HSV-1 and Endogenous Retroviruses as Risk Factors in Demyelination
Abstract
1. Introduction
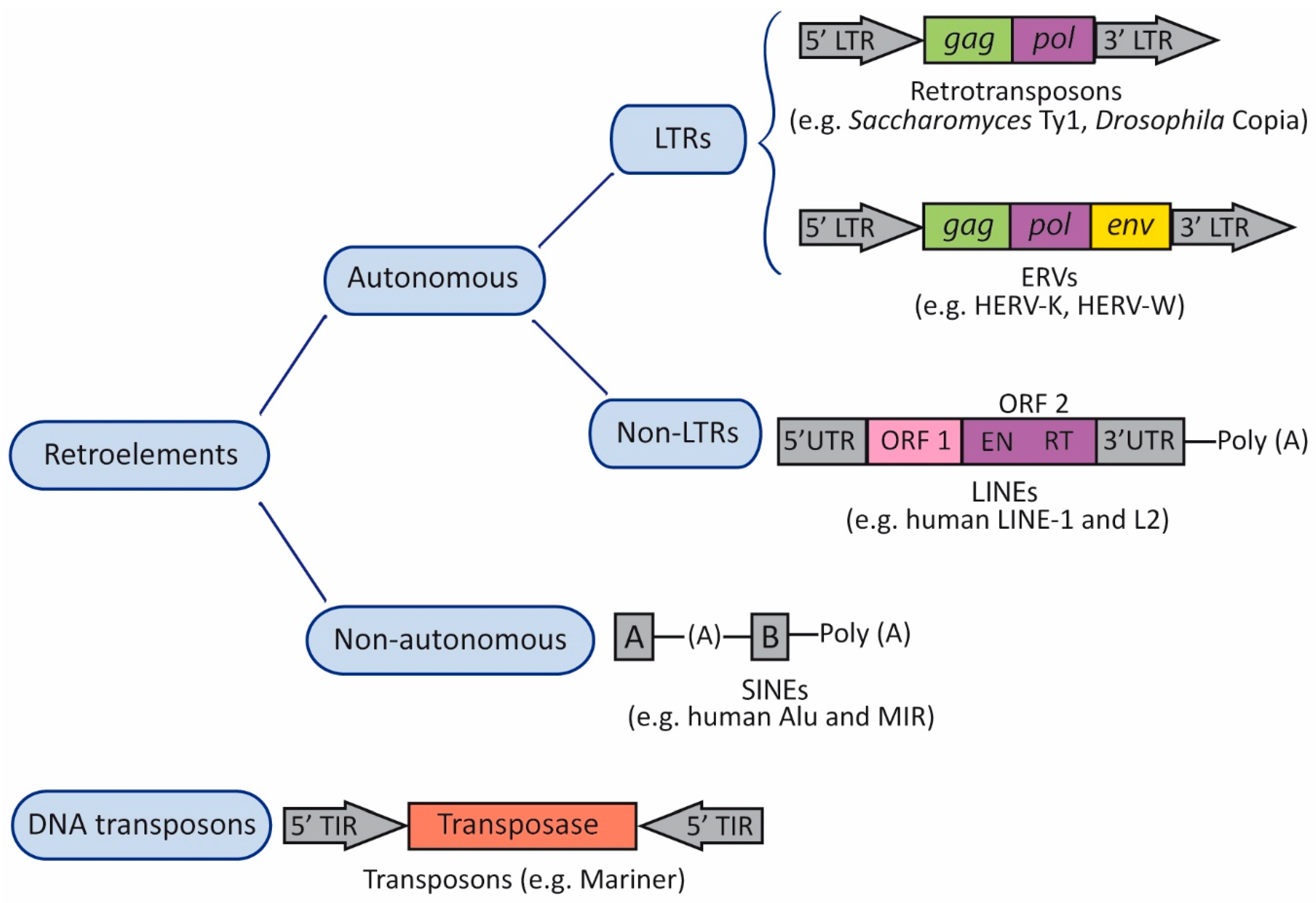
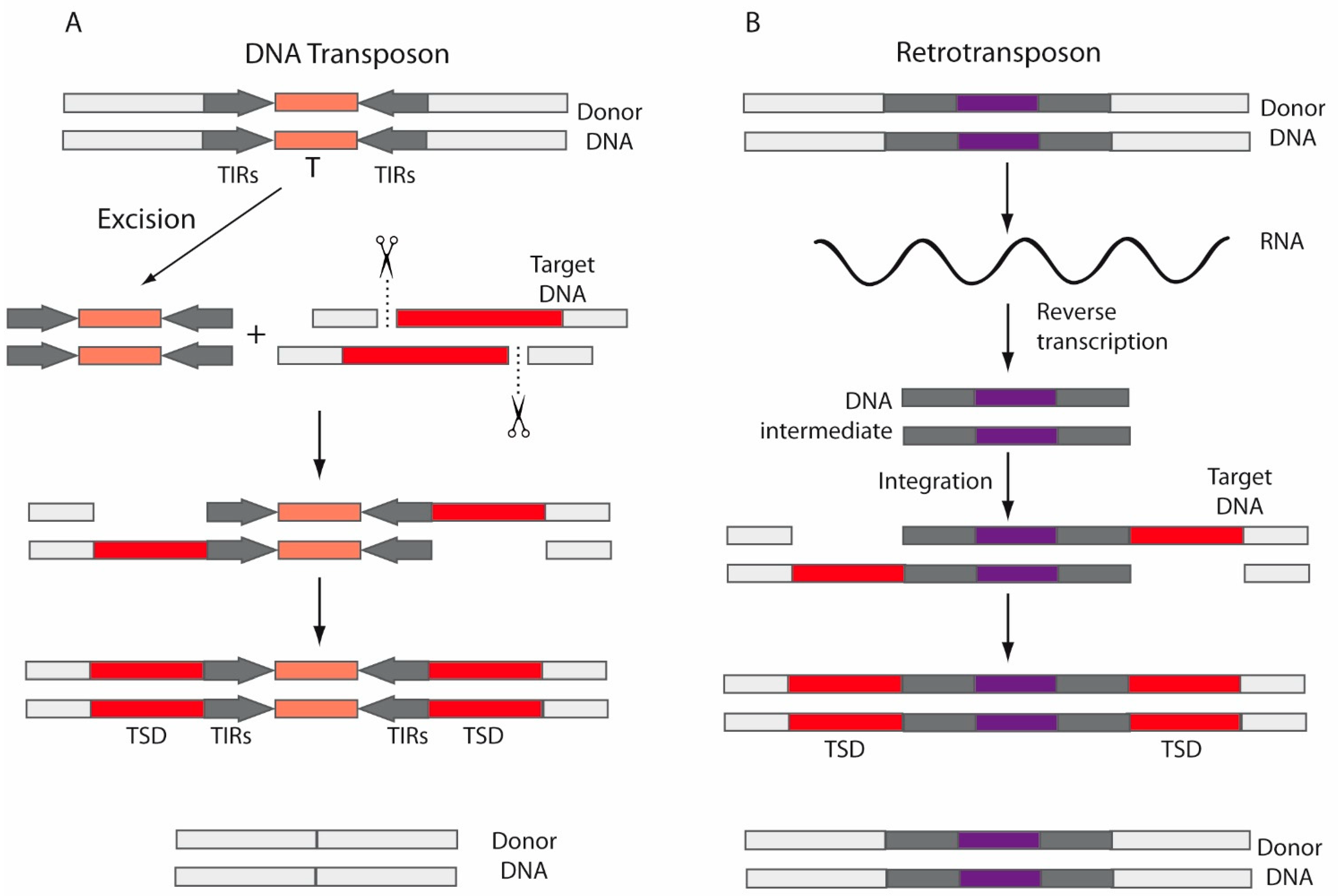
2. Transposable Elements
3. Transposable Elements, Exaptation, and Human Evolution
4. Transposable Elements and Human Disease
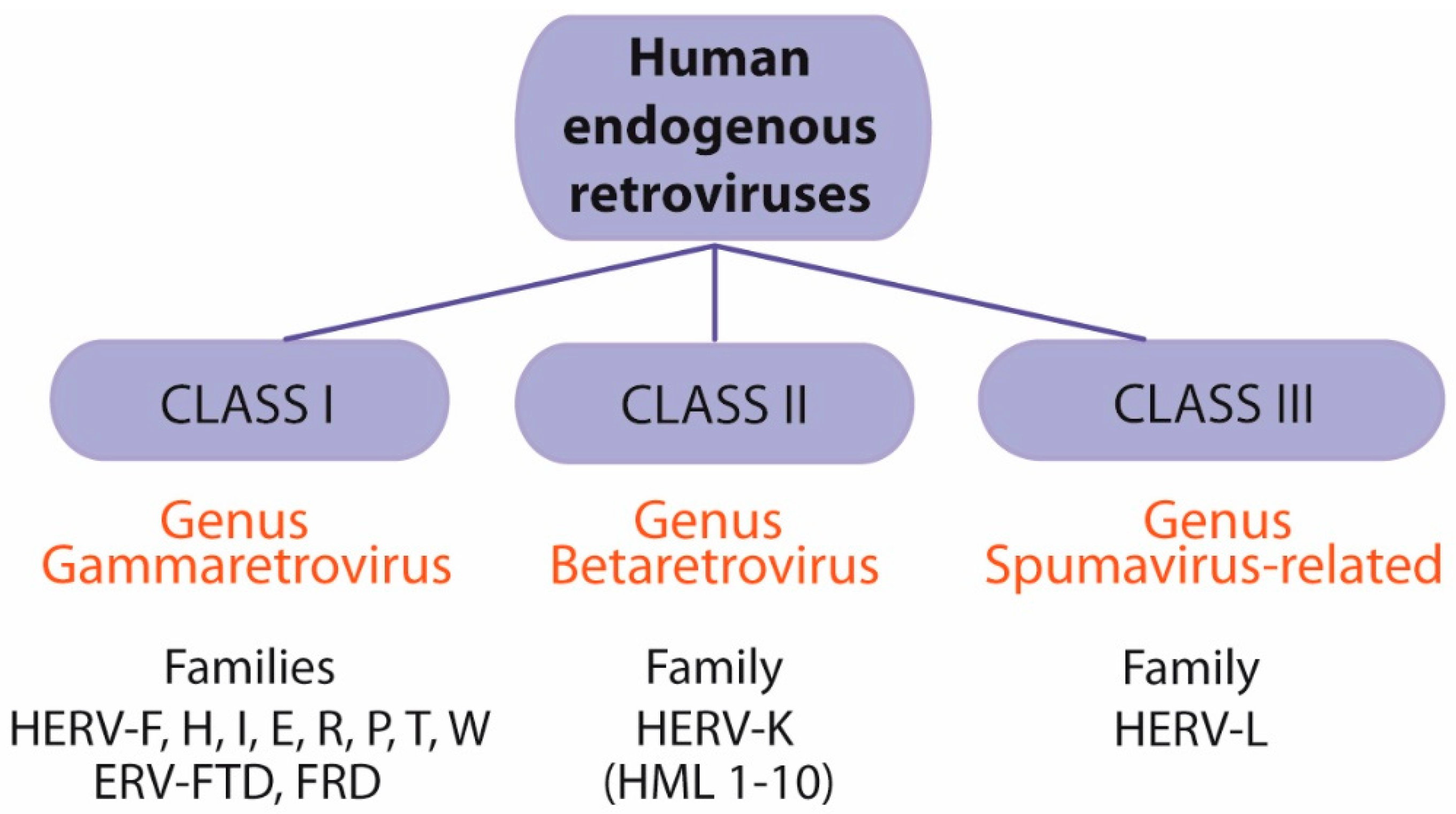
5. Human Endogenous Retroviruses (HERVs)
5.1. Endogenous Retroviruses and Multiple Sclerosis
5.1.1. Multiple Sclerosis-Associated Retrovirus
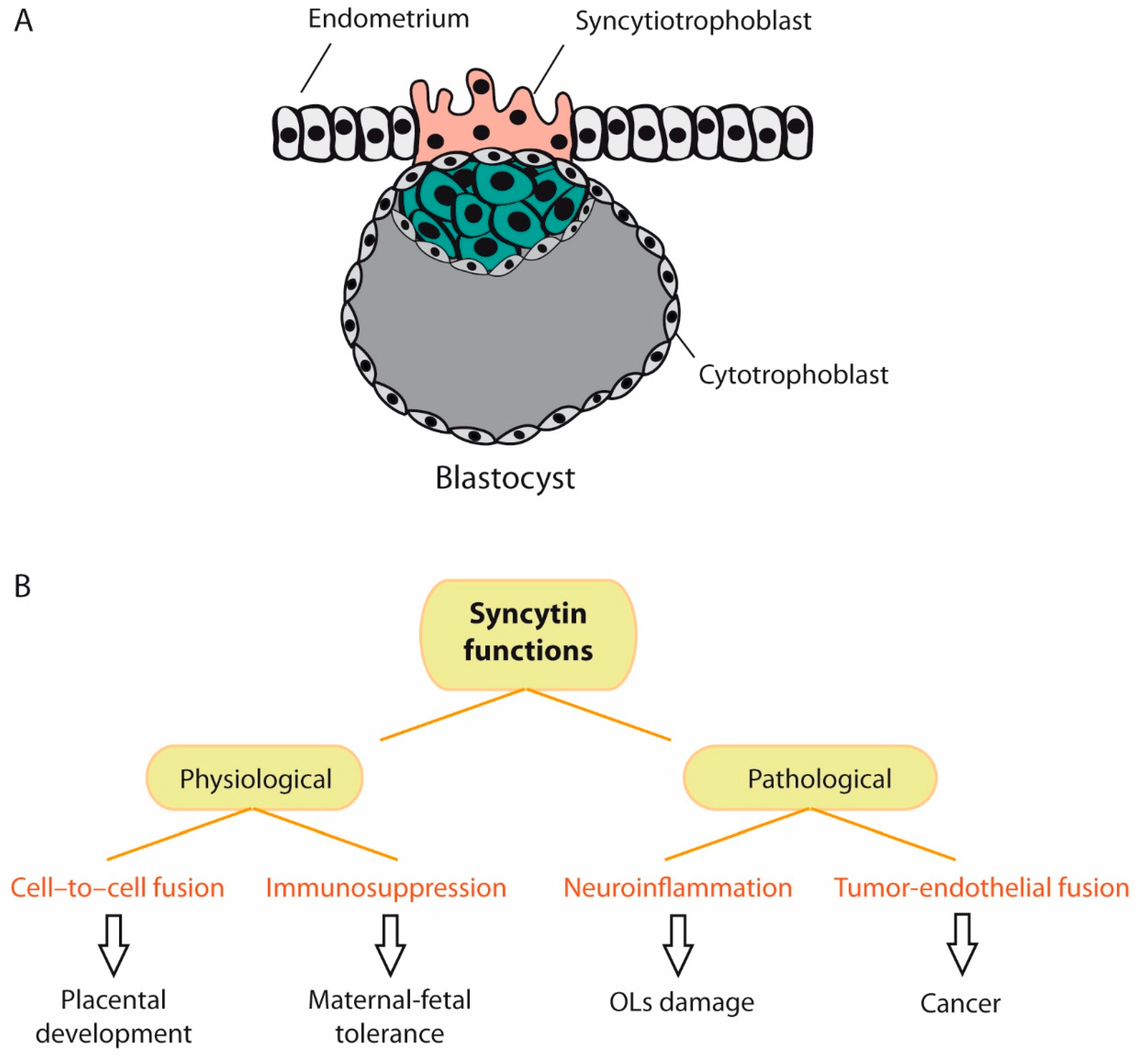
5.1.2. ERVWE1/Syncytin-1
5.1.3. HERV-H
5.2. Herpesviruses and MS
5.3. Herpesviruses, HERVs and MS
6. HSV-1 and HERVs: Implications for MS
6.1. HSV-1 and MSRV
6.2. HSV-1 and ERVWE1/Syncytin-1
6.3. HSV-1 and HERV-K
6.4. HSV-1 and HERV-H
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roizman, B.; Knipe, D.M.; Whitley, R. Herpes simplex viruses. In Fields Virology; Howley, D.M.K.a.P.M., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 2501–2601. [Google Scholar]
- Wald, A.; Corey, L. Persistence in the population: Epidemiology, transmission. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Karasneh, G.A.; Shukla, D. Herpes simplex virus infects most cell types in vitro: Clues to its success. Virol. J. 2011, 8, 481. [Google Scholar] [CrossRef] [PubMed]
- Roizman, B.; Zhou, G.; Du, T. Checkpoints in productive and latent infections with herpes simplex virus 1: Conceptualization of the issues. J. Neurovirology 2011, 17, 512–517. [Google Scholar] [CrossRef]
- Bello-Morales, R.; Andreu, S.; Lopez-Guerrero, J.A. The Role of Herpes Simplex Virus Type 1 Infection in Demyelination of the Central Nervous System. Int. J. Mol. Sci. 2020, 21, 5026. [Google Scholar] [CrossRef]
- Filippi, M.; Bar-Or, A.; Piehl, F.; Preziosa, P.; Solari, A.; Vukusic, S.; Rocca, M.A. Multiple sclerosis. Nat. Rev. Dis. Primers 2018, 4, 43. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, M.; Magliozzi, R.; Ciccarelli, O.; Geurts, J.J.; Reynolds, R.; Martin, R. Exploring the origins of grey matter damage in multiple sclerosis. Nat. Rev. Neurosci. 2015, 16, 147–158. [Google Scholar] [CrossRef]
- Minagar, A.; Alexander, J.S. Blood-brain barrier disruption in multiple sclerosis. Mult. Scler. 2003, 9, 540–549. [Google Scholar] [CrossRef]
- Derada Troletti, C.; de Goede, P.; Kamermans, A.; de Vries, H.E. Molecular alterations of the blood-brain barrier under inflammatory conditions: The role of endothelial to mesenchymal transition. Biochim. Et Biophys. Acta 2016, 1862, 452–460. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, C.; Lucas, R.; Taylor, B. Environmental risk factors for multiple sclerosis: A review with a focus on molecular mechanisms. Int. J. Mol. Sci. 2012, 13, 11718–11752. [Google Scholar] [CrossRef] [PubMed]
- Chan, V.S. Epigenetics in Multiple Sclerosis. Adv. Exp. Med. Biol. 2020, 1253, 309–374. [Google Scholar] [CrossRef]
- Didonna, A.; Oksenberg, J.R. The Genetics of Multiple Sclerosis. In Multiple Sclerosis: Perspectives in Treatment and Pathogenesis; Zagon, I.S., McLaughlin, P.J., Eds.; Codon Publications: Brisbane, Australia, 2017. [Google Scholar] [CrossRef]
- Donati, D. Viral infections and multiple sclerosis. Drug Discov. Today. Dis. Models 2020. [Google Scholar] [CrossRef] [PubMed]
- Tarlinton, R.E.; Martynova, E.; Rizvanov, A.A.; Khaiboullina, S.; Verma, S. Role of Viruses in the Pathogenesis of Multiple Sclerosis. Viruses 2020, 12, 643. [Google Scholar] [CrossRef]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef] [PubMed]
- Lavialle, C.; Cornelis, G.; Dupressoir, A.; Esnault, C.; Heidmann, O.; Vernochet, C.; Heidmann, T. Paleovirology of ′syncytins′, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. Ser. B. Biol. Sci. 2013, 368, 20120507. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Volkman, H.E.; Stetson, D.B. The enemy within: Endogenous retroelements and autoimmune disease. Nat. Immunol. 2014, 15, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, G.; Singh, M.; Ghanbarian, A.T.; Rasko, T.; Szvetnik, A.; Cai, H.; Besser, D.; Prigione, A.; Fuchs, N.V.; et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014, 516, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Medynets, M.; Johnson, K.R.; Doucet-O′Hare, T.T.; DiSanza, B.; Li, W.; Xu, Y.; Bagnell, A.; Tyagi, R.; Sampson, K.; et al. Regulation of stem cell function and neuronal differentiation by HERV-K via mTOR pathway. Proc. Natl. Acad. Sci. USA 2020, 117, 17842–17853. [Google Scholar] [CrossRef]
- Romer, C.; Singh, M.; Hurst, L.D.; Izsvak, Z. How to tame an endogenous retrovirus: HERVH and the evolution of human pluripotency. Curr. Opin. Virol. 2017, 25, 49–58. [Google Scholar] [CrossRef]
- Kury, P.; Nath, A.; Creange, A.; Dolei, A.; Marche, P.; Gold, J.; Giovannoni, G.; Hartung, H.P.; Perron, H. Human Endogenous Retroviruses in Neurological Diseases. Trends Mol. Med. 2018, 24, 379–394. [Google Scholar] [CrossRef]
- Groger, V.; Cynis, H. Human Endogenous Retroviruses and Their Putative Role in the Development of Autoimmune Disorders Such as Multiple Sclerosis. Front. Microbiol. 2018, 9, 265. [Google Scholar] [CrossRef] [PubMed]
- Ryan, F.P. Human endogenous retroviruses in health and disease: A symbiotic perspective. J. R. Soc. Med. 2004, 97, 560–565. [Google Scholar] [CrossRef]
- Chen, J.; Foroozesh, M.; Qin, Z. Transactivation of human endogenous retroviruses by tumor viruses and their functions in virus-associated malignancies. Oncogenesis 2019, 8, 6. [Google Scholar] [CrossRef]
- Morris, G.; Maes, M.; Murdjeva, M.; Puri, B.K. Do Human Endogenous Retroviruses Contribute to Multiple Sclerosis, and if So, How? Mol. Neurobiol. 2019, 56, 2590–2605. [Google Scholar] [CrossRef]
- Morandi, E.; Tanasescu, R.; Tarlinton, R.E.; Constantinescu, C.S.; Zhang, W.; Tench, C.; Gran, B. The association between human endogenous retroviruses and multiple sclerosis: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0172415. [Google Scholar] [CrossRef]
