The Role of HECT-Type E3 Ligase in the Development of Cardiac Disease
Abstract
1. Introduction
2. Ubiquitylation
2.1. Ubiquitylation
2.2. HECT-Type E3 Ligase
2.2.1. NEDD4 Subfamily
2.2.2. HERC Subfamily
2.2.3. Other HECT E3s
3. Importance of Ubiquitylation in Cardiac Disease
4. Cardiac Hypertrophy and HECT-Type E3 Ligase
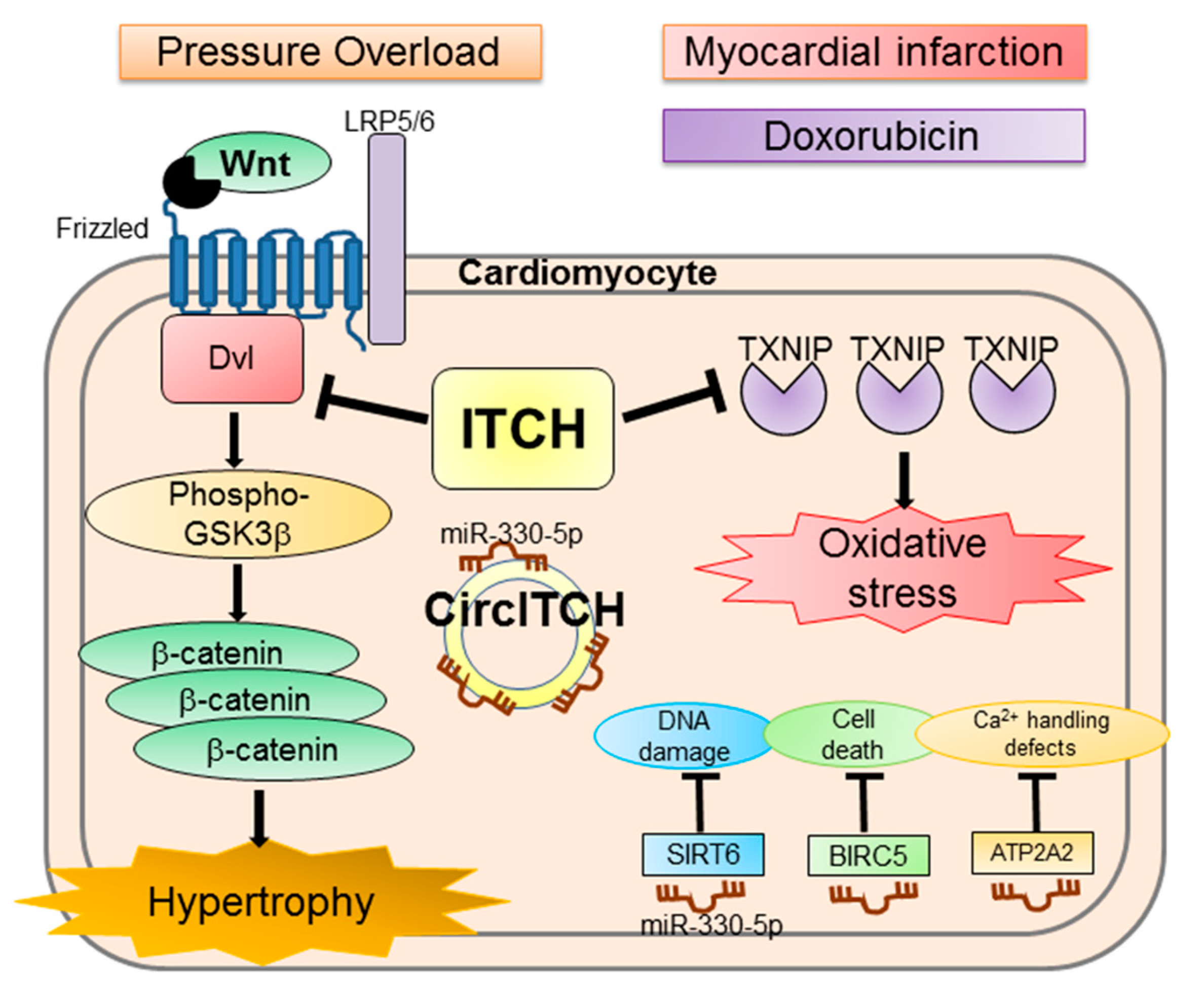
4.1. ITCH
4.2. NEDD4-2
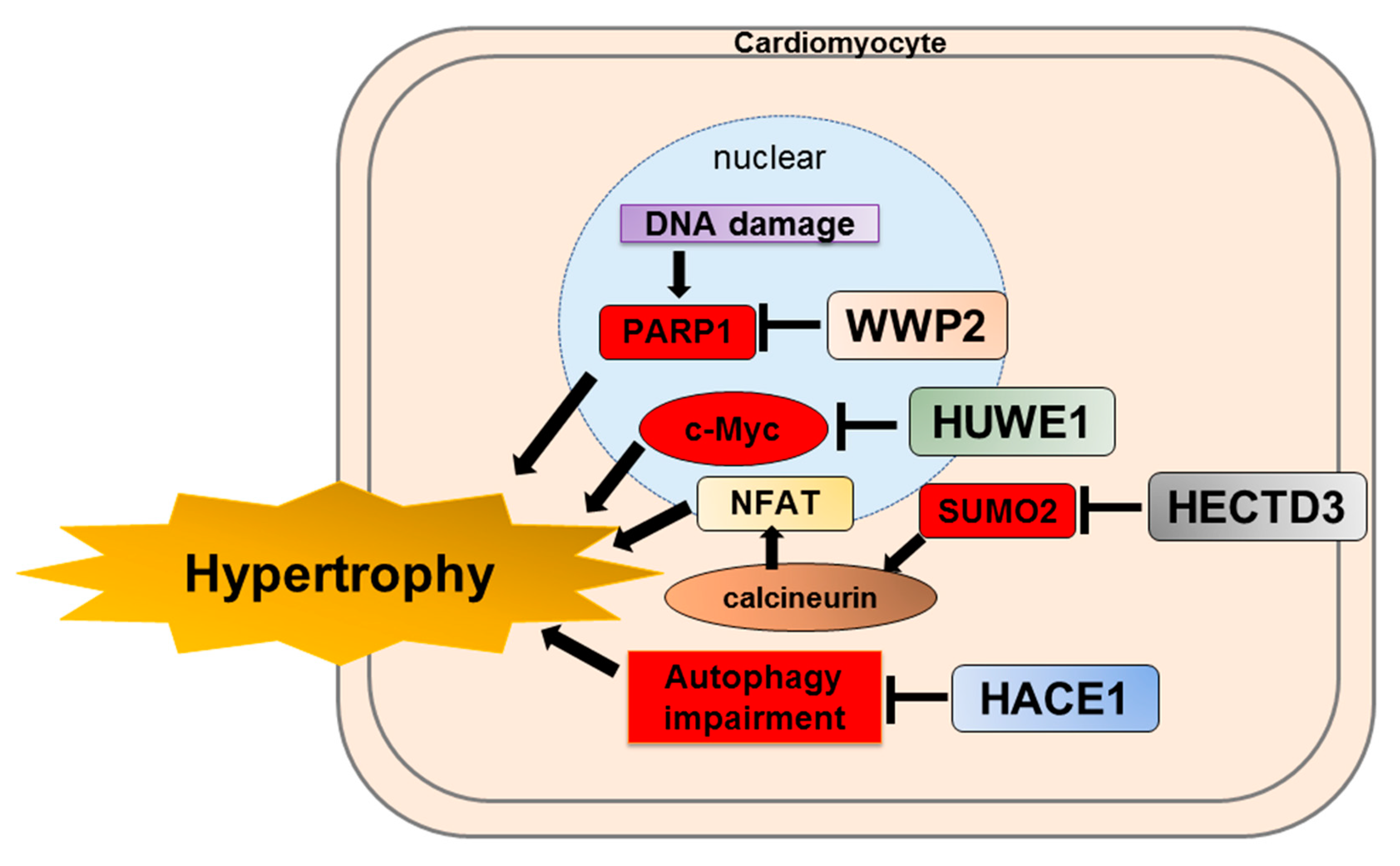
4.3. WWP2
4.4. HUWE1
4.5. HACE1
4.6. HECTD3
5. Cardiac Fibrosis and HECT-Type E3 Ligase
5.1. WWP2
5.2. SMURF1
5.3. SMURF2
6. HECT-Type E3 Ligase and HF with Preserved Ejection Fraction
WWP1
7. Ischemia/Reperfusion Injury and HECT-Type E3 Ligase
7.1. NEDD4-1
7.2. SMURF2
8. Other Cardiac Disease and HECT-Type E3 Ligase
8.1. ITCH and Doxorubicin Cardiotoxicity
8.2. NEDD4-2 and Arrhythmia
8.3. WWP1 and Arrhythmia
9. Drug Discovery
10. Limitation
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| BMP | bone morphogenetic protein |
| CHIP | carboxyl terminus of Hsp70 interacting protein |
| Dvl | dishevelled |
| ECM | extracellular matrix |
| E6AP | E6-associated protein |
| EZH2 | enhancer of zeste homolog 2 |
| HACE | HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase |
| HECT | homologous to E6AP C-terminus |
| HECTD | HECT domain E3 ubiquitin protein ligase |
| HERC | HECT and RLD domain containing E3 ubiquitin protein ligase |
| HF | heart failure |
| HFpEF | heart failure with preserved ejection fraction |
| HUWE | HECT, UBA and WWE domain containing E3 ubiquitin protein ligase |
| I/R | ischemia/reperfusion |
| JAK | Janus kinase |
| MDM2 | mouse double mutant 2 homolog |
| MuRF | muscle ring finger |
| Nav1.5 | voltage-dependent Na+ channel |
| NEDD4 | neural precursor cell expressed developmentally downregulated protein 4 |
| NFAT | nuclear factor of activated T-cells |
| NF- | nuclear factor kappa B |
| PARP1 | poly(ADP-ribose) polymerase-1 |
| PI3K | phosphatidylinositol 3-kinase |
| PTEN | phosphatase and tensin homolog deleted from chromosome 10 |
| RBRs | RING-between-RINGs |
| RING | really interesting new genes |
| RLDs | regulator of chromosome condensation-like domains |
| SERCA2 | sarcoplasmic/endoplasmic reticulum calcium ATPase 2 |
| SMAD | small mother against decapentaplegic |
| SMURF | SMAD specific E3 ubiquitin regulatory factor |
| STAT | signal transduction and activator of transcription |
| SUMO2 | small ubiquitin-like modifier 2 |
| TAC | transverse aortic constriction |
| TGF-β | transforming growth factor beta |
| TRIP12 | thyroid hormone receptor interactor 12 |
| UBA | ubiquitin-associated |
| UBE | ubiquitin–protein ligase E3 |
| UBR5 | ubiquitin protein ligase E3 component N-recognin 5 |
| WT | wild type |
| WWP | WW domain containing E3 ubiquitin protein ligase |
References
- Chen, J.; Normand, S.L.; Wang, Y.; Krumholz, H.M. National and regional trends in heart failure hospitalization and mortality rates for Medicare beneficiaries, 1998–2008. JAMA 2011, 306, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Jones, D.M.; Larson, M.G.; Leip, E.P.; Beiser, A.; D’Agostino, R.B.; Kannel, W.B.; Murabito, J.M.; Vasan, R.S.; Benjamin, E.J.; Levy, D. Lifetime risk for developing congestive heart failure: The Framingham Heart Study. Circulation 2002, 106, 3068–3072. [Google Scholar] [CrossRef]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef]
- Shimizu, I.; Minamino, T. Physiological and pathological cardiac hypertrophy. J. Mol. Cell Cardiol. 2016, 97, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Weekes, J.; Morrison, K.; Mullen, A.; Wait, R.; Barton, P.; Dunn, M.J. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics 2003, 3, 208–216. [Google Scholar] [CrossRef]
- Birks, E.J.; Latif, N.; Enesa, K.; Folkvang, T.; Luong, L.A.; Sarathchandra, P.; Khan, M.; Ovaa, H.; Terracciano, C.M.; Barton, P.J.; et al. Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovasc. Res. 2008, 79, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Kostin, S.; Pool, L.; Elsasser, A.; Hein, S.; Drexler, H.C.; Arnon, E.; Hayakawa, Y.; Zimmermann, R.; Bauer, E.; Klovekorn, W.P.; et al. Myocytes die by multiple mechanisms in failing human hearts. Circ. Res. 2003, 92, 715–724. [Google Scholar] [CrossRef]
- Schlossarek, S.; Frey, N.; Carrier, L. Ubiquitin-proteasome system and hereditary cardiomyopathies. J. Mol. Cell Cardiol. 2014, 71, 25–31. [Google Scholar] [CrossRef]
- Predmore, J.M.; Wang, P.; Davis, F.; Bartolone, S.; Westfall, M.V.; Dyke, D.B.; Pagani, F.; Powell, S.R.; Day, S.M. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation 2010, 121, 997–1004. [Google Scholar] [CrossRef]
- Naito, A.T.; Okada, S.; Minamino, T.; Iwanaga, K.; Liu, M.L.; Sumida, T.; Nomura, S.; Sahara, N.; Mizoroki, T.; Takashima, A.; et al. Promotion of CHIP-mediated p53 degradation protects the heart from ischemic injury. Circ. Res. 2010, 106, 1692–1702. [Google Scholar] [CrossRef] [PubMed]
- Schisler, J.C.; Rubel, C.E.; Zhang, C.; Lockyer, P.; Cyr, D.M.; Patterson, C. CHIP protects against cardiac pressure overload through regulation of AMPK. J. Clin. Investig. 2013, 123, 3588–3599. [Google Scholar] [CrossRef] [PubMed]
- Le, N.T.; Takei, Y.; Shishido, T.; Woo, C.H.; Chang, E.; Heo, K.S.; Lee, H.; Lu, Y.; Morrell, C.; Oikawa, M.; et al. p90RSK targets the ERK5-CHIP ubiquitin E3 ligase activity in diabetic hearts and promotes cardiac apoptosis and dysfunction. Circ. Res. 2012, 110, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Kedar, V.; Hwang, J.R.; McDonough, H.; Li, H.H.; Taylor, J.; Patterson, C. Muscle ring finger protein-1 inhibits PKC{epsilon} activation and prevents cardiomyocyte hypertrophy. J. Cell Biol. 2004, 167, 1147–1159. [Google Scholar] [CrossRef]
- Willis, M.S.; Schisler, J.C.; Li, L.; Rodriguez, J.E.; Hilliard, E.G.; Charles, P.C.; Patterson, C. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circ. Res. 2009, 105, 80–88. [Google Scholar] [CrossRef]
