PODNL1 Methylation Serves as a Prognostic Biomarker and Associates with Immune Cell Infiltration and Immune Checkpoint Blockade Response in Lower-Grade Glioma
Abstract
1. Introduction
2. Results
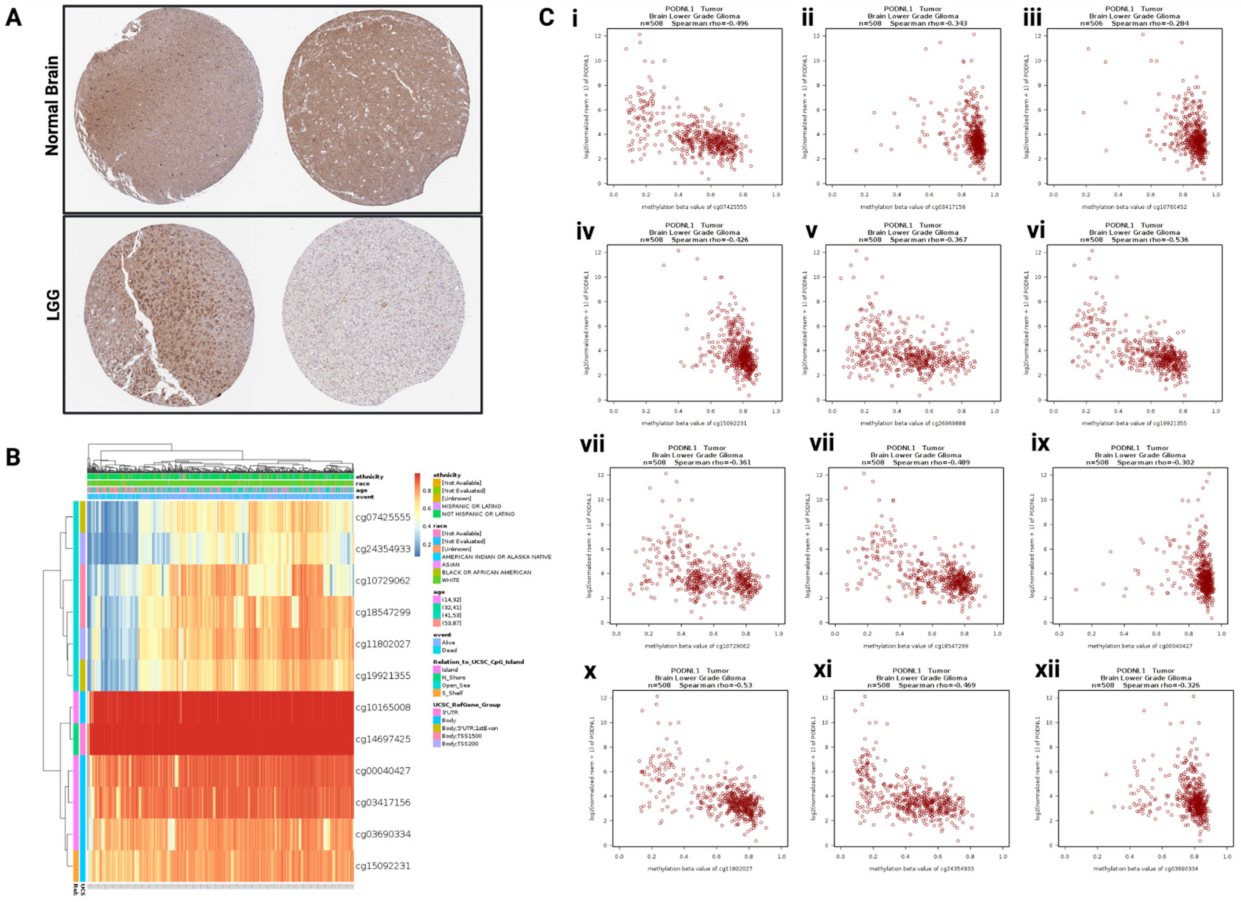
2.1. PODNL1 Expression and Methylation in LGG
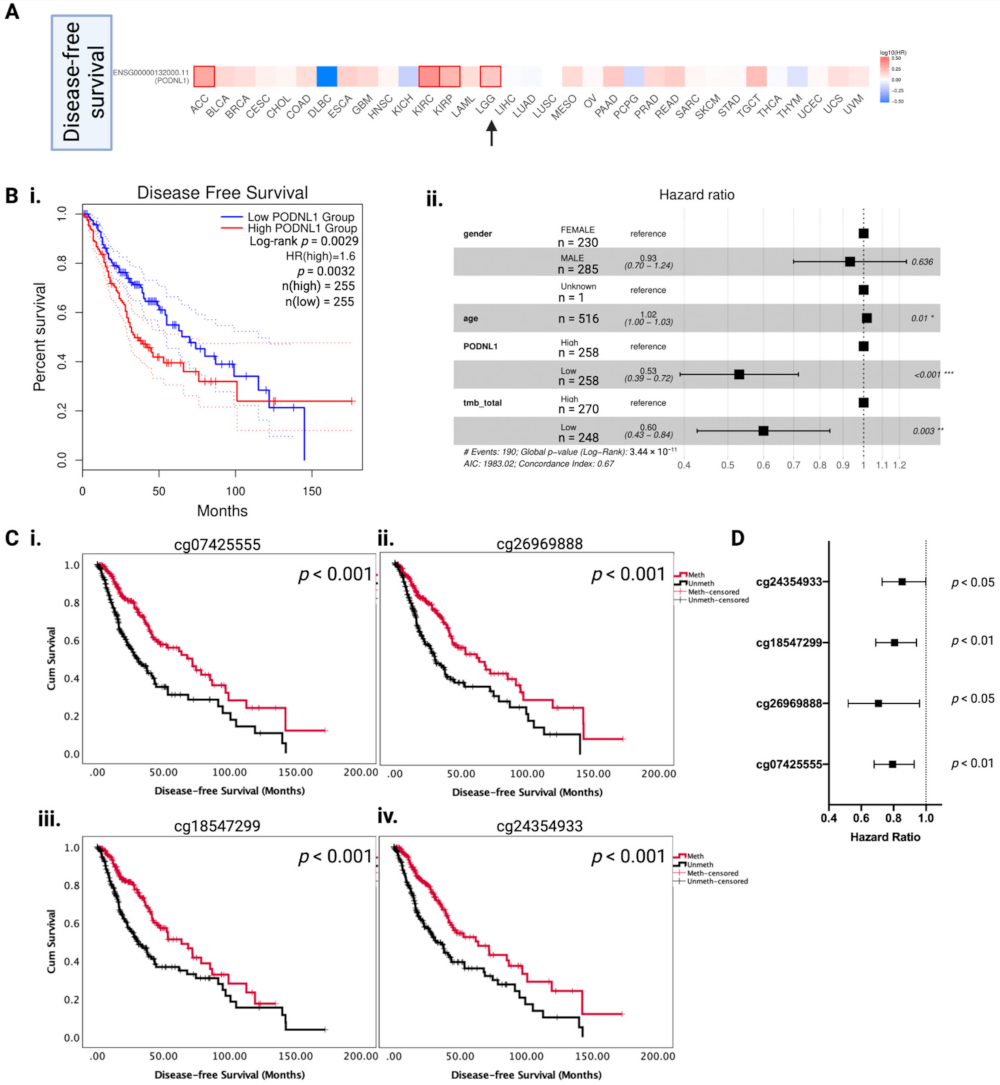
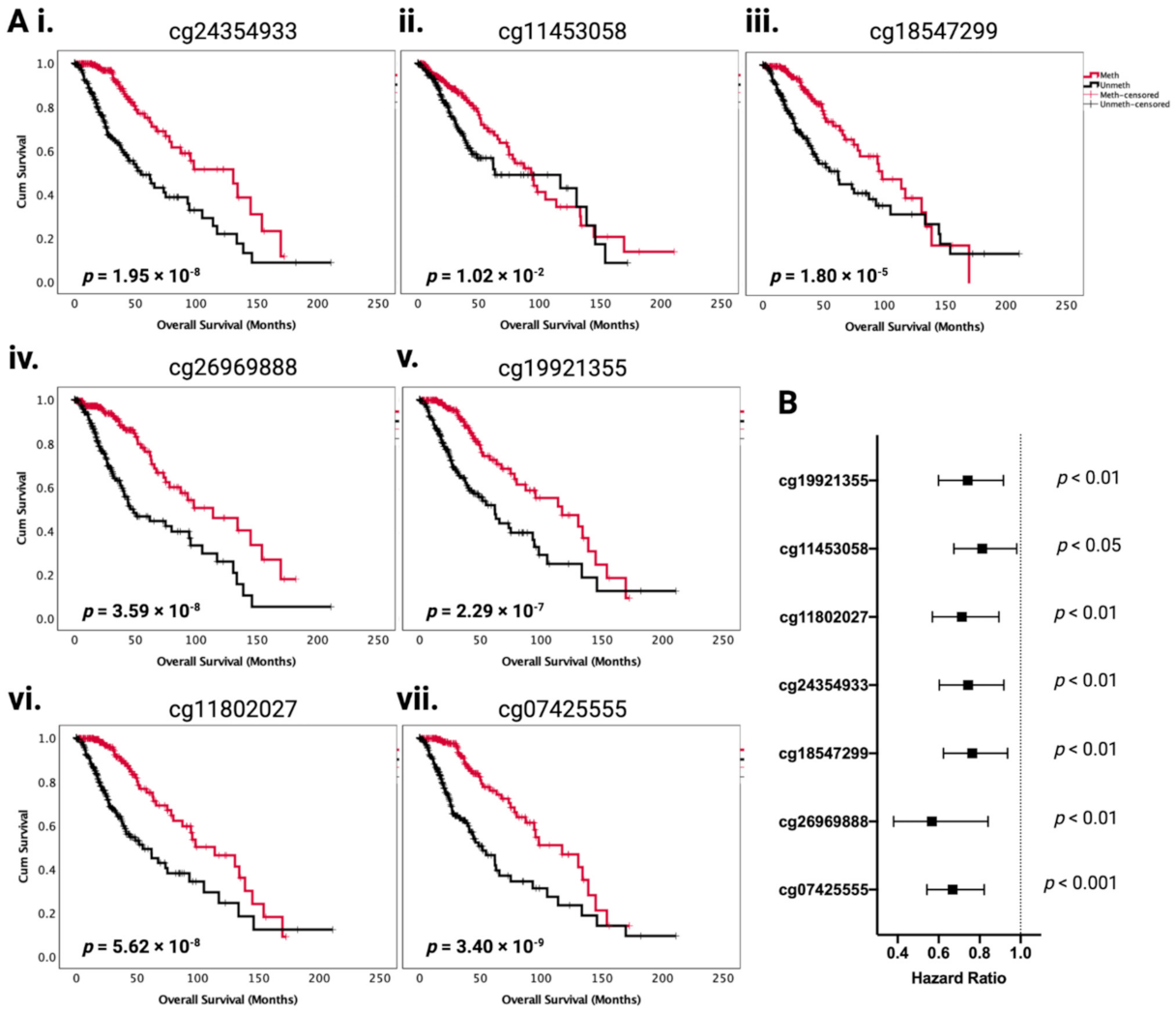
2.2. Association between PODNL1 Aberrations and Tumor Aggressiveness in LGG
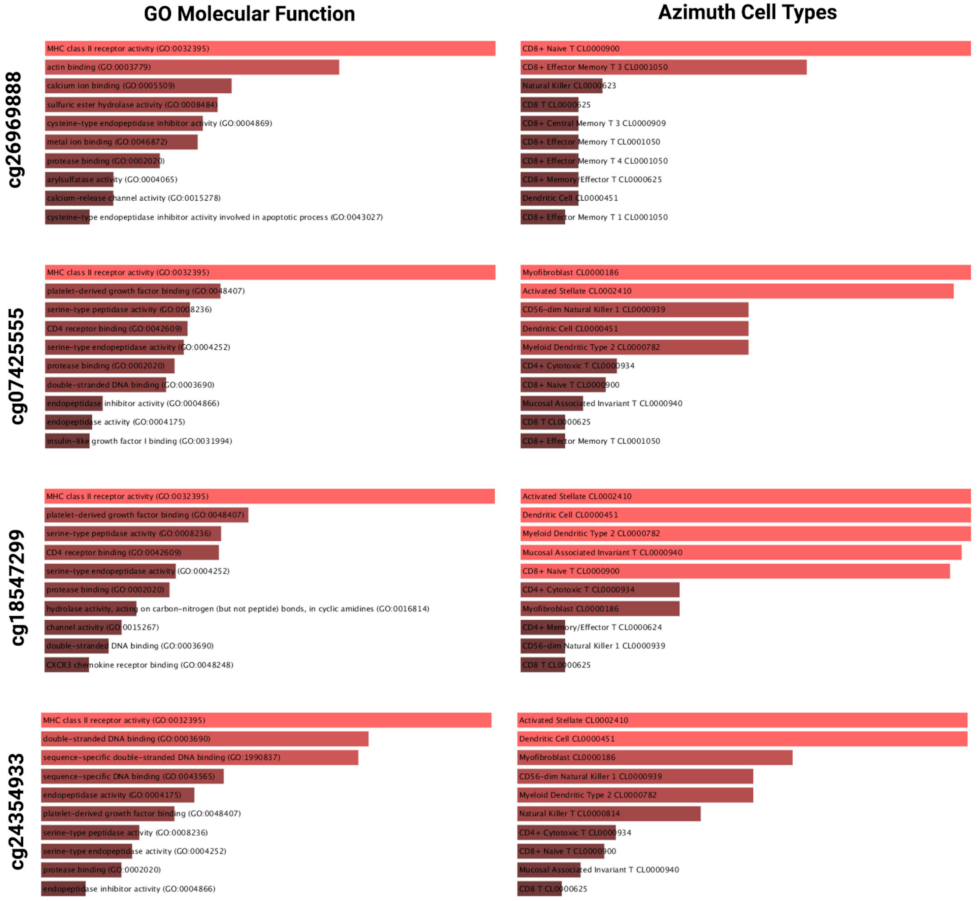
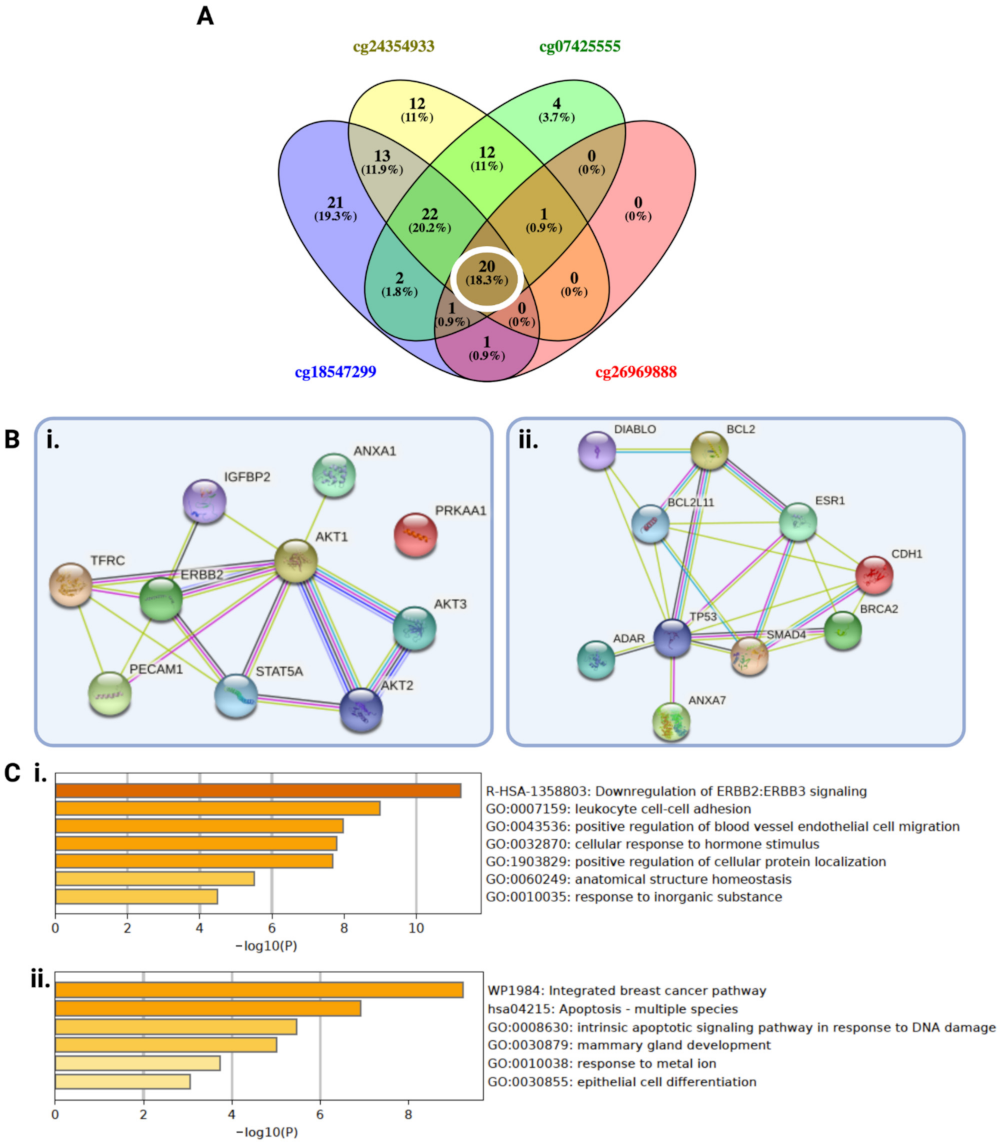
2.3. A Subset of LGGs with Low PODNL1 Methylation Is Associated with Increased Immune Cell Infiltration in the Tumor Microenvironment
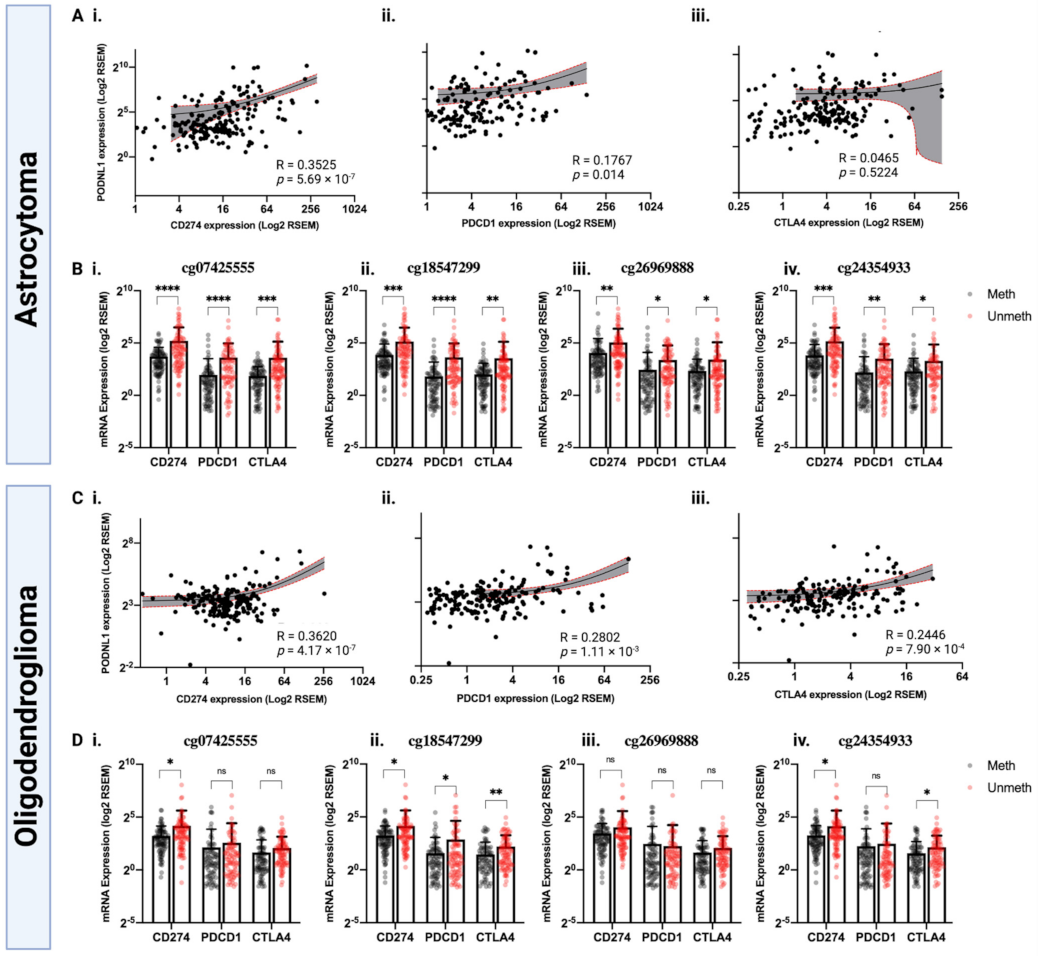
2.4. PODNL1 Methylation May Affect Immune Checkpoint Blockade Response in LGG
3. Discussion
4. Materials and Methods
4.1. Tumor Cohort
4.2. PODNL1 Expression and Methylation Analysis
4.3. Survival Analysis
4.4. Differential Gene and Protein Expression Analysis
4.5. Immune Cell Infiltration Analysis
4.6. Immune Checkpoint Blockade (ICB) Therapy Response Prediction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tom, M.C.; Cahill, D.P.; Buckner, J.C.; Dietrich, J.; Parsons, M.W.; Yu, J.S. Management for Different Glioma Subtypes: Are All Low-Grade Gliomas Created Equal? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 133–145. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Claus, E.B.; Walsh, K.M.; Wiencke, J.K.; Molinaro, A.M.; Wiemels, J.L.; Schildkraut, J.M.; Bondy, M.L.; Berger, M.; Jenkins, R.; Wrensch, M. Survival and low-grade glioma: The emergence of genetic information. Neurosurg. Focus 2015, 38, E6. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.J.; Wen, P.Y. Emerging targeted therapies for glioma. Expert Opin. Emerg. Drugs 2016, 21, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Killela, P.J.; Pirozzi, C.J.; Healy, P.; Reitman, Z.J.; Lipp, E.; Rasheed, B.A.; Yang, R.; Diplas, B.H.; Wang, Z.H.; Greer, P.K.; et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget 2014, 5, 1515–1525. [Google Scholar] [CrossRef]
- Noushmehr, H.; Weisenberger, D.J.; Diefes, K.; Phillips, H.S.; Pujara, K.; Berman, B.P.; Pan, F.; Pelloski, C.E.; Sulman, E.P.; Bhat, K.P.; et al. Cancer Genome Atlas Research, N. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010, 17, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Mur, P.; Mollejo, M.; Ruano, Y.; de Lope, A.R.; Fiano, C.; Garcia, J.F.; Castresana, J.S.; Hernandez-Lain, A.; Rey, J.A.; Melendez, B. Codeletion of 1p and 19q determines distinct gene methylation and expression profiles in IDH-mutated oligodendroglial tumors. Acta Neuropathol. 2013, 126, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar]