- Christensen, T. Association of human endogenous retroviruses with multiple sclerosis and possible interactions with herpes viruses. Rev. Med. Virol. 2005, 15, 179–211. [Google Scholar] [CrossRef]
- Christensen, T. Human endogenous retroviruses in the aetiology of MS. Acta Neurol. Scand. 2017, 136 (Suppl. 201), 18–21. [Google Scholar] [CrossRef]
- Lezhnyova, V.R.; Martynova, E.V.; Khaiboullin, T.I.; Urbanowicz, R.A.; Khaiboullina, S.F.; Rizvanov, A.A. The Relationship of the Mechanisms of the Pathogenesis of Multiple Sclerosis and the Expression of Endogenous Retroviruses. Biology 2020, 9, 464. [Google Scholar] [CrossRef]
- Morandi, E.; Tarlinton, R.E.; Gran, B. Multiple Sclerosis between Genetics and Infections: Human Endogenous Retroviruses in Monocytes and Macrophages. Front. Immunol. 2015, 6, 647. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.M.; Deslauriers, A.M.; Bhat, R.K.; Ellestad, K.K.; Power, C. Human endogenous retroviruses and multiple sclerosis: Innocent bystanders or disease determinants? Biochim. Et Biophys. Acta 2011, 1812, 162–176. [Google Scholar] [CrossRef]
- Tao, C.; Simpson, S., Jr.; Taylor, B.V.; van der Mei, I. Association between human herpesvirus & human endogenous retrovirus and MS onset & progression. J. Neurol. Sci. 2017, 372, 239–249. [Google Scholar] [CrossRef]
- Brudek, T.; Luhdorf, P.; Christensen, T.; Hansen, H.J.; Moller-Larsen, A. Activation of endogenous retrovirus reverse transcriptase in multiple sclerosis patient lymphocytes by inactivated HSV-1, HHV-6 and VZV. J. Neuroimmunol. 2007, 187, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, K.; Obojes, K.; Wengel, V.; Gronen, F.; Kim, K.S.; Perron, H.; Schneider-Schaulies, J.; Rieckmann, P. Regulation of human endogenous retrovirus W protein expression by herpes simplex virus type 1: Implications for multiple sclerosis. J. Neurovirology 2006, 12, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Bergallo, M.; Galliano, I.; Montanari, P.; Gambarino, S.; Mareschi, K.; Ferro, F.; Fagioli, F.; Tovo, P.A.; Ravanini, P. CMV induces HERV-K and HERV-W expression in kidney transplant recipients. J. Clin. Virol. 2015, 68, 28–31. [Google Scholar] [CrossRef]
- Tai, A.K.; Luka, J.; Ablashi, D.; Huber, B.T. HHV-6A infection induces expression of HERV-K18-encoded superantigen. J. Clin. Virol. 2009, 46, 47–48. [Google Scholar] [CrossRef] [PubMed]
- Sutkowski, N.; Conrad, B.; Thorley-Lawson, D.A.; Huber, B.T. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 2001, 15, 579–589. [Google Scholar] [CrossRef]
- Mameli, G.; Poddighe, L.; Mei, A.; Uleri, E.; Sotgiu, S.; Serra, C.; Manetti, R.; Dolei, A. Expression and activation by Epstein Barr virus of human endogenous retroviruses-W in blood cells and astrocytes: Inference for multiple sclerosis. PLoS ONE 2012, 7, e44991. [Google Scholar] [CrossRef]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Pray, L. Transposons, or Jumping Genes: Not Junk DNA? Nat. Educ. 2008, 1, 32. [Google Scholar]
- Piskurek, O.; Jackson, D.J. Transposable elements: From DNA parasites to architects of metazoan evolution. Genes 2012, 3, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Drongitis, D.; Aniello, F.; Fucci, L.; Donizetti, A. Roles of Transposable Elements in the Different Layers of Gene Expression Regulation. Int. J. Mol. Sci. 2019, 20, 5755. [Google Scholar] [CrossRef] [PubMed]
- Cordaux, R.; Batzer, M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009, 10, 691–703. [Google Scholar] [CrossRef]
- Wessler, S.R. Transposable elements and the evolution of eukaryotic genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 17600–17601. [Google Scholar] [CrossRef] [PubMed]
- Peaston, A.E. Retrotransposons of Vertebrates. In Encyclopedia of Virology, 3rd ed.; van Regenmortel, M.H.V., Mahy, B.W.J., Eds.; Academic Press: Cambridge, MA, USA, 2008. [Google Scholar]
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Perez, J.L.; Moran, J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015, 3, MDNA3-0061-2014. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Lopez, M.; Garcia-Perez, J.L. DNA transposons: Nature and applications in genomics. Curr. Genom. 2010, 11, 115–128. [Google Scholar] [CrossRef]
- Bannert, N.; Kurth, R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. USA 2004, 101 (Suppl. 2), 14572–14579. [Google Scholar] [CrossRef]
- Beauregard, A.; Curcio, M.J.; Belfort, M. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008, 42, 587–617. [Google Scholar] [CrossRef]
- Jonsson, M.E.; Garza, R.; Johansson, P.A.; Jakobsson, J. Transposable Elements: A Common Feature of Neurodevelopmental and Neurodegenerative Disorders. Trends Genet. Tig 2020, 36, 610–623. [Google Scholar] [CrossRef]
- Molaro, A.; Malik, H.S. Hide and seek: How chromatin-based pathways silence retroelements in the mammalian germline. Curr. Opin. Genet. Dev. 2016, 37, 51–58. [Google Scholar] [CrossRef]
- Deniz, O.; Frost, J.M.; Branco, M.R. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019, 20, 417–431. [Google Scholar] [CrossRef]
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which transposable elements are active in the human genome? Trends Genet Tig 2007, 23, 183–191. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Doucet-O’Hare, T.; Henderson, L.; Nath, A. Human endogenous retrovirus-K (HML-2): A comprehensive review. Crit. Rev. Microbiol. 2018, 44, 715–738. [Google Scholar] [CrossRef]
- Carducci, F.; Biscotti, M.A.; Barucca, M.; Canapa, A. Transposable elements in vertebrates: Species evolution and environmental adaptation. Eur. Zool. J. 2019, 86, 497–503. [Google Scholar] [CrossRef]
- Serrato-Capuchina, A.; Matute, D.R. The Role of Transposable Elements in Speciation. Genes 2018, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Cosby, R.L.; Chang, N.C.; Feschotte, C. Host-transposon interactions: Conflict, cooperation, and cooption. Genes Dev. 2019, 33, 1098–1116. [Google Scholar] [CrossRef]
- Werren, J.H. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 2), 10863–10870. [Google Scholar] [CrossRef]