- Toth, A.; Nickson, P.; Qin, L.L.; Erhardt, P. Differential regulation of cardiomyocyte survival and hypertrophy by MDM2, an E3 ubiquitin ligase. J. Biol. Chem. 2006, 281, 3679–3689. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Mani, S.; Shiraishi, H.; Johnston, R.K.; Yamane, K.; Willey, C.D.; Cooper, G., 4th; Tuxworth, W.J.; Kuppuswamy, D. Enhanced ubiquitination of cytoskeletal proteins in pressure overloaded myocardium is accompanied by changes in specific E3 ligases. J. Mol. Cell Cardiol. 2006, 41, 669–679. [Google Scholar] [CrossRef]
- Willis, M.S.; Bevilacqua, A.; Pulinilkunnil, T.; Kienesberger, P.; Tannu, M.; Patterson, C. The role of ubiquitin ligases in cardiac disease. J. Mol. Cell Cardiol. 2014, 71, 43–53. [Google Scholar] [CrossRef]
- Zolk, O.; Schenke, C.; Sarikas, A. The ubiquitin-proteasome system: Focus on the heart. Cardiovasc. Res. 2006, 70, 410–421. [Google Scholar] [CrossRef]
- Parry, T.L.; Willis, M.S. Cardiac ubiquitin ligases: Their role in cardiac metabolism, autophagy, cardioprotection and therapeutic potential. Biochim. Biophys. Acta 2016, 1862, 2259–2269. [Google Scholar] [CrossRef]
- Wang, Y.; Argiles-Castillo, D.; Kane, E.I.; Zhou, A.; Spratt, D.E. HECT E3 ubiquitin ligases-emerging insights into their biological roles and disease relevance. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Mao, X.; Sethi, G.; Zhang, Z.; Wang, Q. The Emerging Roles of the HERC Ubiquitin Ligases in Cancer. Curr. Pharm. Des. 2018, 24, 1676–1681. [Google Scholar] [CrossRef]
- Scheffner, M.; Kumar, S. Mammalian HECT ubiquitin-protein ligases: Biological and pathophysiological aspects. Biochim. Biophys. Acta 2014, 1843, 61–74. [Google Scholar] [CrossRef]
- Weber, J.; Polo, S.; Maspero, E. HECT E3 Ligases: A Tale With Multiple Facets. Front. Physiol. 2019, 10, 370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, H.; Wu, B.; You, S.; Wu, S.; Lu, S.; Wang, P.; Cao, L.; Zhang, N.; Sun, Y. E3 Ubiquitin ligase NEDD4 familyregulatory network in cardiovascular disease. Int. J. Biol. Sci. 2020, 16, 2727–2740. [Google Scholar] [CrossRef]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef]
- Varshavsky, A. The ubiquitin system, an immense realm. Annu. Rev. Biochem. 2012, 81, 167–176. [Google Scholar] [CrossRef]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, A.P.; Liu, S.; Diez, D.; Miranda-Saavedra, D. The repertoires of ubiquitinating and deubiquitinating enzymes in eukaryotic genomes. Mol. Biol. Evol. 2013, 30, 1172–1187. [Google Scholar] [CrossRef]
- Hicke, L.; Dunn, R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003, 19, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Lee, D.H.; Goldberg, A.L. Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 1998, 8, 397–403. [Google Scholar] [CrossRef]
- Lyon, R.C.; Lange, S.; Sheikh, F. Breaking down protein degradation mechanisms in cardiac muscle. Trends Mol. Med. 2013, 19, 239–249. [Google Scholar] [CrossRef]
- Portbury, A.L.; Willis, M.S.; Patterson, C. Tearin’ up my heart: Proteolysis in the cardiac sarcomere. J. Biol. Chem. 2011, 286, 9929–9934. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef]
- Schubert, U.; Antón, L.C.; Gibbs, J.; Norbury, C.C.; Yewdell, J.W.; Bennink, J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 2000, 404, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Peralta, D.A.; Araya, A.; Busi, M.V.; Gomez-Casati, D.F. The E3 ubiquitin-ligase SEVEN IN ABSENTIA like 7 mono-ubiquitinates glyceraldehyde-3-phosphate dehydrogenase 1 isoform in vitro and is required for its nuclear localization in Arabidopsis thaliana. Int. J. Biochem. Cell Biol. 2016, 70, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Zhang, Z.; van Dam, H.; Zhang, L.; Zhou, F. Regulation of TGF-β Superfamily Signaling by SMAD Mono-Ubiquitination. Cells 2014, 3, 981–993. [Google Scholar] [CrossRef]
- Welchman, R.L.; Gordon, C.; Mayer, R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005, 6, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Beaudenon, S.; Howley, P.M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 1995, 92, 2563–2567. [Google Scholar] [CrossRef]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef]
- Kim, H.C.; Huibregtse, J.M. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell Biol. 2009, 29, 3307–3318. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991, 10, 4129–4135. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505. [Google Scholar] [CrossRef]
- Scheffner, M.; Staub, O. HECT E3s and human disease. BMC Biochem. 2007, 8 (Suppl. 1), S6. [Google Scholar] [CrossRef] [PubMed]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, K.P.; Sartori, M.A.; Kamadurai, H.B.; Ordureau, A.; Jiang, C.; Mercredi, P.Y.; Murchie, R.; Hu, J.; Persaud, A.; et al. System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes. Mol. Cell 2016, 62, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Rizo, J.; Sudhof, T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998, 273, 15879–15882. [Google Scholar] [CrossRef]
- Ingham, R.J.; Colwill, K.; Howard, C.; Dettwiler, S.; Lim, C.S.; Yu, J.; Hersi, K.; Raaijmakers, J.; Gish, G.; Mbamalu, G.; et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell Biol. 2005, 25, 7092–7106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Riling, C.; Kamadurai, H.; Kumar, S.; O’Leary, C.E.; Wu, K.P.; Manion, E.E.; Ying, M.; Schulman, B.A.; Oliver, P.M. Itch WW Domains Inhibit Its E3 Ubiquitin Ligase Activity by Blocking E2-E3 Ligase Trans-thiolation. J. Biol. Chem. 2015, 290, 23875–23887. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, F.R.; Ponstingl, H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 1991, 354, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Clarke, P.R. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 2000, 288, 1429–1432. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, M.E.; Mizzen, C.A.; Stukenberg, T.; Allis, C.D.; Macara, I.G. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 2001, 292, 1540–1543. [Google Scholar] [CrossRef] [PubMed]