- Popovici-Muller, J.; Lemieux, R.M.; Artin, E.; Saunders, J.O.; Salituro, F.G.; Travins, J.; Cianchetta, G.; Cai, Z.W.; Zhou, D.; Cui, D.W.; et al. Discovery of AG-120 (Ivosidenib): A First-in-Class Mutant IDH1 Inhibitor for the Treatment of IDH1 Mutant Cancers. Acs Med. Chem. Lett. 2018, 9, 300–305. [Google Scholar] [CrossRef]
- Kopinja, J.; Sevilla, R.S.; Levitan, D.; Dai, D.; Vanko, A.; Spooner, E.; Ware, C.; Forget, R.; Hu, K.; Kral, A.; et al. A Brain Penetrant Mutant IDH1 Inhibitor Provides In Vivo Survival Benefit. Sci. Rep. 2017, 7, 13853. [Google Scholar] [CrossRef]
- Hartmann, C.; Meyer, J.; Balss, J.; Capper, D.; Mueller, W.; Christians, A.; Felsberg, J.; Wolter, M.; Mawrin, C.; Wick, W.; et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009, 118, 469–474. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Appay, R.; Dehais, C.; Maurage, C.A.; Alentorn, A.; Carpentier, C.; Colin, C.; Ducray, F.; Escande, F.; Idbaih, A.; Kamoun, A.; et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro-Oncology 2019, 21, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Reis, G.F.; Pekmezci, M.; Hansen, H.M.; Rice, T.; Marshall, R.E.; Molinaro, A.M.; Phillips, J.J.; Vogel, H.; Wiencke, J.K.; Wrensch, M.R.; et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J. Neuropathol. Exp. Neurol. 2015, 74, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Mochida, Y.; Kaku, M.; Yoshida, K.; Katafuchi, M.; Atsawasuwan, P.; Yamauchi, M. Podocan-like protein: A novel small leucine-rich repeat matrix protein in bone. Biochem. Biophys. Res. Commun. 2011, 410, 333–338. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schaefer, L.; Iozzo, R.V. Biological functions of the small leucine-rich proteoglycans: From genetics to signal transduction. J. Biol. Chem. 2008, 283, 21305–21309. [Google Scholar] [CrossRef]
- Shimizu-Hirota, R.; Sasamura, H.; Kuroda, M.; Kobayashi, E.; Saruta, T. Functional characterization of podocan, a member of a new class in the small leucine-rich repeat protein family. FEBS Lett. 2004, 563, 69–74. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, L.; Xu, T.; Zhou, J.; Qin, R.; Chen, C.; Zou, Y.; Fu, D.; Hu, G.; Chen, J. SAMSN1 is highly expressed and associated with a poor survival in glioblastoma multiforme. PLoS ONE 2013, 8, e81905. [Google Scholar]