- Agren, J.A.; Clark, A.G. Selfish genetic elements. PLoS Genet. 2018, 14, e1007700. [Google Scholar] [CrossRef]
- Joly-Lopez, Z.; Bureau, T.E. Exaptation of transposable element coding sequences. Curr. Opin. Genet. Dev. 2018, 49, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Aswad, A.; Katzourakis, A. Paleovirology and virally derived immunity. Trends Ecol. Evol. 2012, 27, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Vrba, E. Exaptation—A Missing Term in the Science of Form. Paleobiology 1982, 8, 4–15. [Google Scholar] [CrossRef]
- Cornelis, G.; Vernochet, C.; Carradec, Q.; Souquere, S.; Mulot, B.; Catzeflis, F.; Nilsson, M.A.; Menzies, B.R.; Renfree, M.B.; Pierron, G.; et al. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc. Natl. Acad. Sci. USA 2015, 112, E487–E496. [Google Scholar] [CrossRef]
- Jangam, D.; Feschotte, C.; Betran, E. Transposable Element Domestication As an Adaptation to Evolutionary Conflicts. Trends Genet. Tig 2017, 33, 817–831. [Google Scholar] [CrossRef]
- Payer, L.M.; Burns, K.H. Transposable elements in human genetic disease. Nat. Rev. Genet. 2019, 20, 760–772. [Google Scholar] [CrossRef]
- Ochoa Thomas, E.; Zuniga, G.; Sun, W.; Frost, B. Awakening the dark side: Retrotransposon activation in neurodegenerative disorders. Curr. Opin. Neurobiol. 2020, 61, 65–72. [Google Scholar] [CrossRef]
- Saleh, A.; Macia, A.; Muotri, A.R. Transposable Elements, Inflammation, and Neurological Disease. Front. Neurol. 2019, 10, 894. [Google Scholar] [CrossRef] [PubMed]
- Dolei, A.; Ibba, G.; Piu, C.; Serra, C. Expression of HERV Genes as Possible Biomarker and Target in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3706. [Google Scholar] [CrossRef]
- Li, W.; Lee, M.H.; Henderson, L.; Tyagi, R.; Bachani, M.; Steiner, J.; Campanac, E.; Hoffman, D.A.; von Geldern, G.; Johnson, K.; et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015, 7, 307ra153. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Liang, G.; Molloy, P.L.; Jones, P.A. DNA methylation enables transposable element-driven genome expansion. Proc. Natl. Acad. Sci. USA 2020, 117, 19359–19366. [Google Scholar] [CrossRef] [PubMed]
- Locke, W.J.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.R.; Fung, K.Y.C.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef]
- Ross, J.P.; Rand, K.N.; Molloy, P.L. Hypomethylation of repeated DNA sequences in cancer. Epigenomics 2010, 2, 245–269. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Gama-Sosa, M.A.; Slagel, V.A.; Trewyn, R.W.; Oxenhandler, R.; Kuo, K.C.; Gehrke, C.W.; Ehrlich, M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983, 11, 6883–6894. [Google Scholar] [CrossRef]
- Anwar, S.L.; Wulaningsih, W.; Lehmann, U. Transposable Elements in Human Cancer: Causes and Consequences of Deregulation. Int. J. Mol. Sci. 2017, 18, 974. [Google Scholar] [CrossRef]
- Mukamel, Z.; Tanay, A. Hypomethylation marks enhancers within transposable elements. Nat. Genet. 2013, 45, 717–718. [Google Scholar] [CrossRef]
- Xie, M.; Hong, C.; Zhang, B.; Lowdon, R.F.; Xing, X.; Li, D.; Zhou, X.; Lee, H.J.; Maire, C.L.; Ligon, K.L.; et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 2013, 45, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Bedrosian, T.A.; Quayle, C.; Novaresi, N.; Gage, F.H. Early life experience drives structural variation of neural genomes in mice. Science 2018, 359, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.A.; Bryan, T.; Rasheed, S.; Khan, A.S. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc. Natl. Acad. Sci. USA 1981, 78, 4892–4896. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.A. The discovery of endogenous retroviruses. Retrovirology 2006, 3, 67. [Google Scholar] [CrossRef]
- Tristem, M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 2000, 74, 3715–3730. [Google Scholar] [CrossRef]
- Patzke, S.; Lindeskog, M.; Munthe, E.; Aasheim, H.C. Characterization of a novel human endogenous retrovirus, HERV-H/F, expressed in human leukemia cell lines. Virology 2002, 303, 164–173. [Google Scholar] [CrossRef][Green Version]
- Barbulescu, M.; Turner, G.; Seaman, M.I.; Deinard, A.S.; Kidd, K.K.; Lenz, J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. Cb 1999, 9, 861–868. [Google Scholar] [CrossRef]
- Subramanian, R.P.; Wildschutte, J.H.; Russo, C.; Coffin, J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 2011, 8, 90. [Google Scholar] [CrossRef]
- Ma, W.; Hong, Z.; Liu, H.; Chen, X.; Ding, L.; Liu, Z.; Zhou, F.; Yuan, Y. Human Endogenous Retroviruses-K (HML-2) Expression Is Correlated with Prognosis and Progress of Hepatocellular Carcinoma. Biomed. Res. Int. 2016, 2016, 8201642. [Google Scholar] [CrossRef]
- Fischer, S.; Echeverria, N.; Moratorio, G.; Landoni, A.I.; Dighiero, G.; Cristina, J.; Oppezzo, P.; Moreno, P. Human endogenous retrovirus np9 gene is over expressed in chronic lymphocytic leukemia patients. Leuk. Res. Rep. 2014, 3, 70–72. [Google Scholar] [CrossRef] [PubMed]
- Serafino, A.; Balestrieri, E.; Pierimarchi, P.; Matteucci, C.; Moroni, G.; Oricchio, E.; Rasi, G.; Mastino, A.; Spadafora, C.; Garaci, E.; et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 2009, 315, 849–862. [Google Scholar] [CrossRef]
- Dolei, A. The aliens inside us: HERV-W endogenous retroviruses and multiple sclerosis. Mult. Scler. 2018, 24, 42–47. [Google Scholar] [CrossRef]
- Vargiu, L.; Rodriguez-Tome, P.; Sperber, G.O.; Cadeddu, M.; Grandi, N.; Blikstad, V.; Tramontano, E.; Blomberg, J. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 2016, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Galindo, R.; Kaplan, M.H.; Dube, D.; Gonzalez-Hernandez, M.J.; Chan, S.; Meng, F.; Dai, M.; Omenn, G.S.; Gitlin, S.D.; Markovitz, D.M. Human Endogenous Retrovirus Type K (HERV-K) Particles Package and Transmit HERV-K-Related Sequences. J. Virol. 2015, 89, 7187–7201. [Google Scholar] [CrossRef]