- Sluimer, J.; Distel, B. Regulating the human HECT E3 ligases. Cell Mol. Life Sci. 2018, 75, 3121–3141. [Google Scholar] [CrossRef] [PubMed]
- Willis, M.S.; Patterson, C. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? N. Engl. J. Med. 2013, 368, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Patterson, C.; Ike, C.; Willis, P.W., 4th; Stouffer, G.A.; Willis, M.S. The bitter end: The ubiquitin-proteasome system and cardiac dysfunction. Circulation 2007, 115, 1456–1463. [Google Scholar] [CrossRef]
- Willis, M.S.; Patterson, C. Into the heart: The emerging role of the ubiquitin-proteasome system. J. Mol. Cell Cardiol. 2006, 41, 567–579. [Google Scholar] [CrossRef]
- Powell, S.R. The ubiquitin-proteasome system in cardiac physiology and pathology. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1–H19. [Google Scholar] [CrossRef]
- Brown, D.I.; Parry, T.L.; Willis, M.S. Ubiquitin Ligases and Posttranslational Regulation of Energy in the Heart: The Hand that Feeds. Compr. Physiol. 2017, 7, 841–862. [Google Scholar] [CrossRef]
- Kelly, S.M.; Vanslyke, J.K.; Musil, L.S. Regulation of ubiquitin-proteasome system mediated degradation by cytosolic stress. Mol. Biol. Cell 2007, 18, 4279–4291. [Google Scholar] [CrossRef]
- Bucciantini, M.; Calloni, G.; Chiti, F.; Formigli, L.; Nosi, D.; Dobson, C.M.; Stefani, M. Prefibrillar amyloid protein aggregates share common features of cytotoxicity. J. Biol. Chem. 2004, 279, 31374–31382. [Google Scholar] [CrossRef] [PubMed]
- Grune, T.; Merker, K.; Sandig, G.; Davies, K.J. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophys. Res. Commun. 2003, 305, 709–718. [Google Scholar] [CrossRef]
- Young, G.W.; Wang, Y.; Ping, P. Understanding proteasome assembly and regulation: Importance to cardiovascular medicine. Trends Cardiovasc. Med. 2008, 18, 93–98. [Google Scholar] [CrossRef][Green Version]
- Su, H.; Wang, X. The ubiquitin-proteasome system in cardiac proteinopathy: A quality control perspective. Cardiovasc. Res. 2010, 85, 253–262. [Google Scholar] [CrossRef]
- Dantuma, N.P.; Lindsten, K. Stressing the ubiquitin-proteasome system. Cardiovasc. Res. 2010, 85, 263–271. [Google Scholar] [CrossRef]
- Tsukamoto, O.; Minamino, T.; Kitakaze, M. Functional alterations of cardiac proteasomes under physiological and pathological conditions. Cardiovasc. Res. 2010, 85, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef]
- Li, H.H.; Kedar, V.; Zhang, C.; McDonough, H.; Arya, R.; Wang, D.Z.; Patterson, C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Investig. 2004, 114, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Aimo, A.; Gaggin, H.K.; Barison, A.; Emdin, M.; Januzzi, J.L., Jr. Imaging, Biomarker, and Clinical Predictors of Cardiac Remodeling in Heart Failure With Reduced Ejection Fraction. JACC Heart Fail. 2019, 7, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Arnon, E.; Kostin, S.; Schönburg, M.; Elsässer, A.; Polyakova, V.; Bauer, E.P.; Klövekorn, W.P.; Schaper, J. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: Structural deterioration and compensatory mechanisms. Circulation 2003, 107, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Razeghi, P.; Baskin, K.K.; Sharma, S.; Young, M.E.; Stepkowski, S.; Essop, M.F.; Taegtmeyer, H. Atrophy, hypertrophy, and hypoxemia induce transcriptional regulators of the ubiquitin proteasome system in the rat heart. Biochem. Biophys. Res. Commun. 2006, 342, 361–364. [Google Scholar] [CrossRef]
- Depre, C.; Wang, Q.; Yan, L.; Hedhli, N.; Peter, P.; Chen, L.; Hong, C.; Hittinger, L.; Ghaleh, B.; Sadoshima, J.; et al. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation 2006, 114, 1821–1828. [Google Scholar] [CrossRef]
- Sugden, P.H.; Clerk, A. Cellular mechanisms of cardiac hypertrophy. J. Mol. Med. 1998, 76, 725–746. [Google Scholar] [CrossRef] [PubMed]
- Allard, M.F.; Schonekess, B.O.; Henning, S.L.; English, D.R.; Lopaschuk, G.D. Contribution of oxidative metabolism and glycolysis to ATP production in hypertrophied hearts. Am. J. Physiol. 1994, 267, H742–H750. [Google Scholar] [CrossRef] [PubMed]
- Friehs, I.; Barillas, R.; Vasilyev, N.V.; Roy, N.; McGowan, F.X.; del Nido, P.J. Vascular endothelial growth factor prevents apoptosis and preserves contractile function in hypertrophied infant heart. Circulation 2006, 114, I290–I295. [Google Scholar] [CrossRef]
- Rohini, A.; Agrawal, N.; Koyani, C.N.; Singh, R. Molecular targets and regulators of cardiac hypertrophy. Pharmacol. Res. 2010, 61, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Booz, G.W.; Day, J.N.; Baker, K.M. Interplay between the cardiac renin angiotensin system and JAK-STAT signaling: Role in cardiac hypertrophy, ischemia/reperfusion dysfunction, and heart failure. J. Mol. Cell Cardiol. 2002, 34, 1443–1453. [Google Scholar] [CrossRef]
- Bergmann, M.W. WNT signaling in adult cardiac hypertrophy and remodeling: Lessons learned from cardiac development. Circ. Res. 2010, 107, 1198–1208. [Google Scholar] [CrossRef]
- Goto, J.; Otaki, Y.; Watanabe, T.; Kobayashi, Y.; Aono, T.; Watanabe, K.; Wanezaki, M.; Kutsuzawa, D.; Kato, S.; Tamura, H.; et al. HECT (Homologous to the E6-AP Carboxyl Terminus)-Type Ubiquitin E3 Ligase ITCH Attenuates Cardiac Hypertrophy by Suppressing the Wnt/beta-Catenin Signaling Pathway. Hypertension 2020, 76, 1868–1878. [Google Scholar] [CrossRef]
- Galiana-Simal, A.; Olivares-Alvaro, E.; Klett-Mingo, M.; Ruiz-Roso, M.B.; Ballesteros, S.; de Las Heras, N.; Fuller, P.J.; Lahera, V.; Martin-Fernandez, B. Proanthocyanidins block aldosterone-dependent up-regulation of cardiac gamma ENaC and Nedd4-2 inactivation via SGK1. J. Nutr. Biochem. 2016, 37, 13–19. [Google Scholar] [CrossRef]
