2009, 276, 5571–5578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naidu, S.; Shi, L.; Magee, P.; Middleton, J.D.; Laganá, A.; Sahoo, S.; Leong, H.S.; Galvin, M.; Frese, K.; Dive, C.; et al. PDGFR-modulated miR-23b cluster and miR-125a-5p suppress lung tumorigenesis by targeting multiple components of KRAS and NF-kB pathways. Sci. Rep. 2017, 7, 15441. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Sun, S.; Shi, J.; Cao, F.; Han, X.; Chen, Z. MicroRNA-125a-5p plays a role as a tumor suppressor in lung carcinoma cells by directly targeting STAT3. Tumor Biol. 2017, 39, 1010428317697579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, Z.; Liu, N.; Lin, L.; Guo, X.; Yang, D.; Zhang, Q. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed. Pharmacother. 2015, 75, 129–136. [Google Scholar] [CrossRef]
- Yan, L.; Yu, M.; Gao, G.; Liang, H.; Zhou, X.; Zhu, Z.; Zhang, C.; Wang, Y.; Chen, X. MiR-125a-5p functions as a tumour suppressor in breast cancer by downregulating BAP1. J. Cell. Biochem. 2018, 119, 8773–8783. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Mimori, K.; Fabbri, M.; Yokobori, T.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Doki, Y.; Mori, M. Mi-croRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin. Cancer Res. 2011, 17, 2725–2733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kappelmann, M.; Kuphal, S.; Meister, G.; Vardimon, L.; Bosserhoff, A.-K. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2012, 32, 2984–2991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyholm, A.M.; Lerche, C.M.; Manfé, V.; Biskup, E.; Johansen, P.; Morling, N.; Thomsen, B.M.; Glud, M.; Gniadecki, R. miR-125b induces cellular senescence in malignant melanoma. BMC Dermatol. 2014, 14, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerloff, D.; Lützkendorf, J.; Moritz, R.K.C.; Wersig, T.; Mäder, K.; Müller, L.P.; Sunderkötter, C. Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA). Cancers 2020, 12, 464. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.C.; Kirschner, M.B.; Cheng, Y.Y.; Van Zandwijk, N.; Reid, G. MicroRNA gene expression signatures in long-surviving malignant pleural mesothelioma patients. Genom. Data 2016, 9, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Terashima, Y.; Toda, E.; Itakura, M.; Otsuji, M.; Yoshinaga, S.; Okumura, K.; Shand, F.H.W.; Komohara, Y.; Takeda, M.; Kokubo, K.; et al. Targeting FROUNT with disulfiram suppresses macrophage accumulation and its tumor-promoting properties. Nat. Commun. 2020, 11, 609. [Google Scholar] [CrossRef] [Green Version]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 1–15. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [Green Version]