- Perron, H.; Jouvin-Marche, E.; Michel, M.; Ounanian-Paraz, A.; Camelo, S.; Dumon, A.; Jolivet-Reynaud, C.; Marcel, F.; Souillet, Y.; Borel, E.; et al. Multiple sclerosis retrovirus particles and recombinant envelope trigger an abnormal immune response in vitro, by inducing polyclonal Vbeta16 T-lymphocyte activation. Virology 2001, 287, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Gifford, R.J.; Blomberg, J.; Coffin, J.M.; Fan, H.; Heidmann, T.; Mayer, J.; Stoye, J.; Tristem, M.; Johnson, W.E. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 2018, 15, 59. [Google Scholar] [CrossRef]
- Groger, V.; Emmer, A.; Staege, M.S.; Cynis, H. Endogenous Retroviruses in Nervous System Disorders. Pharmaceuticals 2021, 14, 70. [Google Scholar] [CrossRef]
- Garson, J.A.; Usher, L.; Al-Chalabi, A.; Huggett, J.; Day, E.F.; McCormick, A.L. Quantitative analysis of human endogenous retrovirus-K transcripts in postmortem premotor cortex fails to confirm elevated expression of HERV-K RNA in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 2019, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Montojo, M.; Li, W.; Nath, A. Technical considerations in detection of HERV-K in amyotrophic lateral sclerosis: Selection of controls and the perils of qPCR. Acta Neuropathol. Commun. 2019, 7, 101. [Google Scholar] [CrossRef]
- Garson, J.A.; Usher, L.; Al-Chalabi, A.; Huggett, J.; Day, E.F.; McCormick, A.L. Response to the Letter from Garcia-Montojo and colleagues concerning our paper entitled, Quantitative analysis of human endogenous retrovirus-K transcripts in postmortem premotor cortex fails to confirm elevated expression of HERV-K RNA in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 2019, 7, 102. [Google Scholar] [CrossRef]
- Christensen, T.; Dissing Sorensen, P.; Riemann, H.; Hansen, H.J.; Moller-Larsen, A. Expression of sequence variants of endogenous retrovirus RGH in particle form in multiple sclerosis. Lancet 1998, 352, 1033. [Google Scholar] [CrossRef]
- Christensen, T.; Sorensen, P.D.; Hansen, H.J.; Moller-Larsen, A. Antibodies against a human endogenous retrovirus and the preponderance of env splice variants in multiple sclerosis patients. Mult. Scler. 2003, 9, 6–15. [Google Scholar] [CrossRef]
- Arru, G.; Mameli, G.; Astone, V.; Serra, C.; Huang, Y.M.; Link, H.; Fainardi, E.; Castellazzi, M.; Granieri, E.; Fernandez, M.; et al. Multiple Sclerosis and HERV-W/MSRV: A Multicentric Study. Int. J. Biomed. Sci. 2007, 3, 292–297. [Google Scholar]
- Serra, C.; Mameli, G.; Arru, G.; Sotgiu, S.; Rosati, G.; Dolei, A. In vitro modulation of the multiple sclerosis (MS)-associated retrovirus by cytokines: Implications for MS pathogenesis. J. Neurovirology 2003, 9, 637–643. [Google Scholar] [CrossRef]
- Petersen, T.; Moller-Larsen, A.; Thiel, S.; Brudek, T.; Hansen, T.K.; Christensen, T. Effects of interferon-beta therapy on innate and adaptive immune responses to the human endogenous retroviruses HERV-H and HERV-W, cytokine production, and the lectin complement activation pathway in multiple sclerosis. J. Neuroimmunol. 2009, 215, 108–116. [Google Scholar] [CrossRef]
- Mameli, G.; Serra, C.; Astone, V.; Castellazzi, M.; Poddighe, L.; Fainardi, E.; Neri, W.; Granieri, E.; Dolei, A. Inhibition of multiple-sclerosis-associated retrovirus as biomarker of interferon therapy. J. Neurovirology 2008, 14, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Rolland, A.; Jouvin-Marche, E.; Viret, C.; Faure, M.; Perron, H.; Marche, P.N. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J. Immunol. 2006, 176, 7636–7644. [Google Scholar] [CrossRef] [PubMed]
- Rolland, A.; Jouvin-Marche, E.; Saresella, M.; Ferrante, P.; Cavaretta, R.; Creange, A.; Marche, P.; Perron, H. Correlation between disease severity and in vitro cytokine production mediated by MSRV (multiple sclerosis associated retroviral element) envelope protein in patients with multiple sclerosis. J. Neuroimmunol. 2005, 160, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Kriesel, J.D.; Bhetariya, P.J.; Chan, B.K.; Wilson, T.; Fischer, K.F. Enrichment of Retroviral Sequences in Brain Tissue from Patients with Severe Demyelinating Diseases. J. Emerg. Dis. Virol. 2017, 3. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Rodriguez-Martin, E.; Ramos-Mozo, P.; Ortega-Madueno, I.; Dominguez-Mozo, M.I.; Arias-Leal, A.; Garcia-Martinez, M.A.; Casanova, I.; Galan, V.; Arroyo, R.; et al. Syncytin-1/HERV-W envelope is an early activation marker of leukocytes and is upregulated in multiple sclerosis patients. Eur. J. Immunol. 2020, 50, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Van Horssen, J.; van der Pol, S.; Nijland, P.; Amor, S.; Perron, H. Human endogenous retrovirus W in brain lesions: Rationale for targeted therapy in multiple sclerosis. Mult. Scler. Relat. Disord. 2016, 8, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Tramontano, E. Type W Human Endogenous Retrovirus (HERV-W) Integrations and Their Mobilization by L1 Machinery: Contribution to the Human Transcriptome and Impact on the Host Physiopathology. Viruses 2017, 9, 162. [Google Scholar] [CrossRef]
- Blond, J.L.; Beseme, F.; Duret, L.; Bouton, O.; Bedin, F.; Perron, H.; Mandrand, B.; Mallet, F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999, 73, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Mameli, G.; Astone, V.; Arru, G.; Marconi, S.; Lovato, L.; Serra, C.; Sotgiu, S.; Bonetti, B.; Dolei, A. Brains and peripheral blood mononuclear cells of multiple sclerosis (MS) patients hyperexpress MS-associated retrovirus/HERV-W endogenous retrovirus, but not Human herpesvirus 6. J. Gen. Virol. 2007, 88, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Tarlinton, R.; Wang, B.; Morandi, E.; Gran, B.; Khaiboullin, T.; Martynova, E.; Rizvanov, A.; Khaiboullina, S. Differential Expression of HERV-W in Peripheral Blood in Multiple Sclerosis and Healthy Patients in Two Different Ethnic Groups. Front. Pharmacol. 2019, 10, 1645. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.; Schichel, T.; Forster, M.; Tzekova, N.; Bernard, C.; van der Valk, P.; van Horssen, J.; Hartung, H.P.; Perron, H.; Kury, P. Human endogenous retrovirus type W envelope protein inhibits oligodendroglial precursor cell differentiation. Ann. Neurol. 2013, 74, 721–732. [Google Scholar] [CrossRef]