- Shi, P.P.; Cao, X.R.; Sweezer, E.M.; Kinney, T.S.; Williams, N.R.; Husted, R.F.; Nair, R.; Weiss, R.M.; Williamson, R.A.; Sigmund, C.D.; et al. Salt-sensitive hypertension and cardiac hypertrophy in mice deficient in the ubiquitin ligase Nedd4-2. Am. J. Physiol. Renal. Physiol. 2008, 295, F462–F470. [Google Scholar] [CrossRef]
- Yang, M.H.; Wang, H.; Han, S.N.; Jia, X.; Zhang, S.; Dai, F.F.; Zhou, M.J.; Yin, Z.; Wang, T.Q.; Zang, M.X.; et al. Circular RNA expression in isoproterenol hydrochloride-induced cardiac hypertrophy. Aging 2020, 12, 2530–2544. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, Y.; Qian, H.; Wu, S.; Cao, L.; Sun, Y. Selective targeting of ubiquitination and degradation of PARP1 by E3 ubiquitin ligase WWP2 regulates isoproterenol-induced cardiac remodeling. Cell Death Differ. 2020, 27, 2605–2619. [Google Scholar] [CrossRef] [PubMed]
- Dadson, K.; Hauck, L.; Hao, Z.; Grothe, D.; Rao, V.; Mak, T.W.; Billia, F. The E3 ligase Mule protects the heart against oxidative stress and mitochondrial dysfunction through Myc-dependent inactivation of Pgc-1alpha and Pink1. Sci. Rep. 2017, 7, 41490. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, X.; Sharma, P.; Moon, M.; Sheftel, A.D.; Dawood, F.; Nghiem, M.P.; Wu, J.; Li, R.K.; Gramolini, A.O.; et al. HACE1-dependent protein degradation provides cardiac protection in response to haemodynamic stress. Nat. Commun. 2014, 5, 3430. [Google Scholar] [CrossRef]
- Rangrez, A.Y.; Borlepawar, A.; Schmiedel, N.; Deshpande, A.; Remes, A.; Kumari, M.; Bernt, A.; Christen, L.; Helbig, A.; Jungmann, A.; et al. The E3 ubiquitin ligase HectD3 attenuates cardiac hypertrophy and inflammation in mice. Commun. Biol. 2020, 3, 562. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Moreno-Moral, A.; Pesce, F.; Devapragash, N.; Mancini, M.; Heng, E.L.; Rotival, M.; Srivastava, P.K.; Harmston, N.; Shkura, K.; et al. WWP2 regulates pathological cardiac fibrosis by modulating SMAD2 signaling. Nat. Commun. 2019, 10, 3616. [Google Scholar] [CrossRef]
- Wang, S.; Sun, A.; Li, L.; Zhao, G.; Jia, J.; Wang, K.; Ge, J.; Zou, Y. Up-regulation of BMP-2 antagonizes TGF-β1/ROCK-enhanced cardiac fibrotic signalling through activation of Smurf1/Smad6 complex. J. Cell Mol. Med. 2012, 16, 2301–2310. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, H.; Mai, J.; Chen, Z.; Huang, T.; Wang, S.; Chen, Y.; Wang, J. Distinct Anti-Fibrotic Effects of Exosomes Derived from Endothelial Colony-Forming Cells Cultured Under Normoxia and Hypoxia. Med. Sci. Monit. 2018, 24, 6187–6199. [Google Scholar] [CrossRef]
- Cunnington, R.H.; Nazari, M.; Dixon, I.M. c-Ski, Smurf2, and Arkadia as regulators of TGF-beta signaling: New targets for managing myofibroblast function and cardiac fibrosis. Can. J. Physiol. Pharmacol. 2009, 87, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Qin, Y.; Chen, J.; Wei, L.; Huang, X.R.; Yu, X.; Lan, H.Y. Treatment of Hypertensive Heart Disease by Targeting Smad3 Signaling in Mice. Mol. Ther. Methods Clin. Dev. 2020, 18, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Matesic, L.E.; Freeburg, L.A.; Snyder, L.B.; Duncan, L.A.; Moore, A.; Perreault, P.E.; Zellars, K.N.; Goldsmith, E.C.; Spinale, F.G. The ubiquitin ligase WWP1 contributes to shifts in matrix proteolytic profiles and a myocardial aging phenotype with diastolic heart. Am. J. Physiol. Heart Circ. Physiol. 2020, 319, H765–H774. [Google Scholar] [CrossRef]
- Perry, W.L.; Hustad, C.M.; Swing, D.A.; O’Sullivan, T.N.; Jenkins, N.A.; Copeland, N.G. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 1998, 18, 143–146. [Google Scholar] [CrossRef]
- Infante, P.; Lospinoso Severini, L.; Bernardi, F.; Bufalieri, F.; Di Marcotullio, L. Targeting Hedgehog Signalling through the Ubiquitylation Process: The Multiple Roles of the HECT-E3 Ligase Itch. Cells 2019, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Melino, G.; Gallagher, E.; Aqeilan, R.I.; Knight, R.; Peschiaroli, A.; Rossi, M.; Scialpi, F.; Malatesta, M.; Zocchi, L.; Browne, G.; et al. Itch: A HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008, 15, 1103–1112. [Google Scholar] [CrossRef]
- Aki, D.; Zhang, W.; Yun-Cai, L. The E3 ligase Itch in immune regulation and beyond. Immunol. Rev. 2015, 266, 6–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Fan, X.; Yang, W.; Yu, B.; Kou, J.; Li, F. Captopril attenuates TAC-induced heart failure via inhibiting Wnt3a/beta-catenin and Jak2/Stat3 pathways. Biomed. Pharmacother. 2019, 113, 108780. [Google Scholar] [CrossRef] [PubMed]
- van de Schans, V.A.; van den Borne, S.W.; Strzelecka, A.E.; Janssen, B.J.; van der Velden, J.L.; Langen, R.C.; Wynshaw-Boris, A.; Smits, J.F.; Blankesteijn, W.M. Interruption of Wnt signaling attenuates the onset of pressure overload-induced cardiac hypertrophy. Hypertension 2007, 49, 473–480. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, C.; Hong, X.; Miao, J.; Liao, Y.; Hou, F.F.; Zhou, L.; Liu, Y. Wnt/beta-catenin signaling mediates both heart and kidney injury in type 2 cardiorenal syndrome. Kidney Int. 2019, 95, 815–829. [Google Scholar] [CrossRef]
- Gao, C.; Chen, Y.G. Dishevelled: The hub of Wnt signaling. Cell Signal 2010, 22, 717–727. [Google Scholar] [CrossRef]
- Mossinger, J.; Wieffer, M.; Krause, E.; Freund, C.; Gerth, F.; Krauss, M.; Haucke, V. Phosphatidylinositol 4-kinase IIalpha function at endosomes is regulated by the ubiquitin ligase Itch. EMBO Rep. 2012, 13, 1087–1094. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Doan, J.; Murray, J.; Molkentin, J.D.; Liu, Q. Transforming growth factor beta-activated kinase 1 signaling pathway critically regulates myocardial survival and remodeling. Circulation 2014, 130, 2162–2172. [Google Scholar] [CrossRef]
- Kumar, S.; Tomooka, Y.; Noda, M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 1992, 185, 1155–1161. [Google Scholar] [CrossRef]