- Geng, Y.; Zuo, P.; Li, X.O.; Zhang, L. PODNL1 promotes cell proliferation and migration in glioma via regulating Akt/mTOR pathway. J. Cancer 2020, 11, 6234–6242. [Google Scholar] [CrossRef]
- Teng, C.; Zheng, H. Low expression of microRNA-1908 predicts a poor prognosis for patients with ovarian cancer. Oncol. Lett. 2017, 14, 4277–4281. [Google Scholar] [CrossRef]
- Feng, Q.; Li, L.; Li, M.; Wang, X. Immunological classification of gliomas based on immunogenomic profiling. J. Neuroinflamm. 2020, 17, 360. [Google Scholar] [CrossRef]
- Kang, K.; Xie, F.; Wu, Y.; Han, C.; Bai, Y.; Long, J.; Lian, X.; Zhang, F. Genomic instability in lower-grade glioma: Prediction of prognosis based on lncRNA and immune infiltration. Mol. Ther. Oncolyt. 2021, 22, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [PubMed]
- Marinari, E.; Allard, M.; Gustave, R.; Widmer, V.; Philippin, G.; Merkler, D.; Tsantoulis, P.; Dutoit, V.; Dietrich, P.Y. Inflammation and lymphocyte infiltration are associated with shorter survival in patients with high-grade glioma. Oncoimmunology 2020, 9, 1779990. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yan, X.; Liu, W.; Yin, W.; Xu, H.; Cheng, D.; Jiang, X.; Ren, C. Three Immune-Associated Subtypes of Diffuse Glioma Differ in Immune Infiltration, Immune Checkpoint Molecules, and Prognosis. Front. Oncol. 2020, 10, 586019. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 6352. [Google Scholar] [CrossRef]
- Deng, G.; Chen, A.; Hong, J.; Chae, H.S.; Kim, Y.S. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999, 59, 2029–2033. [Google Scholar]
- Lapidus, R.G.; Ferguson, A.T.; Ottaviano, Y.L.; Parl, F.F.; Smith, H.S.; Weitzman, S.A.; Baylin, S.B.; Issa, J.P.; Davidson, N.E. Methylation of estrogen and progesterone receptor gene 5′ CpG islands correlates with lack of estrogen and progesterone receptor gene expression in breast tumors. Clin. Cancer Res. 1996, 2, 805–810. [Google Scholar]
- Strathdee, G.; Davies, B.R.; Vass, J.K.; Siddiqui, N.; Brown, R. Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis 2004, 25, 693–701. [Google Scholar] [CrossRef]
- Zochbauer-Muller, S.; Fong, K.M.; Maitra, A.; Lam, S.; Geradts, J.; Ashfaq, R.; Virmani, A.K.; Milchgrub, S.; Gazdar, A.F.; Minna, J.D. 5′ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001, 61, 3581–3585. [Google Scholar] [PubMed]