- Madeira, A.; Burgelin, I.; Perron, H.; Curtin, F.; Lang, A.B.; Faucard, R. MSRV envelope protein is a potent, endogenous and pathogenic agonist of human toll-like receptor 4: Relevance of GNbAC1 in multiple sclerosis treatment. J. Neuroimmunol. 2016, 291, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Kremer, D.; Gruchot, J.; Weyers, V.; Oldemeier, L.; Gottle, P.; Healy, L.; Ho Jang, J.; Kang, T.X.Y.; Volsko, C.; Dutta, R.; et al. pHERV-W envelope protein fuels microglial cell-dependent damage of myelinated axons in multiple sclerosis. Proc. Natl. Acad. Sci. USA 2019, 116, 15216–15225. [Google Scholar] [CrossRef] [PubMed]
- Brudek, T.; Christensen, T.; Aagaard, L.; Petersen, T.; Hansen, H.J.; Moller-Larsen, A. B cells and monocytes from patients with active multiple sclerosis exhibit increased surface expression of both HERV-H Env and HERV-W Env, accompanied by increased seroreactivity. Retrovirology 2009, 6, 104. [Google Scholar] [CrossRef]
- Lopez-Guerrero, J.A.; Ripa, I.; Andreu, S.; Bello-Morales, R. The Role of Extracellular Vesicles in Demyelination of the Central Nervous System. Int. J. Mol. Sci. 2020, 21, 9111. [Google Scholar] [CrossRef]
- Duperray, A.; Barbe, D.; Raguenez, G.; Weksler, B.B.; Romero, I.A.; Couraud, P.O.; Perron, H.; Marche, P.N. Inflammatory response of endothelial cells to a human endogenous retrovirus associated with multiple sclerosis is mediated by TLR4. Int. Immunol. 2015, 27, 545–553. [Google Scholar] [CrossRef]
- Kornmann, G.; Curtin, F. Temelimab, an IgG4 Anti-Human Endogenous Retrovirus Monoclonal Antibody: An Early Development Safety Review. Drug Saf. 2020, 43, 1287–1296. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Cornelis, G.; Funk, M.; Vernochet, C.; Leal, F.; Tarazona, O.A.; Meurice, G.; Heidmann, O.; Dupressoir, A.; Miralles, A.; Ramirez-Pinilla, M.P.; et al. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc. Natl. Acad. Sci. USA 2017, 114, E10991–E11000. [Google Scholar] [CrossRef]
- Zhuang, X.W.; Li, J.; Brost, B.C.; Xia, X.Y.; Chen, H.B.; Wang, C.X.; Jiang, S.W. Decreased expression and altered methylation of syncytin-1 gene in human placentas associated with preeclampsia. Curr. Pharm. Des. 2014, 20, 1796–1802. [Google Scholar] [CrossRef]
- Antony, J.M.; van Marle, G.; Opii, W.; Butterfield, D.A.; Mallet, F.; Yong, V.W.; Wallace, J.L.; Deacon, R.M.; Warren, K.; Power, C. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat. Neurosci. 2004, 7, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, J.; Zhu, F. Human Endogenous Retroviral Envelope Protein Syncytin-1 and Inflammatory Abnormalities in Neuropsychological Diseases. Front. Psychiatry 2018, 9, 422. [Google Scholar] [CrossRef]
- Bjerregaard, B.; Holck, S.; Christensen, I.J.; Larsson, L.I. Syncytin is involved in breast cancer-endothelial cell fusions. Cell. Mol. Life Sci. 2006, 63, 1906–1911. [Google Scholar] [CrossRef]
- Liu, C.; Xu, J.; Wen, F.; Yang, F.; Li, X.; Geng, D.; Li, L.; Chen, J.; Zheng, J. Upregulation of syncytin-1 promotes invasion and metastasis by activating epithelial-mesenchymal transition-related pathway in endometrial carcinoma. Oncotargets Ther. 2019, 12, 31–40. [Google Scholar] [CrossRef]
- Strick, R.; Ackermann, S.; Langbein, M.; Swiatek, J.; Schubert, S.W.; Hashemolhosseini, S.; Koscheck, T.; Fasching, P.A.; Schild, R.L.; Beckmann, M.W.; et al. Proliferation and cell-cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-beta. J. Mol. Med. 2007, 85, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.L.; Wang, M.; Xu, Z.; Huang, C.M.; Zhou, X.C.; Jiang, E.H.; Zhao, X.P.; Song, Y.; Song, K.; Shao, Z.; et al. Up-regulation of syncytin-1 contributes to TNF-alpha-enhanced fusion between OSCC and HUVECs partly via Wnt/beta-catenin-dependent pathway. Sci. Rep. 2017, 7, 40983. [Google Scholar] [CrossRef] [PubMed]
- Blaise, S.; de Parseval, N.; Benit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.P.; Chen, L.F.; Yang, S.R.; Chen, C.Y.; Ko, C.C.; Chang, G.D.; Chen, H. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biol. Reprod. 2008, 79, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Laufer, G.; Mayer, J.; Mueller, B.F.; Mueller-Lantzsch, N.; Ruprecht, K. Analysis of transcribed human endogenous retrovirus W env loci clarifies the origin of multiple sclerosis-associated retrovirus env sequences. Retrovirology 2009, 6, 37. [Google Scholar] [CrossRef]
- Antony, J.M.; Zhu, Y.; Izad, M.; Warren, K.G.; Vodjgani, M.; Mallet, F.; Power, C. Comparative expression of human endogenous retrovirus-W genes in multiple sclerosis. Aids Res. Hum. Retrovir. 2007, 23, 1251–1256. [Google Scholar] [CrossRef]
- Dolei, A.; Perron, H. The multiple sclerosis-associated retrovirus and its HERV-W endogenous family: A biological interface between virology, genetics, and immunology in human physiology and disease. J. Neurovirology 2009, 15, 4–13. [Google Scholar] [CrossRef]
- Christensen, T.; Dissing Sorensen, P.; Riemann, H.; Hansen, H.J.; Munch, M.; Haahr, S.; Moller-Larsen, A. Molecular characterization of HERV-H variants associated with multiple sclerosis. Acta Neurol. Scand. 2000, 101, 229–238. [Google Scholar] [CrossRef]
- Christensen, T.; Petersen, T.; Thiel, S.; Brudek, T.; Ellermann-Eriksen, S.; Moller-Larsen, A. Gene-environment interactions in multiple sclerosis: Innate and adaptive immune responses to human endogenous retrovirus and herpesvirus antigens and the lectin complement activation pathway. J. Neuroimmunol. 2007, 183, 175–188. [Google Scholar] [CrossRef]
- Alvarez-Lafuente, R.; Garcia-Montojo, M.; De Las Heras, V.; Dominguez-Mozo, M.I.; Bartolome, M.; Benito-Martin, M.S.; Arroyo, R. Herpesviruses and human endogenous retroviral sequences in the cerebrospinal fluid of multiple sclerosis patients. Mult. Scler. 2008, 14, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Kjellman, C.; Sjogren, H.O.; Widegren, B. HERV-F, a new group of human endogenous retrovirus sequences. J. Gen. Virol. 1999, 80 Pt 9, 2383–2392. [Google Scholar] [CrossRef]