- Leung, K.L.; Verma, D.; Azam, Y.J.; Bakker, E. The use of multi-omics data and approaches in breast cancer immunotherapy: A review. Future Oncol. 2020, 16, 2101–2119. [Google Scholar] [CrossRef]
- Tan, A.; Porcher, R.; Crequit, P.; Ravaud, P.; Dechartres, A. Differences in Treatment Effect Size Between Overall Survival and Progression-Free Survival in Immunotherapy Trials: A Meta-Epidemiologic Study of Trials With Results Posted at ClinicalTri-als.gov. J. Clin. Oncol. 2017, 35, 1686–1694. [Google Scholar] [CrossRef]
- Wessely, A.; Steeb, T.; Erdmann, M.; Heinzerling, L.; Vera, J.; Schlaak, M.; Berking, C.; Heppt, M.V. The Role of Immune Checkpoint Blockade in Uveal Melanoma. Int. J. Mol. Sci. 2020, 21, 879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, C.F.; Ricketts, C.J.; Wang, M.; Yang, L.; Cherniack, A.D.; Shen, H.; Buhay, C.; Kang, H.; Kim, S.C.; Fahey, C.C.; et al. The Somatic Genomic Landscape of Chromophobe Renal Cell Carcinoma. Cancer Cell 2014, 26, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e25. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.H. Biostatistics 104: Correlational analysis. Singap. Med. J. 2003, 44, 614–619. [Google Scholar]
- Pinzón, N.; Li, B.; Martinez, L.; Sergeeva, A.; Presumey, J.; Apparailly, F.; Seitz, H. microRNA target prediction programs predict many false positives. Genome Res. 2017, 27, 234–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [Green Version]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yue, D.; Chen, Y.; Gao, S.-J.; Huang, Y. Improving performance of mammalian microRNA target prediction. BMC Bioinform. 2010, 11, 476. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Ursu, O.; Holmes, J.; Knockel, J.; Bologa, C.G.; Yang, J.J.; Mathias, S.L.; Nelson, S.J.; Oprea, T.I. DrugCentral: Online drug compendium. Nucleic Acids Res. 2017, 45, D932–D939. [Google Scholar] [CrossRef]

| Positively Correlated with CD274 | Negatively Correlated with CD274 | Positively Correlated with PDCD1 |
|---|---|---|
| PDCD1LG2 (12) | GATD3A (2) | SH2D1A (13) |
| GBP5 (11) | COX19 (2) | ACAP1 (12) |
| CD80 (9) | DNASE1 (2) | ARHGAP9 (12) |
| GBP1 (8) | DUS1L (2) | CXCR6 (12) |
| JAK2 (8) | PTPRCAP (12) | |
| SAMD9L (8) | RASAL3 (12) | |
| LCP2 (8) | GZMK (11) | |
| CCR5 (8) | NKG7 (11) | |
| PYHIN1 (11) | ||
| TBC1D10C (11) | ||
| ZNF831 (11) | ||
| CD96 (10) | ||
| CST7 (10) | ||
| GPR171 (10) | ||
| GZMH (10) | ||
| GZMM (10) | ||
| TRAF3IP3 (10) | ||
| GZMA (9) | ||
| HCST (9) | ||
| IL21R (9) | ||
| PSTPIP1 (9) | ||
| GIMAP5 (8) | ||
| IKZF1 (8) | ||
| JAKMIP1 (8) | ||
| LTA (8) | ||
| MAP4K1 (8) | ||
| P2RY10 (8) | ||
| SEPTIN1 (8) |
| Cancer | Positively Correlated Genes Where Low Expression Is Beneficial for Patients (p ≤ 0.05) | Positively Correlated Genes Where High Expression Is Beneficial for Patients (p ≤ 0.05) | Negatively Correlated Genes Where Low Expression Is Beneficial for Patients (p ≤ 0.05) | Negatively Correlated Genes Where High Expression Is Beneficial for Patients (p ≤ 0.05) |