- de Souza, C.F.; Sabedot, T.S.; Malta, T.M.; Stetson, L.; Morozova, O.; Sokolov, A.; Laird, P.W.; Wiznerowicz, M.; Iavarone, A.; Snyder, J.; et al. A Distinct DNA Methylation Shift in a Subset of Glioma CpG Island Methylator Phenotypes during Tumor Recurrence. Cell Rep. 2018, 23, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Mazor, T.; Pankov, A.; Johnson, B.E.; Hong, C.; Hamilton, E.G.; Bell, R.J.A.; Smirnov, I.V.; Reis, G.F.; Phillips, J.J.; Barnes, M.J.; et al. DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer Cell 2015, 28, 307–317. [Google Scholar] [CrossRef]
- Issa, J.P. DNA methylation as a therapeutic target in cancer. Clin. Cancer Res. 2007, 13, 1634–1637. [Google Scholar] [CrossRef] [PubMed]
- Mellai, M.; Piazzi, A.; Caldera, V.; Annovazzi, L.; Monzeglio, O.; Senetta, R.; Cassoni, P.; Schiffer, D. Promoter hypermethylation of the EMP3 gene in a series of 229 human gliomas. Biomed. Res. Int. 2013, 2013, 756302. [Google Scholar] [CrossRef] [PubMed]
- Ohka, F.; Natsume, A.; Motomura, K.; Kishida, Y.; Kondo, Y.; Abe, T.; Nakasu, Y.; Namba, H.; Wakai, K.; Fukui, T.; et al. The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS ONE 2011, 6, e23332. [Google Scholar] [CrossRef]
- Qi, C.; Lei, L.; Hu, J.; Wang, G.; Liu, J.; Ou, S. Serine Incorporator 2 (SERINC2) Expression Predicts an Unfavorable Prognosis of Low-Grade Glioma (LGG): Evidence from Bioinformatics Analysis. J. Mol. Neurosci. 2020, 70, 1521–1532. [Google Scholar] [CrossRef]
- Sharma, N.; Saxena, S.; Agrawal, I.; Singh, S.; Srinivasan, V.; Arvind, S.; Epari, S.; Paul, S.; Jha, S. Differential Expression Profile of NLRs and AIM2 in Glioma and Implications for NLRP12 in Glioblastoma. Sci. Rep. 2019, 9, 8480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, M.; Wang, W.; Mo, J. Incidence and prognostic value of multiple gene promoter methylations in gliomas. J. Neurooncol. 2014, 116, 349–356. [Google Scholar] [CrossRef]
- Bacolod, M.D.; Barany, F.; Fisher, P.B. Can CpG methylation serve as surrogate markers for immune infiltration in cancer? Adv. Cancer Res. 2019, 143, 351–384. [Google Scholar]
- Chen, F.; Fan, Y.; Hou, J.; Liu, B.; Zhang, B.; Shang, Y.; Chang, Y.; Cao, P.; Tan, K. Integrated analysis identifies TfR1 as a prognostic biomarker which correlates with immune infiltration in breast cancer. Aging 2021, 13, 21671–21699. [Google Scholar] [CrossRef] [PubMed]
- Koirala, P.; Roth, M.E.; Gill, J.; Piperdi, S.; Chinai, J.M.; Geller, D.S.; Hoang, B.H.; Park, A.; Fremed, M.A.; Zang, X.; et al. Immune infiltration and PD-L1 expression in the tumor microenvironment are prognostic in osteosarcoma. Sci. Rep. 2016, 6, 30093. [Google Scholar] [CrossRef]