- Laska, M.J.; Brudek, T.; Nissen, K.K.; Christensen, T.; Moller-Larsen, A.; Petersen, T.; Nexo, B.A. Expression of HERV-Fc1, a human endogenous retrovirus, is increased in patients with active multiple sclerosis. J. Virol. 2012, 86, 3713–3722. [Google Scholar] [CrossRef] [PubMed]
- Nexo, B.A.; Christensen, T.; Frederiksen, J.; Moller-Larsen, A.; Oturai, A.B.; Villesen, P.; Hansen, B.; Nissen, K.K.; Laska, M.J.; Petersen, T.S.; et al. The etiology of multiple sclerosis: Genetic evidence for the involvement of the human endogenous retrovirus HERV-Fc1. PLoS ONE 2011, 6, e16652. [Google Scholar] [CrossRef]
- Nexo, B.A.; Villesen, P.; Nissen, K.K.; Lindegaard, H.M.; Rossing, P.; Petersen, T.; Tarnow, L.; Hansen, B.; Lorenzen, T.; Horslev-Petersen, K.; et al. Are human endogenous retroviruses triggers of autoimmune diseases? Unveiling associations of three diseases and viral loci. Immunol. Res. 2016, 64, 55–63. [Google Scholar] [CrossRef] [PubMed]
- De la Hera, B.; Varade, J.; Garcia-Montojo, M.; Alcina, A.; Fedetz, M.; Alloza, I.; Astobiza, I.; Leyva, L.; Fernandez, O.; Izquierdo, G.; et al. Human endogenous retrovirus HERV-Fc1 association with multiple sclerosis susceptibility: A meta-analysis. PLoS ONE 2014, 9, e90182. [Google Scholar] [CrossRef] [PubMed]
- Ascherio, A.; Munger, K.L. Environmental risk factors for multiple sclerosis. Part I: The role of infection. Ann. Neurol. 2007, 61, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Correale, J.; Gaitan, M.I. Multiple sclerosis and environmental factors: The role of vitamin D, parasites, and Epstein-Barr virus infection. Acta Neurol. Scand. 2015, 132, 46–55. [Google Scholar] [CrossRef]
- Leibovitch, E.C.; Jacobson, S. Evidence linking HHV-6 with multiple sclerosis: An update. Curr. Opin. Virol. 2014, 9, 127–133. [Google Scholar] [CrossRef]
- Manouchehrinia, A.; Tanasescu, R.; Kareem, H.; Jerca, O.P.; Jabeen, F.; Shafei, R.; Breuer, J.; Neal, K.; Irving, W.; Constantinescu, C.S. Prevalence of a history of prior varicella/herpes zoster infection in multiple sclerosis. J. Neurovirology 2017, 23, 839–844. [Google Scholar] [CrossRef]
- Sotelo, J.; Ordonez, G.; Pineda, B.; Flores, J. The participation of varicella zoster virus in relapses of multiple sclerosis. Clin. Neurol. Neurosurg. 2014, 119, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, J.O.; Wohler, J.; Fenton, K.; Reich, D.S.; Jacobson, S. Oligoclonal bands in multiple sclerosis reactive against two herpesviruses and association with magnetic resonance imaging findings. Mult. Scler. 2014, 20, 27–34. [Google Scholar] [CrossRef]
- Rostrom, B.; Link, H.; Laurenzi, M.A.; Kam-Hansen, S.; Norrby, E.; Wahren, B. Viral antibody activity of oligoclonal and polyclonal immunoglobulins synthesized within the central nervous system in multiple sclerosis. Ann. Neurol. 1981, 9, 569–574. [Google Scholar] [CrossRef]
- Virtanen, J.O.; Pietilainen-Nicklen, J.; Uotila, L.; Farkkila, M.; Vaheri, A.; Koskiniemi, M. Intrathecal human herpesvirus 6 antibodies in multiple sclerosis and other demyelinating diseases presenting as oligoclonal bands in cerebrospinal fluid. J. Neuroimmunol. 2011, 237, 93–97. [Google Scholar] [CrossRef]
- Kristensson, K.; Svennerholm, B.; Persson, L.; Vahlne, A.; Lycke, E. Latent herpes simplex virus trigeminal ganglionic infection in mice and demyelination in the central nervous system. J. Neurol. Sci. 1979, 43, 253–263. [Google Scholar] [CrossRef]
- Kastrukoff, L.F.; Lau, A.S.; Kim, S.U. Multifocal CNS demyelination following peripheral inoculation with herpes simplex virus type 1. Ann. Neurol. 1987, 22, 52–59. [Google Scholar] [CrossRef]
- Kastrukoff, L.F.; Lau, A.S.; Leung, G.Y.; Walker, D.G.; Thomas, E.E. Herpes simplex virus type I (HSV I)-induced multifocal central nervous system (CNS) demyelination in mice. J. Neuropathol. Exp. Neurol. 1992, 51, 432–439. [Google Scholar] [CrossRef]
- Kastrukoff, L.F.; Lau, A.S.; Thomas, E.E. The effect of mouse strain on herpes simplex virus type 1 (HSV-1) infection of the central nervous system (CNS). Herpesviridae 2012, 3, 4. [Google Scholar] [CrossRef]
- Kristensson, K.; Svennerholm, B.; Vahlne, A.; Nilheden, E.; Persson, L.; Lycke, E. Virus-induced demyelination in herpes simplex virus-infected mice. J. Neurol. Sci. 1982, 53, 205–216. [Google Scholar] [CrossRef]
- Lee, D.H.; Zandian, M.; Kuo, J.; Mott, K.R.; Chen, S.; Arditi, M.; Ghiasi, H. Suppression of IL-12p70 formation by IL-2 or following macrophage depletion causes T-cell autoreactivity leading to CNS demyelination in HSV-1-infected mice. PLoS Pathog. 2017, 13, e1006401. [Google Scholar] [CrossRef]
- Wakisaka, H.; Hato, N.; Honda, N.; Takahashi, H.; Kisaki, H.; Murakami, S.; Gyo, K.; Mominoki, K.; Kobayashi, N.; Matsuda, S. Demyelination associated with HSV-1-induced facial paralysis. Exp. Neurol. 2002, 178, 68–79. [Google Scholar] [CrossRef]
- Boukhvalova, M.S.; Mortensen, E.; Mbaye, A.; Lopez, D.; Kastrukoff, L.; Blanco, J.C.G. Herpes Simplex Virus 1 Induces Brain Inflammation and Multifocal Demyelination in the Cotton Rat Sigmodon hispidus. J. Virol. 2019, 94. [Google Scholar] [CrossRef]
- Gudnadottir, M.; Helgadottir, H.; Bjarnason, O.; Jonsdottir, K. Virus Isolated from the Brain of a Patient with Multiple Sclerosis. Exp. Neurol. 1964, 9, 85–95. [Google Scholar] [CrossRef]
- Bergstrom, T.; Andersen, O.; Vahlne, A. Isolation of herpes simplex virus type 1 during first attack of multiple sclerosis. Ann. Neurol. 1989, 26, 283–285. [Google Scholar] [CrossRef]
- Sanders, V.J.; Felisan, S.; Waddell, A.; Tourtellotte, W.W. Detection of herpesviridae in postmortem multiple sclerosis brain tissue and controls by polymerase chain reaction. J. Neurovirology 1996, 2, 249–258. [Google Scholar] [CrossRef]
- Sanders, V.J.; Waddell, A.E.; Felisan, S.L.; Li, X.; Conrad, A.J.; Tourtellotte, W.W. Herpes simplex virus in postmortem multiple sclerosis brain tissue. Arch. Neurol. 1996, 53, 125–133. [Google Scholar] [CrossRef]