- Ishigami, T.; Kino, T.; Minegishi, S.; Araki, N.; Umemura, M.; Ushio, H.; Saigoh, S.; Sugiyama, M. Regulators of Epithelial Sodium Channels in Aldosterone-Sensitive Distal Nephrons (ASDN): Critical Roles of Nedd4L/Nedd4-2 and Salt-Sensitive Hypertension. Int. J. Mol. Sci. 2020, 21, 3871. [Google Scholar] [CrossRef]
- Manning, J.A.; Kumar, S. Physiological Functions of Nedd4-2: Lessons from Knockout Mouse Models. Trends Biochem. Sci. 2018, 43, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, T.; Membrez, M.; Nicolas, C.S.; Pitard, B.; Staub, O.; Olesen, S.P.; Baro, I.; Abriel, H. The KCNQ1 potassium channel is down-regulated by ubiquitylating enzymes of the Nedd4/Nedd4-like family. Cardiovasc. Res. 2007, 74, 64–74. [Google Scholar] [CrossRef]
- Guo, J.; Wang, T.; Li, X.; Shallow, H.; Yang, T.; Li, W.; Xu, J.; Fridman, M.D.; Yang, X.; Zhang, S. Cell surface expression of human ether-a-go-go-related gene (hERG) channels is regulated by caveolin-3 protein via the ubiquitin ligase Nedd4-2. J. Biol. Chem. 2012, 287, 33132–33141. [Google Scholar] [CrossRef] [PubMed]
- Goel, P.; Manning, J.A.; Kumar, S. NEDD4-2 (NEDD4L): The ubiquitin ligase for multiple membrane proteins. Gene 2015, 557, 1–10. [Google Scholar] [CrossRef]
- Pirozzi, G.; McConnell, S.J.; Uveges, A.J.; Carter, J.M.; Sparks, A.B.; Kay, B.K.; Fowlkes, D.M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J. Biol. Chem. 1997, 272, 14611–14616. [Google Scholar] [CrossRef]
- Wood, J.D.; Yuan, J.; Margolis, R.L.; Colomer, V.; Duan, K.; Kushi, J.; Kaminsky, Z.; Kleiderlein, J.J.; Sharp, A.H.; Ross, C.A. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol. Cell Neurosci. 1998, 11, 149–160. [Google Scholar] [CrossRef]
- Chen, W.; Jiang, X.; Luo, Z. WWP2: A multifunctional ubiquitin ligase gene. Pathol. Oncol. Res. 2014, 20, 799–803. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.; Wang, B.; Zhang, J.; Zhao, Y.; Jin, Y. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol. Cell Biol. 2007, 27, 5296–5305. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Yamamoto, K.; He, X.; Otsuki, B.; Kim, Y.; Murao, H.; Soeda, T.; Tsumaki, N.; Deng, J.M.; Zhang, Z.; et al. Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat. Commun. 2011, 2, 251. [Google Scholar] [CrossRef]
- Yang, Y.; Liao, B.; Wang, S.; Yan, B.; Jin, Y.; Shu, H.B.; Wang, Y.Y. E3 ligase WWP2 negatively regulates TLR3-mediated innate immune response by targeting TRIF for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2013, 110, 5115–5120. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Jiang, H.; Xu, W.; Li, X.; Dempsey, D.R.; Zhang, X.; Devreotes, P.; Wolberger, C.; Amzel, L.M.; Gabelli, S.B.; et al. A Tunable Brake for HECT Ubiquitin Ligases. Mol. Cell 2017, 66, 345–357.e6. [Google Scholar] [CrossRef]
- Maddika, S.; Kavela, S.; Rani, N.; Palicharla, V.R.; Pokorny, J.L.; Sarkaria, J.N.; Chen, J. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 2011, 13, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, C.; Nakashima, D.; Kasamatsu, A.; Unozawa, M.; Shida-Sakazume, T.; Higo, M.; Ogawara, K.; Yokoe, H.; Shiiba, M.; Tanzawa, H.; et al. WWP2 is overexpressed in human oral cancer, determining tumor size and poor prognosis in patients: Downregulation of WWP2 inhibits the AKT signaling and tumor growth in mice. Oncoscience 2014, 1, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, J.; Luo, W.; Luo, Z.; Shi, S. WWP2 Is One Promising Novel Oncogene. Pathol. Oncol. Res. 2019, 25, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Pillai, J.B.; Russell, H.M.; Raman, J.; Jeevanandam, V.; Gupta, M.P. Increased expression of poly(ADP-ribose) polymerase-1 contributes to caspase-independent myocyte cell death during heart failure. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H486–H496. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhang, R.; Hong, H.; Yang, Z.; Sun, D.; Sun, S.; Guo, X.; Ye, J.; Li, Z.; Liu, P. The poly(ADP-ribosyl)ation of FoxO3 mediated by PARP1 participates in isoproterenol-induced cardiac hypertrophy. Biochim. Biophys. Acta 2016, 1863, 3027–3039. [Google Scholar] [CrossRef] [PubMed]
- Virag, L.; Robaszkiewicz, A.; Rodriguez-Vargas, J.M.; Oliver, F.J. Poly(ADP-ribose) signaling in cell death. Mol. Asp. Med. 2013, 34, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Gao, W.; Du, F.; Wang, X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 2005, 121, 1085–1095. [Google Scholar] [CrossRef]
- Kao, S.H.; Wu, H.T.; Wu, K.J. Ubiquitination by HUWE1 in tumorigenesis and beyond. J. Biomed. Sci. 2018, 25, 67. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Du, D.; Deng, Y.; Zhou, Y.; Sun, L.; Yuan, S. The structure and regulation of the E3 ubiquitin ligase HUWE1 and its biological functions in cancer. Investig. New Drugs 2020, 38, 515–524. [Google Scholar] [CrossRef]
- Chen, D.; Kon, N.; Li, M.; Zhang, W.; Qin, J.; Gu, W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 2005, 121, 1071–1083. [Google Scholar] [CrossRef]
- Bernassola, F.; Karin, M.; Ciechanover, A.; Melino, G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell 2008, 14, 10–21. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, P.; Fukuda, R.; Kumar, G.; Krishnamachary, B.; Zeller, K.I.; Dang, C.V.; Semenza, G.L. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 2007, 11, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Y.; Zeller, K.I.; Potter, J.J.; Wonsey, D.R.; O’Donnell, K.A.; Kim, J.W.; Yustein, J.T.; Lee, L.A.; Dang, C.V. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell Biol. 2005, 25, 6225–6234. [Google Scholar] [CrossRef]
- Ahuja, P.; Zhao, P.; Angelis, E.; Ruan, H.; Korge, P.; Olson, A.; Wang, Y.; Jin, E.S.; Jeffrey, F.M.; Portman, M.; et al. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J. Clin. Investig. 2010, 120, 1494–1505. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Evdokimova, V.; Melnyk, N.; Zhang, L.; Fernandez, C.V.; Grundy, P.E.; Leach, S.; Marra, M.A.; Brooks-Wilson, A.R.; Penninger, J.; et al. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms’ tumor versus normal kidney. Hum. Mol. Genet. 2004, 13, 2061–2074. [Google Scholar] [CrossRef]