|---|---|---|---|---|
| Cervical | N/A | N/A | N/A | N/A |
| Liver | N/A | CCR5 | DNASE1 | N/A |
| Ovarian | N/A | CD80 | N/A | DUS1L |
| Esophageal | N/A | N/A | N/A | N/A |
| Pancreatic | N/A | N/A | N/A | GATD3A |
| Gastric | N/A | N/A | N/A | DUS1L |
| Head and Neck | N/A | CCR5 | GATD3A | N/A |
| Lymphoma | SAMD9L | N/A | N/A | N/A |
| Breast | N/A | JAK2, LCP2, SAMD9L | N/A | DUS1L |
| Colorectal | N/A | N/A | COX19 | N/A |
| Melanoma | N/A | CCR5, CD80, GBP1, GBP5, JAK2, LCP2, PDCD1LG2, SAMD9L | DUS1L | N/A |
| Lung | N/A | N/A | N/A | N/A |
| Renal | CD80, GBP1, LCP2 | JAK2 | COX19, DNASE1, DUS1L | N/A |
| Bladder | N/A | JAK2 | N/A | DNASE1 |
| Mesothelioma | N/A | N/A | N/A | N/A |
| Cancer | Positively Correlated Genes Where Low Expression Is Beneficial for Patients (p ≤ 0.05) | Positively Correlated Genes Where High Expression Is Beneficial for Patients (p ≤ 0.05) |
|---|---|---|
| Cervical | N/A | ACAP1, CST7, CXCR6, GPR171, GZMH, GZMK, GZMM, JAKMIP1, MAP4K1, P2YR10, PSTPIP1, RASAL3, SH2D1A, TBC1D10C, ZNF831 |
| Liver | N/A | ACAP1, CD96, CST7, CXCR6, GIMAP5, GPR171, GZMH, GZMK, IKZF1, NKG7, P2RY10, PYHIN1, SH2D1A, TBC1D10C, TRAF3IP3, ZNF831 |
| Ovarian | N/A | N/A |
| Esophageal | RASAL3 | N/A |
| Pancreatic | N/A | SEPTIN1, ZNF831 |
| Gastric | N/A | IL21R, JAKMIP1 |
| Head and Neck | N/A | ACAP1, CD96, CST7, CXCR6, GPR171, GZMK, GZMM, IKZF1, IL21R, LTA, MAP4K1, NKG7, P2RY10, PTPRCAP, PYHIN1, RASAL3, SEPTIN1, SH2D1A, TBC1D10C, TRAF3IP3, ZNF831 |
| Lymphoma | ACAP1 | |
| Breast | N/A | ACAP1, CD96, CST7, CXCR6, GPR171, GZMA, GZMH, GZMK, GZMM, HCST, IKZF1, MAP4K1, PSTPIP1, PYHIN1, SEPTIN1, SH2D1A, TBC1D10C, TRAF3IP3, ZNF831 |
| Colorectal | N/A | N/A |
| Melanoma | N/A | ACAP1, ARHGAP9, CD96, CST7, CXCR6, GIMAP5, GPR171, GZMA, GZMH, GZMK, GZMM, HCST, IKZF1, IL21R, JAKMIP1, LTA, MAP4K1, NKG7, P2RY10, PSTPIP1, PTPRCAP, PYHIN1, RASAL3, SEPTIN1, SH2D1A, TBC1D10C, TRAF3IP3, ZNF831 |
| Lung | N/A | ARHGAP9, CXCR6, GPR171, IKZF1, LTA, MAP4K1, PSTPIP1, PTPRCAP, PYHIN1, RASAL3, SEPTIN1, TBC1D10C, TRAF3IP3 |
| Renal | ACAP1, ARHGAP9, CXCR6, GPR171, GZMH, GZMM, HCST, IL21R, LTA, MAP4K1, PTPRCAP, RASAL3, TBC1D10C | GIMAP5 |
| Bladder | N/A | CD96, CXCR6, GPR171, GZMA, GZMH, MAP4K1, PTPRCAP, PYHIN1, SEPTIN1, SH2D1A |
| Mesothelioma | N/A | N/A |
| microRNA Name | Gene Symbol of Target |
|---|---|
| hsa-miR-142-5p | GBP5 |
| ZNF831 | |
| hsa-miR-340-5p | GBP5 |
| ZNF831 | |
| hsa-miR-106b-5p | PDCD1LG2 |
| hsa-miR-128-3p | SAMD9L |
| hsa-miR-1284 | JAK2 |
| hsa-miR-1297 | GBP1 |
| hsa-miR-1323 | ZNF831 |
| hsa-miR-135a-5p | JAK2 |
| hsa-miR-135b-5p | JAK2 |
| hsa-miR-146a-5p | CD80 |
| hsa-miR-146b-5p | CD80 |