- Ferro, M.T.; Franciotta, D.; Prelle, A.; Bestetti, A.; Cinque, P. Active intrathecal herpes simplex virus type 1 (HSV-1) and human herpesvirus-6 (HHV-6) infection at onset of multiple sclerosis. J. Neurovirology 2012, 18, 437–440. [Google Scholar] [CrossRef]
- Buscarinu, M.C.; Fornasiero, A.; Romano, S.; Ferraldeschi, M.; Renié, R.; Trasimeni, G.; Salvetti, M.; Ristori, G. Coincident onset of multiple sclerosis and Herpes simplex virus 1 encephalitis: A case report. Mult. Scler. Demyelinating Disord. 2017, 2, 6. [Google Scholar] [CrossRef][Green Version]
- Bech, E.; Lycke, J.; Gadeberg, P.; Hansen, H.J.; Malmestrom, C.; Andersen, O.; Christensen, T.; Ekholm, S.; Haahr, S.; Hollsberg, P.; et al. A randomized, double-blind, placebo-controlled MRI study of anti-herpes virus therapy in MS. Neurology 2002, 58, 31–36. [Google Scholar] [CrossRef]
- Ferrante, P.; Mancuso, R.; Pagani, E.; Guerini, F.R.; Calvo, M.G.; Saresella, M.; Speciale, L.; Caputo, D. Molecular evidences for a role of HSV-1 in multiple sclerosis clinical acute attack. J. Neurovirology 2000, 6 (Suppl. 2), S109–S114. [Google Scholar]
- Pietropaolo, V.; Fioriti, D.; Mischitelli, M.; Anzivino, E.; Santini, M.; Millefiorini, E.; Di Rezze, S.; Degener, A.M. Detection of human herpesviruses and polyomaviruses DNA in a group of patients with relapsing-remitting multiple sclerosis. New Microbiol. 2005, 28, 199–203. [Google Scholar] [PubMed]
- Czarnowska, A.; Kapica-Topczewska, K.; Zajkowska, O.; Swierzbinska, R.; Chorazy, M.; Tarasiuk, J.; Zajkowska, J.; Kochanowicz, J.; Kulakowska, A. Herpesviridae Seropositivity in Patients with Multiple Sclerosis: First Polish Study. Eur. Neurol. 2018, 80, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Merelli, E.; Bedin, R.; Sola, P.; Barozzi, P.; Mancardi, G.L.; Ficarra, G.; Franchini, G. Human herpes virus 6 and human herpes virus 8 DNA sequences in brains of multiple sclerosis patients, normal adults and children. J. Neurol. 1997, 244, 450–454. [Google Scholar] [CrossRef]
- Liedtke, W.; Malessa, R.; Faustmann, P.M.; Eis-Hubinger, A.M. Human herpesvirus 6 polymerase chain reaction findings in human immunodeficiency virus associated neurological disease and multiple sclerosis. J. Neurovirology 1995, 1, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Challoner, P.B.; Smith, K.T.; Parker, J.D.; MacLeod, D.L.; Coulter, S.N.; Rose, T.M.; Schultz, E.R.; Bennett, J.L.; Garber, R.L.; Chang, M.; et al. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc. Natl. Acad. Sci. USA 1995, 92, 7440–7444. [Google Scholar] [CrossRef]
- Pormohammad, A.; Azimi, T.; Falah, F.; Faghihloo, E. Relationship of human herpes virus 6 and multiple sclerosis: A systematic review and meta-analysis. J. Cell. Physiol. 2018, 233, 2850–2862. [Google Scholar] [CrossRef]
- Fotheringham, J.; Jacobson, S. Human herpesvirus 6 and multiple sclerosis: Potential mechanisms for virus-induced disease. Herpes 2005, 12, 4–9. [Google Scholar] [PubMed]
- Guan, Y.; Jakimovski, D.; Ramanathan, M.; Weinstock-Guttman, B.; Zivadinov, R. The role of Epstein-Barr virus in multiple sclerosis: From molecular pathophysiology to in vivo imaging. Neural Regen. Res. 2019, 14, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, K.; Wildemann, B.; Jarius, S. Low intrathecal antibody production despite high seroprevalence of Epstein-Barr virus in multiple sclerosis: A review of the literature. J. Neurol. 2018, 265, 239–252. [Google Scholar] [CrossRef]
- Lang, H.L.; Jacobsen, H.; Ikemizu, S.; Andersson, C.; Harlos, K.; Madsen, L.; Hjorth, P.; Sondergaard, L.; Svejgaard, A.; Wucherpfennig, K.; et al. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat. Immunol. 2002, 3, 940–943. [Google Scholar] [CrossRef]
- Ascherio, A.; Munger, K.L. Epstein-barr virus infection and multiple sclerosis: A review. J. Neuroimmune Pharmacol. 2010, 5, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Angelini, D.F.; Serafini, B.; Piras, E.; Severa, M.; Coccia, E.M.; Rosicarelli, B.; Ruggieri, S.; Gasperini, C.; Buttari, F.; Centonze, D.; et al. Increased CD8+ T cell response to Epstein-Barr virus lytic antigens in the active phase of multiple sclerosis. PLoS Pathog. 2013, 9, e1003220. [Google Scholar] [CrossRef]
- Pender, M.P.; Csurhes, P.A.; Burrows, J.M.; Burrows, S.R. Defective T-cell control of Epstein-Barr virus infection in multiple sclerosis. Clin. Transl. Immunol. 2017, 6, e126. [Google Scholar] [CrossRef]
- Lunemann, J.D.; Jelcic, I.; Roberts, S.; Lutterotti, A.; Tackenberg, B.; Martin, R.; Munz, C. EBNA1-specific T cells from patients with multiple sclerosis cross react with myelin antigens and co-produce IFN-gamma and IL-2. J. Exp. Med. 2008, 205, 1763–1773. [Google Scholar] [CrossRef]
- Marashi, S.M.; Mostafa, A.; Shoja, Z.; Nejati, A.; Shahmahmoodi, S.; Mollaei-Kandelous, Y.; Sahraian, M.A.; Jalilvand, S. Human herpesvirus 8 DNA detection and variant analysis in patients with multiple sclerosis. Virusdisease 2018, 29, 540–543. [Google Scholar] [CrossRef] [PubMed]
- Voisset, C.; Bouton, O.; Bedin, F.; Duret, L.; Mandrand, B.; Mallet, F.; Paranhos-Baccala, G. Chromosomal distribution and coding capacity of the human endogenous retrovirus HERV-W family. Aids Res. Hum. Retrovir. 2000, 16, 731–740. [Google Scholar] [CrossRef]
- Douville, R.N.; Nath, A. Human endogenous retroviruses and the nervous system. Handb. Clin. Neurol. 2014, 123, 465–485. [Google Scholar] [CrossRef]
- Perron, H.; Bernard, C.; Bertrand, J.B.; Lang, A.B.; Popa, I.; Sanhadji, K.; Portoukalian, J. Endogenous retroviral genes, Herpesviruses and gender in Multiple Sclerosis. J. Neurol. Sci. 2009, 286, 65–72. [Google Scholar] [CrossRef]
- Sutkowski, N.; Chen, G.; Calderon, G.; Huber, B.T. Epstein-Barr virus latent membrane protein LMP-2A is sufficient for transactivation of the human endogenous retrovirus HERV-K18 superantigen. J. Virol. 2004, 78, 7852–7860. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, F.C.; Lin, M.; Tai, A.; Chen, G.; Huber, B.T. Cutting edge: Epstein-Barr virus transactivates the HERV-K18 superantigen by docking to the human complement receptor 2 (CD21) on primary B cells. J. Immunol 2006, 177, 2056–2060. [Google Scholar] [CrossRef] [PubMed]