- Liao, G.; Lv, J.; Ji, A.; Meng, S.; Chen, C. TLR3 Serves as a Prognostic Biomarker and Associates with Immune Infiltration in the Renal Clear Cell Carcinoma Microenvironment. J. Oncol. 2021, 2021, 3336770. [Google Scholar] [CrossRef]
- Sawe, R.T.; Kerper, M.; Badve, S.; Li, J.; Sandoval-Cooper, M.; Xie, J.; Shi, Z.; Patel, K.; Chumba, D.; Ofulla, A.; et al. Aggressive breast cancer in western Kenya has early onset, high proliferation, and immune cell infiltration. BMC Cancer 2016, 16, 204. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Zhang, X.D.; Liu, X.Y.; Lu, P. YAP1 is a Prognostic Biomarker and Correlated with Immune Cell Infiltration in Pancreatic Cancer. Front. Mol. Biosci. 2021, 8, 625731. [Google Scholar] [CrossRef]
- Tan, P.; Chen, H.; Huang, Z.; Huang, M.; Du, Y.; Li, T.; Chen, Z.; Liu, Y.; Fu, W. MMP25-AS1/hsa-miR-10a-5p/SERPINE1 axis as a novel prognostic biomarker associated with immune cell infiltration in KIRC. Mol. Ther. Oncol. 2021, 22, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, D.; Hua, H. TBC1D3 family is a prognostic biomarker and correlates with immune infiltration in kidney renal clear cell carcinoma. Mol. Ther. Oncol. 2021, 22, 528–538. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Y.; Shen, P.; Gong, L. Identification of Immune Cell Infiltration Landscape and Their Prognostic Significance in Uveal Melanoma. Front. Cell Dev. Biol. 2021, 9, 713569. [Google Scholar] [CrossRef]
- Fan, Y.; Peng, X.; Wang, Y.; Li, B.; Zhao, G. Comprehensive Analysis of HDAC Family Identifies HDAC1 as a Prognostic and Immune Infiltration Indicator and HDAC1-Related Signature for Prognosis in Glioma. Front. Mol. Biosci. 2021, 8, 720020. [Google Scholar] [CrossRef]
- Ge, X.; Wang, Z.; Jiang, R.; Ren, S.; Wang, W.; Wu, B.; Zhang, Y.; Liu, Q. SCAMP4 is a novel prognostic marker and correlated with the tumor progression and immune infiltration in glioma. Int. J. Biochem. Cell Biol. 2021, 139, 106054. [Google Scholar] [CrossRef]
- Gong, X.; Liu, L.; Xiong, J.; Li, X.; Xu, J.; Xiao, Y.; Li, J.; Luo, X.; Mao, D.; Liu, L. Construction of a Prognostic Gene Signature Associated with Immune Infiltration in Glioma: A Comprehensive Analysis Based on the CGGA. J. Oncol. 2021, 2021, 6620159. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Guo, D. EGFR mutation: Novel prognostic factor associated with immune infiltration in lower-grade glioma; an exploratory study. BMC Cancer 2019, 19, 1184. [Google Scholar] [CrossRef]
- Hu, Z.; Qu, S. EVA1C Is a Potential Prognostic Biomarker and Correlated With Immune Infiltration Levels in WHO Grade II/III Glioma. Front. Immunol. 2021, 12, 683572. [Google Scholar] [CrossRef] [PubMed]