- Zhang, L.; Anglesio, M.S.; O’Sullivan, M.; Zhang, F.; Yang, G.; Sarao, R.; Mai, P.N.; Cronin, S.; Hara, H.; Melnyk, N.; et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat. Med. 2007, 13, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Torrino, S.; Visvikis, O.; Doye, A.; Boyer, L.; Stefani, C.; Munro, P.; Bertoglio, J.; Gacon, G.; Mettouchi, A.; Lemichez, E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev. Cell 2011, 21, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Seipel, K.; O’Brien, S.P.; Iannotti, E.; Medley, Q.G.; Streuli, M. Tara, a novel F-actin binding protein, associates with the Trio guanine nucleotide exchange factor and regulates actin cytoskeletal organization. J. Cell Sci. 2001, 114, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, F.; Cheng, Z.; Kong, Y.; Chen, C. The role of E3 ubiquitin ligase HECTD3 in cancer and beyond. Cell Mol. Life Sci. 2020, 77, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Bernt, A.; Rangrez, A.Y.; Eden, M.; Jungmann, A.; Katz, S.; Rohr, C.; Muller, O.J.; Katus, H.A.; Sossalla, S.T.; Williams, T.; et al. Sumoylation-independent activation of Calcineurin-NFAT-signaling via SUMO2 mediates cardiomyocyte hypertrophy. Sci. Rep. 2016, 6, 35758. [Google Scholar] [CrossRef]
- Kuusisto, J.; Karja, V.; Sipola, P.; Kholova, I.; Peuhkurinen, K.; Jaaskelainen, P.; Naukkarinen, A.; Yla-Herttuala, S.; Punnonen, K.; Laakso, M. Low-grade inflammation and the phenotypic expression of myocardial fibrosis in hypertrophic cardiomyopathy. Heart 2012, 98, 1007–1013. [Google Scholar] [CrossRef]
- Kishore, R.; Verma, S.K. Roles of STATs signaling in cardiovascular diseases. JAKSTAT 2012, 1, 118–124. [Google Scholar] [CrossRef]
- Travers, J.G.; Kamal, F.A.; Robbins, J.; Yutzey, K.E.; Blaxall, B.C. Cardiac Fibrosis: The Fibroblast Awakens. Circ. Res. 2016, 118, 1021–1040. [Google Scholar] [CrossRef]
- Berk, B.C.; Fujiwara, K.; Lehoux, S. ECM remodeling in hypertensive heart disease. J. Clin. Investig. 2007, 117, 568–575. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Cardiac fibrosis. Cardiovasc. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ashrafian, H.; McKenna, W.J.; Watkins, H. Disease pathways and novel therapeutic targets in hypertrophic cardiomyopathy. Circ. Res. 2011, 109, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Bernaba, B.N.; Chan, J.B.; Lai, C.K.; Fishbein, M.C. Pathology of late-onset anthracycline cardiomyopathy. Cardiovasc. Pathol. 2010, 19, 308–311. [Google Scholar] [CrossRef]
- Asbun, J.; Villarreal, F.J. The pathogenesis of myocardial fibrosis in the setting of diabetic cardiomyopathy. J. Am. Coll Cardiol. 2006, 47, 693–700. [Google Scholar] [CrossRef]
- Kong, P.; Christia, P.; Frangogiannis, N.G. The pathogenesis of cardiac fibrosis. Cell Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef]
- Baum, J.; Duffy, H.S. Fibroblasts and myofibroblasts: What are we talking about? J. Cardiovasc. Pharmacol. 2011, 57, 376–379. [Google Scholar] [CrossRef]
- Watanabe, K.; Narumi, T.; Watanabe, T.; Otaki, Y.; Takahashi, T.; Aono, T.; Goto, J.; Toshima, T.; Sugai, T.; Wanezaki, M.; et al. The association between microRNA-21 and hypertension-induced cardiac remodeling. PLoS ONE 2020, 15, e0226053. [Google Scholar] [CrossRef]
- Walton, K.L.; Johnson, K.E.; Harrison, C.A. Targeting TGF-beta Mediated SMAD Signaling for the Prevention of Fibrosis. Front. Pharmacol. 2017, 8, 461. [Google Scholar] [CrossRef]
- Hu, H.H.; Chen, D.Q.; Wang, Y.N.; Feng, Y.L.; Cao, G.; Vaziri, N.D.; Zhao, Y.Y. New insights into TGF-beta/Smad signaling in tissue fibrosis. Chem. Biol. Interact. 2018, 292, 76–83. [Google Scholar] [CrossRef]
- Ross, S.; Hill, C.S. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 2008, 40, 383–408. [Google Scholar] [CrossRef]
- Moreno-Moral, A.; Pesce, F.; Behmoaras, J.; Petretto, E. Systems Genetics as a Tool to Identify Master Genetic Regulators in Complex Disease. Methods Mol. Biol. 2017, 1488, 337–362. [Google Scholar] [CrossRef]
- Zhu, H.; Kavsak, P.; Abdollah, S.; Wrana, J.L.; Thomsen, G.H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 1999, 400, 687–693. [Google Scholar] [CrossRef]
- Lin, X.; Liang, M.; Feng, X.H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J. Biol. Chem. 2000, 275, 36818–36822. [Google Scholar] [CrossRef]
- David, D.; Nair, S.A.; Pillai, M.R. Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochim. Biophys. Acta 2013, 1835, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Li, Y.; Han, D.; Dong, L. SMURF1, a promoter of tumor cell progression? Cancer Gene Ther. 2020. [Google Scholar] [CrossRef]
- Duenas, A.; Exposito, A.; Munoz, M.D.M.; de Manuel, M.J.; Camara-Morales, A.; Serrano-Osorio, F.; Garcia-Padilla, C.; Hernandez-Torres, F.; Dominguez, J.N.; Aranega, A.; et al. MiR-195 enhances cardiomyogenic differentiation of the proepicardium/septum transversum by Smurf1 and Foxp1 modulation. Sci. Rep. 2020, 10, 9334. [Google Scholar] [CrossRef] [PubMed]
- Koefoed, K.; Skat-Rørdam, J.; Andersen, P.; Warzecha, C.B.; Pye, M.; Andersen, T.A.; Ajbro, K.D.; Bendsen, E.; Narimatsu, M.; Vilhardt, F.; et al. The E3 ubiquitin ligase SMURF1 regulates cell-fate specification and outflow tract septation during mammalian heart development. Sci. Rep. 2018, 8, 9542. [Google Scholar] [CrossRef]
- Molina-Navarro, M.M.; Trivino, J.C.; Martinez-Dolz, L.; Lago, F.; Gonzalez-Juanatey, J.R.; Portoles, M.; Rivera, M. Functional networks of nucleocytoplasmic transport-related genes differentiate ischemic and dilated cardiomyopathies. A new therapeutic opportunity. PLoS ONE 2014, 9, e104709. [Google Scholar] [CrossRef]