| hsa-miR-15a-5p | CD80 |
| hsa-miR-15b-5p | CD80 |
| hsa-miR-16-5p | CD80 |
| hsa-miR-17-5p | PDCD1LG2 |
| hsa-miR-19b-3p | ZNF831 |
| hsa-miR-204-5p | JAK2 |
| hsa-miR-20a-5p | PDCD1LG2 |
| hsa-miR-20b-5p | PDCD1LG2 |
| hsa-miR-211-5p | JAK2 |
| hsa-miR-216a-5p | JAK2 |
| hsa-miR-218-5p | ZNF831 |
| hsa-miR-219a-1-3p | JAK2 |
| hsa-miR-22-3p | CD80 |
| hsa-miR-27b-3p | TRAF3IP3 |
| hsa-miR-302f | GBP1 |
| hsa-miR-337-3p | JAK2 |
| hsa-miR-377-3p | GBP1 |
| hsa-miR-380-3p | JAK2 |
| hsa-miR-452-5p | LCP2 |
| hsa-miR-485-5p | ZNF831 |
| hsa-miR-497-5p | CD80 |
| hsa-miR-513a-5p | SAMD9L |
| hsa-miR-519b-3p | PDCD1LG2 |
| hsa-miR-519c-3p | PDCD1LG2 |
| hsa-miR-519d-3p | PDCD1LG2 |
| hsa-miR-543 | GBP1 |
| hsa-miR-552-3p | SAMD9L |
| hsa-miR-561-3p | GBP5 |
| hsa-miR-568 | JAK2 |
| hsa-miR-579-3p | LCP2 |
| hsa-miR-580-3p | SAMD9L |
| hsa-miR-584-5p | GBP5 |
| hsa-miR-629-5p | ZNF831 |
| hsa-miR-636 | ZNF831 |
| hsa-miR-665 | IKZF1 |
| hsa-miR-93-5p | PDCD1LG2 |
| hsa-miR-125b-5p * | DUS1L |
| hsa-miR-125a-5p * | DUS1L |
| Indication | ||||
|---|---|---|---|---|
| Drug | Target | Cancer | Other | Cancer Trial |
| Cytarabine | JAK2 | Leukemia | Yes | |
| Pyrimethamine | JAK2 | No | Toxoplasmosis, acute malaria | Yes |
| Fluorouracil | JAK2 | Multiple (including colon, esophageal, gastric, breast, stomach, head and neck, cervical, pancreas, renal cell) | Yes | |
| Sunitinib | JAK2, MAP4K1 | Renal cell carcinoma; gastrointestinal stromal tumor | Yes | |
| Azathioprine | JAK2 | No | Rheumatoid arthritis, transplant rejection | Yes |
| Floxuridine | JAK2 | Liver cancer and metastases | Yes | |
| Cladribine | JAK2 | Leukemia, lymphoma | ||
| Erlotinib | JAK2 | Non-small cell lung cancer, pancreatic cancer | Yes | |
| Albendazole | JAK2 | No | Anthelmintic | |
| Triamterene | JAK2 | No | Edema | |
| Podofilox | JAK2 | No | Genital warts | |
| Dasatinib | JAK2, MAP4K1 | Chronic myelogenous leukemia, acute lymphoblastic leukemia | Yes | |
| Astemizole | JAK2 | No | Allergy | |
| Trifluridine | JAK2 | Colorectal | Keratoconjunctivitis and recurrent epithelial keratitis due to herpes simplex virus | Yes |
| Disulfiram | CCR5, CXCR6 | No | Chronic alcoholism | Yes |
| Terfenadine | CCR5 | No | Allergic rhinitis, hay fever, and allergic skin disorders | |
| Maraviroc | CCR5 | No | HIV-1 | Yes |
| Clioquinol | CXCR6 | No | Antifungal | Terminated (Phase 1) |
| Chloroxine | CXCR6 | No | Dandruff and seborrheic dermatitis | |
| Oxyphenbutazone | CXCR6 | No | ||
| Etanercept | LTA | No | Rheumatoid arthritis, plaque psoriasis, polyarticular idiopathic arthritis, psoriatic arthritis, ankylosing spondylitis | |
| Nilotinib | MAP4K1 | Leukemia | Yes | |
| Sorafenib | MAP4K1 | Liver, renal | Yes | |
| Cancer Type | Study 1 | Study 2 | Study 3 | Study 4 | Study 5 |