- Ju, Q.; Li, X.; Zhang, H.; Yan, S.; Li, Y.; Zhao, Y. NFE2L2 Is a Potential Prognostic Biomarker and Is Correlated with Immune Infiltration in Brain Lower Grade Glioma: A Pan-Cancer Analysis. Oxid. Med. Cell Longev. 2020, 2020, 3580719. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Song, Y.; Song, Z.; Huang, C. CKMT1B is a potential prognostic biomarker and associated with immune infiltration in Lower-grade glioma. PLoS ONE 2021, 16, e0245524. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhou, H.; Tian, L.; Yan, T.; Han, X.; Chen, P.; Li, H.; Wang, W.; Xiao, Z.; Hou, L.; et al. A Prognostic DNA Damage Repair Genes Signature and Its Impact on Immune Cell Infiltration in Glioma. Front. Oncol. 2021, 11, 682932. [Google Scholar] [CrossRef]
- Xiao, M.; Du, C.; Zhang, C.; Zhang, X.; Li, S.; Zhang, D.; Jia, W. Bioinformatics analysis of the prognostic value of NEK8 and its effects on immune cell infiltration in glioma. J. Cell Mol. Med. 2021, 25, 8748–8763. [Google Scholar] [CrossRef]
- Annovazzi, L.; Mellai, M.; Caldera, V.; Valente, G.; Tessitore, L.; Schiffer, D. mTOR, S6 and AKT expression in relation to proliferation and apoptosis/autophagy in glioma. Anticancer Res. 2009, 29, 3087–3094. [Google Scholar]
- Bhaskara, V.K.; Sundaram, C.; Babu, P.P. pERK, pAkt and pBad: A possible role in cell proliferation and sustained cellular survival during tumorigenesis and tumor progression in ENU induced transplacental glioma rat model. Neurochem. Res. 2006, 31, 1163–1170. [Google Scholar] [CrossRef]
- Suzuki, Y.; Shirai, K.; Oka, K.; Mobaraki, A.; Yoshida, Y.; Noda, S.E.; Okamoto, M.; Suzuki, Y.; Itoh, J.; Itoh, H.; et al. Higher pAkt expression predicts a significant worse prognosis in glioblastomas. J. Rad. Res. 2010, 51, 343–348. [Google Scholar] [CrossRef]
- Antonelli, M.; Massimino, M.; Morra, I.; Garre, M.L.; Gardiman, M.P.; Buttarelli, F.R.; Arcella, A.; Giangaspero, F. Expression of pERK and pAKT in pediatric high grade astrocytomas: Correlation with YKL40 and prognostic significance. Neuropathology 2012, 32, 133–138. [Google Scholar] [CrossRef]
- Modhukur, V.; Iljasenko, T.; Metsalu, T.; Lokk, K.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Diez-Villanueva, A.; Mallona, I.; Peinado, M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenet. Chrom. 2015, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, D.; Lu, C. The SMART App: An interactive web application for comprehensive DNA methylation analysis and visualization. Epigenet. Chrom. 