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.F.; Coats, A.J.S.; Falk, V.; Gonzalez-Juanatey, J.R.; Harjola, V.P.; Jankowska, E.A.; et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2016, 37, 2129–2200. [Google Scholar] [CrossRef] [PubMed]
- Andersson, C.; Vasan, R.S. Epidemiology of heart failure with preserved ejection fraction. Heart Fail. Clin. 2014, 10, 377–388. [Google Scholar] [CrossRef]
- Dunlay, S.M.; Roger, V.L.; Redfield, M.M. Epidemiology of heart failure with preserved ejection fraction. Nat. Rev. Cardiol. 2017, 14, 591–602. [Google Scholar] [CrossRef]
- Lu, L.; Guo, J.; Hua, Y.; Huang, K.; Magaye, R.; Cornell, J.; Kelly, D.J.; Reid, C.; Liew, D.; Zhou, Y.; et al. Cardiac fibrosis in the ageing heart: Contributors and mechanisms. Clin. Exp. Pharmacol. Physiol. 2017, 44 (Suppl. 1), 55–63. [Google Scholar] [CrossRef]
- Upadhya, B.; Taffet, G.E.; Cheng, C.P.; Kitzman, D.W. Heart failure with preserved ejection fraction in the elderly: Scope of the problem. J. Mol. Cell Cardiol. 2015, 83, 73–87. [Google Scholar] [CrossRef]
- Butler, J.; Fonarow, G.C.; Zile, M.R.; Lam, C.S.; Roessig, L.; Schelbert, E.B.; Shah, S.J.; Ahmed, A.; Bonow, R.O.; Cleland, J.G.; et al. Developing therapies for heart failure with preserved ejection fraction: Current state and future directions. JACC Heart Fail. 2014, 2, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.; Zhang, H.; Boyce, B.F.; Xing, L. Ubiquitin E3 ligase Wwp1 negatively regulates osteoblast function by inhibiting osteoblast differentiation and migration. J. Bone Miner. Res. 2013, 28, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Chen, C. WWP1: A versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol. Life Sci. 2012, 69, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Yeung, B.; Ho, K.C.; Yang, X. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS ONE 2013, 8, e61027. [Google Scholar] [CrossRef] [PubMed]
- Komuro, A.; Imamura, T.; Saitoh, M.; Yoshida, Y.; Yamori, T.; Miyazono, K.; Miyazawa, K. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1). Oncogene 2004, 23, 6914–6923. [Google Scholar] [CrossRef]
- Mosser, E.A.; Kasanov, J.D.; Forsberg, E.C.; Kay, B.K.; Ney, P.A.; Bresnick, E.H. Physical and functional interactions between the transactivation domain of the hematopoietic transcription factor NF-E2 and WW domains. Biochemistry 1998, 37, 13686–13695. [Google Scholar] [CrossRef]
- Hahn, V.S.; Knutsdottir, H.; Luo, X.; Bedi, K.; Margulies, K.B.; Haldar, S.M.; Stolina, M.; Yin, J.; Khakoo, A.Y.; Vaishnav, J.; et al. Myocardial Gene Expression Signatures in Human Heart Failure With Preserved Ejection Fraction. Circulation 2021, 143, 120–134. [Google Scholar] [CrossRef]
- Kadowaki, S.; Shishido, T.; Sasaki, T.; Sugai, T.; Narumi, T.; Honda, Y.; Otaki, Y.; Kinoshita, D.; Takahashi, T.; Nishiyama, S.; et al. Deficiency of Senescence Marker Protein 30 Exacerbates Cardiac Injury after Ischemia/Reperfusion. Int. J. Mol. Sci. 2016, 17, 542. [Google Scholar] [CrossRef] [PubMed]
- Kukan, M. Emerging roles of proteasomes in ischemia-reperfusion injury of organs. J. Physiol. Pharmacol. 2004, 55, 3–15. [Google Scholar]
- Yellon, D.M.; Hausenloy, D.J. Myocardial reperfusion injury. N. Engl. J. Med. 2007, 357, 1121–1135. [Google Scholar] [CrossRef]
- Eefting, F.; Rensing, B.; Wigman, J.; Pannekoek, W.J.; Liu, W.M.; Cramer, M.J.; Lips, D.J.; Doevendans, P.A. Role of apoptosis in reperfusion injury. Cardiovasc. Res. 2004, 61, 414–426. [Google Scholar] [CrossRef]
- Zhao, Z. Inhibition of myocardial apoptosis reduces infarct size and improves regional contractile dysfunction during reperfusion. Cardiovasc. Res. 2003, 59, 132–142. [Google Scholar] [CrossRef]
- Shukla, S.K.; Rafiq, K. Proteasome biology and therapeutics in cardiac diseases. Transl. Res. 2019, 205, 64–76. [Google Scholar] [CrossRef]
- Alva, N.; Panisello-Roselló, A.; Flores, M.; Roselló-Catafau, J. Teresa Carbonell Ubiquitin-proteasome system and oxidative stress in liver transplantation. World J. Gastroenterol. 2018, 21, 3521–3530. [Google Scholar] [CrossRef] [PubMed]
- Hochrainer, K. Protein Modifications with Ubiquitin as Response to Cerebral Ischemia-Reperfusion Injury. Transl. Stroke Res. 2018, 9, 157–173. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, Z.; He, X.R.; Michael, L.H.; Patterson, C. CHIP, a cochaperone/ubiquitin ligase that regulates protein quality control, is required for maximal cardioprotection after myocardial infarction in mice. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H2836–H2842. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhang, P.; Gu, J.; Yu, Q.; Zhang, D. NEDD4-1 protects against ischaemia/reperfusion-induced cardiomyocyte apoptosis via the PI3K/Akt pathway. Apoptosis 2017, 22, 437–448. [Google Scholar] [CrossRef]
- Dong, W.; Xie, F.; Chen, X.Y.; Huang, W.L.; Zhang, Y.Z.; Luo, W.B.; Chen, J.; Xie, M.T.; Peng, X.P. Inhibition of Smurf2 translation by miR-322/503 protects from ischemia-reperfusion injury by modulating EZH2/Akt/GSK3beta signaling. Am. J. Physiol. Cell Physiol. 2019, 317, C253–C261. [Google Scholar] [CrossRef]
- Otaki, Y.; Takahashi, H.; Watanabe, T.; Funayama, A.; Netsu, S.; Honda, Y.; Narumi, T.; Kadowaki, S.; Hasegawa, H.; Honda, S.; et al. HECT-Type Ubiquitin E3 Ligase ITCH Interacts With Thioredoxin-Interacting Protein and Ameliorates Reactive Oxygen Species-Induced Cardiotoxicity. J. Am. Heart Assoc. 2016, 5. [Google Scholar] [CrossRef]
- Zhang, W.C.; Yang, J.H.; Liu, G.H.; Yang, F.; Gong, J.L.; Jia, M.G.; Zhang, M.J.; Zhao, L.S. miR-34b/c regulates doxorubicin-induced myocardial cell injury through ITCH. Cell Cycle 2019, 18, 3263–3274. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, X. Circular RNA ITCH mediates H(2)O(2)-induced myocardial cell apoptosis by targeting miR-17-5p via wnt/β-catenin signalling pathway. Int. J. Exp. Pathol. 2020, 102, 22–31. [Google Scholar] [CrossRef]