- Lafon, M.; Jouvin-Marche, E.; Marche, P.N.; Perron, H. Human viral superantigens: To be or not to be transactivated? Trends Immunol. 2002, 23, 238–239. [Google Scholar] [CrossRef]
- Perron, H.; Geny, C.; Laurent, A.; Mouriquand, C.; Pellat, J.; Perret, J.; Seigneurin, J.M. Leptomeningeal cell line from multiple sclerosis with reverse transcriptase activity and viral particles. Res. Virol. 1989, 140, 551–561. [Google Scholar] [CrossRef]
- Perron, H.; Lalande, B.; Gratacap, B.; Laurent, A.; Genoulaz, O.; Geny, C.; Mallaret, M.; Schuller, E.; Stoebner, P.; Seigneurin, J.M. Isolation of retrovirus from patients with multiple sclerosis. Lancet 1991, 337, 862–863. [Google Scholar] [CrossRef]
- Perron, H.; Gratacap, B.; Lalande, B.; Genoulaz, O.; Laurent, A.; Geny, C.; Mallaret, M.; Innocenti, P.; Schuller, E.; Stoebner, P.; et al. In vitro transmission and antigenicity of a retrovirus isolated from a multiple sclerosis patient. Res. Virol. 1992, 143, 337–350. [Google Scholar] [CrossRef]
- Perron, H.; Suh, M.; Lalande, B.; Gratacap, B.; Laurent, A.; Stoebner, P.; Seigneurin, J.M. Herpes simplex virus ICP0 and ICP4 immediate early proteins strongly enhance expression of a retrovirus harboured by a leptomeningeal cell line from a patient with multiple sclerosis. J. Gen. Virol. 1993, 74 Pt 1, 65–72. [Google Scholar] [CrossRef]
- Perron, H.; Garson, J.A.; Bedin, F.; Beseme, F.; Paranhos-Baccala, G.; Komurian-Pradel, F.; Mallet, F.; Tuke, P.W.; Voisset, C.; Blond, J.L.; et al. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The Collaborative Research Group on Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 1997, 94, 7583–7588. [Google Scholar] [CrossRef]
- Haahr, S.; Sommerlund, M.; Moller-Larsen, A.; Nielsen, R.; Hansen, H.J. Just another dubious virus in cells from a patient with multiple sclerosis? Lancet 1991, 337, 863–864. [Google Scholar] [CrossRef]
- Garson, J.A.; Tuke, P.W.; Giraud, P.; Paranhos-Baccala, G.; Perron, H. Detection of virion-associated MSRV-RNA in serum of patients with multiple sclerosis. Lancet 1998, 351, 33. [Google Scholar] [CrossRef]
- Dolei, A.; Serra, C.; Mameli, G.; Pugliatti, M.; Sechi, G.; Cirotto, M.C.; Rosati, G.; Sotgiu, S. Multiple sclerosis-associated retrovirus (MSRV) in Sardinian MS patients. Neurology 2002, 58, 471–473. [Google Scholar] [CrossRef]
- Nowak, J.; Januszkiewicz, D.; Pernak, M.; Liwen, I.; Zawada, M.; Rembowska, J.; Nowicka, K.; Lewandowski, K.; Hertmanowska, H.; Wender, M. Multiple sclerosis-associated virus-related pol sequences found both in multiple sclerosis and healthy donors are more frequently expressed in multiple sclerosis patients. J. Neurovirology 2003, 9, 112–117. [Google Scholar] [CrossRef]
- Perron, H.; Germi, R.; Bernard, C.; Garcia-Montojo, M.; Deluen, C.; Farinelli, L.; Faucard, R.; Veas, F.; Stefas, I.; Fabriek, B.O.; et al. Human endogenous retrovirus type W envelope expression in blood and brain cells provides new insights into multiple sclerosis disease. Mult. Scler. 2012, 18, 1721–1736. [Google Scholar] [CrossRef] [PubMed]
- Sotgiu, S.; Mameli, G.; Serra, C.; Zarbo, I.R.; Arru, G.; Dolei, A. Multiple sclerosis-associated retrovirus and progressive disability of multiple sclerosis. Mult. Scler. 2010, 16, 1248–1251. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Kwun, H.J.; Kim, H.S.; Jang, K.L. Activation of the human endogenous retrovirus W long terminal repeat by herpes simplex virus type 1 immediate early protein 1. Mol. Cells 2003, 15, 75–80. [Google Scholar] [PubMed]
- Mayer, J.; Sauter, M.; Racz, A.; Scherer, D.; Mueller-Lantzsch, N.; Meese, E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 1999, 21, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Tai, A.K.; O’Reilly, E.J.; Alroy, K.A.; Simon, K.C.; Munger, K.L.; Huber, B.T.; Ascherio, A. Human endogenous retrovirus-K18 Env as a risk factor in multiple sclerosis. Mult. Scler. 2008, 14, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Kwun, H.J.; Han, H.J.; Lee, W.J.; Kim, H.S.; Jang, K.L. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res. 2002, 86, 93–100. [Google Scholar] [CrossRef]
- Brudek, T.; Christensen, T.; Hansen, H.J.; Bobecka, J.; Moller-Larsen, A. Simultaneous presence of endogenous retrovirus and herpes virus antigens has profound effect on cell-mediated immune responses: Implications for multiple sclerosis. Aids Res. Hum. Retrovir. 2004, 20, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Brudek, T.; Christensen, T.; Hansen, H.J.; Petersen, T.; Moller-Larsen, A. Synergistic immune responses induced by endogenous retrovirus and herpesvirus antigens result in increased production of inflammatory cytokines in multiple sclerosis patients. Scand. J. Immunol. 2008, 67, 295–303. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bello-Morales, R.; Andreu, S.; Ripa, I.; López-Guerrero, J.A. HSV-1 and Endogenous Retroviruses as Risk Factors in Demyelination. Int. J. Mol. Sci. 2021, 22, 5738. https://doi.org/10.3390/ijms22115738
Bello-Morales R, Andreu S, Ripa I, López-Guerrero JA. HSV-1 and Endogenous Retroviruses as Risk Factors in Demyelination. International Journal of Molecular Sciences. 2021; 22(11):5738. https://doi.org/10.3390/ijms22115738
Chicago/Turabian StyleBello-Morales, Raquel, Sabina Andreu, Inés Ripa, and José Antonio López-Guerrero. 2021. "HSV-1 and Endogenous Retroviruses as Risk Factors in Demyelination" International Journal of Molecular Sciences 22, no. 11: 5738. https://doi.org/10.3390/ijms22115738
APA StyleBello-Morales, R., Andreu, S., Ripa, I., & López-Guerrero, J. A. (2021). HSV-1 and Endogenous Retroviruses as Risk Factors in Demyelination. International Journal of Molecular Sciences, 22(11), 5738. https://doi.org/10.3390/ijms22115738