2019, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucl. Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Cai, M.; Du, Y.; Yang, E.; Ji, J.; Wu, J. CVCDAP: An integrated platform for molecular and clinical analysis of cancer virtual cohorts. Nucl. Acids Res. 2020, 48, W463–W471. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013, 14, 128. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef]
- Miao, Y.R.; Zhang, Q.; Lei, Q.; Luo, M.; Xie, G.Y.; Wang, H.; Guo, A.Y. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv. Sci. 2020, 7, 1902880. [Google Scholar] [CrossRef]

| Univariate Analysis | Multivariate Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Overall Survival | Disease-Free Survival | Overall Survival | Disease-Free Survival | ||||||
| CpG loci | Position | HR [95% CI] | p-Value | HR [95% CI] | p-Value | HR [95% CI] | p-Value | HR [95% CI] | p-Value |
| cg26498537 | 13929563 | 0.68 [0.48–0.97] | 3.40 × 10−2 | 0.75 [0.56–1.00] | 5.26 × 10−2 | 1.01 [0.69–1.47] | 9.60 ×10−1 | 0.95 [0.70–1.28] | 7.68 × 10−1 |
| cg21993464 | 13930493 | 0.60 [0.42–0.87] | 6.56 × 10−3 | 0.82 [0.62–1.09] | 1.89 × 10−1 | 0.91 [0.63–1.33] | 6.57 × 10−1 | 0.99 [0.74–1.33] | 9.82 × 10−1 |
| cg14697425 | 13932027 | 1.02 [0.72–1.46] | 8.81 × 10−1 | 1.05 [0.79–1.39] | 7.23 × 10−1 | 1.20 [0.83–1.73] | 3.12 × 10−1 | 1.19 [0.89–1.60] | 2.25 × 10−1 |
| cg03417156 | 13932833 | 0.66 [0.46–0.94] | 2.34 × 10−2 | 0.78 [0.58–1.04] | 9.42 × 10−2 | 1.09 [0.73–1.62] | 6.50 × 10−1 | 1.07 [0.79–1.45] | 6.47 × 10−1 |
| cg03690334 | 13933004 | 0.57 [0.40–0.82] | 2.43 × 10−3 | 0.72 [0.54–0.96] | 2.62 × 10−2 | 0.72 [0.49–1.04] | 8.64 × 10−2 | 0.85 [0.63–1.14] | 2.93 × 10−1 |
| cg10165008 | 13933098 | 0.60 [0.42–0.86] | 6.25 × 10−3 | 0.72 [0.54–0.96] | 2.83 × 10−2 | 0.83 [0.57–1.21] | 3.45 × 10−1 | 0.83 [0.61–1.12] | 2.40 × 10−1 |
| cg00040427 | 13933384 | 0.43 [0.29–0.62] | 6.00 × 10−6 | 0.59 [0.44–0.79] | 4.20 × 10−4 | 0.71 [0.48–1.06] | 1.01 × 10−1 | 0.79 [0.58–1.08] | 1.54 × 10−1 |
| cg15092231 | 13935853 | 0.49 [0.34–0.71] | 1.49 × 10−4 | 0.65 [0.49–0.87] | 3.66 × 10−3 | 0.79 [0.53–1.18] | 2.62 × 10−1 | 0.87 [0.64–1.18] | 3.82 × 10−1 |
| cg07425555 | 13938206 | 0.32 [0.22–0.46] | 3.40 × 10−9 | 0.47 [0.35–0.63] | 5.45 × 10−7 | 0.44 [0.29–0.67] | 1.39 × 10−4 | 0.62 [0.46–0.85] | 3.53 × 10−3 |
| cg19921355 | 13938280 | 0.37 [0.25–0.54] | 2.29 × 10−7 | 0.56 [0.42–0.74] | 8.30 × 10−5 | 0.54 [0.35–0.84] | 5.70 × 10−3 | 0.76 [0.55–1.05] | 1.03 × 10−1 |