|---|---|---|---|---|---|
| Cervical | Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA, Firehose Legacy) | ||||
| Liver | Liver Hepatocellular Carcinoma (TCGA, Firehose Legacy) | ||||
| Ovarian | Ovarian Serous Cystadenocarcinoma (TCGA, Firehose Legacy) | ||||
| Esophageal | Esophageal Carcinoma (TCGA, Firehose Legacy) | ||||
| Pancreatic | Pancreatic Adenocarcinoma (TCGA, Firehose Legacy) | ||||
| Gastric | Stomach Adenocarcinoma (TCGA, Firehose Legacy) | Stomach Adenocarcinoma (TCGA, Nature 2014) [53] | |||
| Head and Neck | Head and Neck Squamous Cell Carcinoma (TCGA, Firehose Legacy) | Head and Neck Squamous Cell Carcinoma (TCGA, Nature 2015) [54] | |||
| Lymphoma | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma (TCGA, Firehose Legacy) | ||||
| Breast | Breast Invasive Carcinoma (TCGA, Firehose Legacy) | Breast Invasive Carcinoma (TCGA, Cell 2015) [55] | |||
| Colorectal | Colorectal Adenocarcinoma (TCGA, Firehose Legacy) | ||||
| Melanoma | Skin Cutaneous Melanoma (TCGA, Firehose Legacy) | ||||
| Lung | Lung Adenocarcinoma (TCGA, Firehose Legacy) | Lung Squamous Cell Carcinoma (TCGA, Firehose Legacy) | Lung Adenocarcinoma (TCGA, Nature 2014) [56] | ||
| Renal | Kidney Renal Clear Cell Carcinoma (TCGA, Firehose Legacy) | Kidney Renal Clear Cell Carcinoma (TCGA, Nature 2013) [57] | Kidney Renal Papillary Cell Carcinoma (TCGA, Firehose Legacy) | Kidney Chromophobe (TCGA, Cancer Cell 2014) [58] | Kidney Chromophobe (TCGA, Firehose Legacy) |
| Bladder | Bladder Urothelial Carcinoma (TCGA, Firehose Legacy) | Bladder Cancer (TCGA, Cell 2017) [59] | Bladder Urothelial Carcinoma (TCGA, Nature 2014) [60] | ||
| Mesothelioma | Mesothelioma (TCGA, Firehose Legacy) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kannan, S.; O’Connor, G.M.; Bakker, E.Y. Molecular Mechanisms of PD-1 and PD-L1 Activity on a Pan-Cancer Basis: A Bioinformatic Exploratory Study. Int. J. Mol. Sci. 2021, 22, 5478. https://doi.org/10.3390/ijms22115478
Kannan S, O’Connor GM, Bakker EY. Molecular Mechanisms of PD-1 and PD-L1 Activity on a Pan-Cancer Basis: A Bioinformatic Exploratory Study. International Journal of Molecular Sciences. 2021; 22(11):5478. https://doi.org/10.3390/ijms22115478
Chicago/Turabian StyleKannan, Siddarth, Geraldine Martina O’Connor, and Emyr Yosef Bakker. 2021. "Molecular Mechanisms of PD-1 and PD-L1 Activity on a Pan-Cancer Basis: A Bioinformatic Exploratory Study" International Journal of Molecular Sciences 22, no. 11: 5478. https://doi.org/10.3390/ijms22115478
APA StyleKannan, S., O’Connor, G. M., & Bakker, E. Y. (2021). Molecular Mechanisms of PD-1 and PD-L1 Activity on a Pan-Cancer Basis: A Bioinformatic Exploratory Study. International Journal of Molecular Sciences, 22(11), 5478. https://doi.org/10.3390/ijms22115478