| cg24354933 | 13938482 | 0.33 [0.22–0.49] | 1.95 × 10−8 | 0.51 [0.38–0.68] | 7.00 × 10−6 | 0.55 [0.36–0.84] | 5.93 × 10−3 | 0.72 [0.53–0.99] | 4.74 × 10−2 |
| cg11802027 | 13938630 | 0.35 [0.24–0.51] | 5.62 × 10−8 | 0.53 [0.39–0.70] | 1.70 × 10−5 | 0.50 [0.32–0.79] | 3.32 × 10−3 | 0.76 [0.54–1.07] | 1.25 × 10−1 |
| cg18547299 | 13938766 | 0.44 [0.30–0.64] | 1.80 × 10−5 | 0.52 [0.39–0.70] | 1.50 × 10−5 | 0.58 [0.38–0.87] | 9.25 × 10−3 | 0.64 [0.47–0.88] | 6.52 × 10−3 |
| cg10729062 | 13939010 | 0.48 [0.33–0.69] | 1.05 × 10−4 | 0.50 [0.37–0.67] | 5.00 × 10−6 | 0.73 [0.48–1.10] | 1.37 × 10−1 | 0.73 [0.53–1.01] | 5.73 × 10−2 |
| cg11453058 | 13953364 | 0.62 [0.44–0.89] | 1.02 × 10−2 | 0.70 [0.53–0.94] | 1.73 × 10−2 | 0.66 [0.45–0.96] | 3.03 × 10−2 | 0.79 [0.59–1.06] | 1.18 × 10−1 |
| cg26969888 | 13953442 | 0.34 [0.24–0.50] | 3.59 × 10−8 | 0.51 [0.38–0.68] | 7.00 × 10−6 | 0.56 [0.38–0.84] | 4.85 × 10−3 | 0.70 [0.51–0.96] | 2.65 × 10−2 |
| cg10760452 | 13953776 | 0.72 [0.50–1.02] | 7.00 × 10−2 | 0.83 [0.62–1.10] | 2.07 × 10−1 | 0.79 [0.55–1.13] | 2.06 ×10−1 | 0.94 [0.71–1.27] | 7.26 × 10−1 |
| cg10609371 | 13954060 | 0.94 [0.66–1.34] | 7.38 × 10−1 | 1.04 [0.78–1.39] | 7.50 × 10−1 | 1.14 [0.79–1.65] | 4.60 × 10−1 | 1.14 [0.85–1.52] | 3.68 × 10−1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noor, H.; Zaman, A.; Teo, C.; Sughrue, M.E. PODNL1 Methylation Serves as a Prognostic Biomarker and Associates with Immune Cell Infiltration and Immune Checkpoint Blockade Response in Lower-Grade Glioma. Int. J. Mol. Sci. 2021, 22, 12572. https://doi.org/10.3390/ijms222212572
Noor H, Zaman A, Teo C, Sughrue ME. PODNL1 Methylation Serves as a Prognostic Biomarker and Associates with Immune Cell Infiltration and Immune Checkpoint Blockade Response in Lower-Grade Glioma. International Journal of Molecular Sciences. 2021; 22(22):12572. https://doi.org/10.3390/ijms222212572
Chicago/Turabian StyleNoor, Humaira, Ashraf Zaman, Charles Teo, and Michael E. Sughrue. 2021. "PODNL1 Methylation Serves as a Prognostic Biomarker and Associates with Immune Cell Infiltration and Immune Checkpoint Blockade Response in Lower-Grade Glioma" International Journal of Molecular Sciences 22, no. 22: 12572. https://doi.org/10.3390/ijms222212572
APA StyleNoor, H., Zaman, A., Teo, C., & Sughrue, M. E. (2021). PODNL1 Methylation Serves as a Prognostic Biomarker and Associates with Immune Cell Infiltration and Immune Checkpoint Blockade Response in Lower-Grade Glioma. International Journal of Molecular Sciences, 22(22), 12572. https://doi.org/10.3390/ijms222212572