- Han, D.; Wang, Y.; Wang, Y.; Dai, X.; Zhou, T.; Chen, J.; Tao, B.; Zhang, J.; Cao, F. The Tumor-Suppressive Human Circular RNA CircITCH Sponges miR-330-5p to Ameliorate Doxorubicin-Induced Cardiotoxicity Through Upregulating SIRT6, Survivin, and SERCA2a. Circ. Res. 2020, 127, e108–e125. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Ning, F.; Du, Y.; Song, B.; Yang, D.; Salvage, S.C.; Wang, Y.; Fraser, J.A.; Zhang, S.; Ma, A.; et al. Calcium-dependent Nedd4-2 upregulation mediates degradation of the cardiac sodium channel Nav1.5: Implications for heart failure. Acta Physiol. 2017, 221, 44–58. [Google Scholar] [CrossRef]
- Basheer, W.A.; Harris, B.S.; Mentrup, H.L.; Abreha, M.; Thames, E.L.; Lea, J.B.; Swing, D.A.; Copeland, N.G.; Jenkins, N.A.; Price, R.L.; et al. Cardiomyocyte-specific overexpression of the ubiquitin ligase Wwp1 contributes to reduction in Connexin 43 and arrhythmogenesis. J. Mol. Cell Cardiol. 2015, 88, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Trotman, L.C.; Koppie, T.; Alimonti, A.; Chen, Z.; Gao, Z.; Wang, J.; Erdjument-Bromage, H.; Tempst, P.; Cordon-Cardo, C.; et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 2007, 128, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Y.; Xu, N.; Malyukova, A.; Scarlett, C.J.; Sun, Y.T.; Zhang, X.D.; Ling, D.; Su, S.P.; Nelson, C.; Chang, D.K.; et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013, 20, 503–514. [Google Scholar] [CrossRef]
- Platta, H.W.; Abrahamsen, H.; Thoresen, S.B.; Stenmark, H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012, 441, 399–406. [Google Scholar] [CrossRef]
- Tofaris, G.K.; Kim, H.T.; Hourez, R.; Jung, J.W.; Kim, K.P.; Goldberg, A.L. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 17004–17009. [Google Scholar] [CrossRef]
- Huang, X.; Chen, J.; Cao, W.; Yang, L.; Chen, Q.; He, J.; Yi, Q.; Huang, H.; Zhang, E.; Cai, Z. The many substrates and functions of NEDD4-1. Cell Death Dis. 2019, 10, 904. [Google Scholar] [CrossRef]
- Fouladkou, F.; Lu, C.; Jiang, C.; Zhou, L.; She, Y.; Walls, J.R.; Kawabe, H.; Brose, N.; Henkelman, R.M.; Huang, A.; et al. The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. J. Biol. Chem. 2010, 285, 6770–6780. [Google Scholar] [CrossRef]
- Duan, Q.; Madan, N.D.; Wu, J.; Kalisz, J.; Doshi, K.Y.; Haldar, S.M.; Liu, L.; Pierre, S.V. Role of phosphoinositide 3-kinase IA (PI3K-IA) activation in cardioprotection induced by ouabain preconditioning. J. Mol. Cell Cardiol. 2015, 80, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Guo, S.; Zhang, G.; Liu, Y.; Bi, S.; Wang, X.; Lu, Q. miR-26a-5p protects against myocardial ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT signaling pathway. Braz. J. Med. Biol. Res. 2020, 53, e9106. [Google Scholar] [CrossRef] [PubMed]
- Abdulkareem, I.H.; Blair, M. Phosphatase and tensin homologue deleted on chromosome. Niger. Med. J. 2013, 54, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.D.; Lum, M.A.; Xu, C.; Black, J.D.; Wang, X. Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4-1, in the insulin-like growth factor-1 response. J. Biol. Chem. 2013, 288, 1674–1684. [Google Scholar] [CrossRef]
- Kim, H.K.; Kang, S.W.; Jeong, S.H.; Kim, N.; Ko, J.H.; Bang, H.; Park, W.S.; Choi, T.H.; Ha, Y.R.; Lee, Y.S.; et al. Identification of potential target genes of cardioprotection against ischemia-reperfusion injury by express sequence tags analysis in rat hearts. J. Cardiol. 2012, 60, 98–110. [Google Scholar] [CrossRef]
- Li, C.H.; Chen, Y. Targeting EZH2 for cancer therapy: Progress and perspective. Curr. Protein Pept. Sci. 2015, 16, 559–570. [Google Scholar]
- Gan, L.; Xu, M.; Hua, R.; Tan, C.; Zhang, J.; Gong, Y.; Wu, Z.; Weng, W.; Sheng, W.; Guo, W. The polycomb group protein EZH2 induces epithelial-mesenchymal transition and pluripotent phenotype of gastric cancer cells by binding to PTEN promoter. J. Hematol. Oncol. 2018, 11, 9. [Google Scholar] [CrossRef]
- Bertrand Coiffier, E.L.; Briere, J.; Herbrecht, R.; Tilly, H.; Bouabdallah, R.; Morel, P.; van den Neste, E.; Salles, G.; Gaulard, P.; Reyes, F.; et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002, 346, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hortobagyi, G.N. Treatment of breast cancer. N. Engl. J. Med. 1998, 339, 974–984. [Google Scholar] [CrossRef] [PubMed]
- van Wijk, F.H.; Aapro, M.S.; Bolis, G.; Chevallier, B.; van der Burg, M.E.; Poveda, A.; de Oliveira, C.F.; Tumolo, S.; Scotto di Palumbo, V.; Piccart, M.; et al. Doxorubicin versus doxorubicin and cisplatin in endometrial carcinoma: Definitive results of a randomised study (55872) by the EORTC Gynaecological Cancer Group. Ann. Oncol. 2003, 14, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Zamorano, J.L.; Lancellotti, P.; Rodriguez Munoz, D.; Aboyans, V.; Asteggiano, R.; Galderisi, M.; Habib, G.; Lenihan, D.J.; Lip, G.Y.H.; Lyon, A.R.; et al. 2016 ESC Position Paper on cancer treatments and cardiovascular toxicity developed under the auspices of the ESC Committee for Practice Guidelines: The Task Force for cancer treatments and cardiovascular toxicity of the European Society of Cardiology (ESC). Eur. Heart J. 2016, 37, 2768–2801. [Google Scholar] [CrossRef]
- Prathumsap, N.; Shinlapawittayatorn, K.; Chattipakorn, S.C.; Chattipakorn, N. Effects of doxorubicin on the heart: From molecular mechanisms to intervention strategies. Eur. J. Pharmacol. 2020, 866, 172818. [Google Scholar] [CrossRef]
- Escobedo, A.; Gomes, T.; Aragon, E.; Martin-Malpartida, P.; Ruiz, L.; Macias, M.J. Structural basis of the activation and degradation mechanisms of the E3 ubiquitin ligase Nedd4L. Structure 2014, 22, 1446–1457. [Google Scholar] [CrossRef]
- Rook, M.B.; Evers, M.M.; Vos, M.A.; Bierhuizen, M.F. Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc. Res. 2012, 93, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.L.; Pfahnl, A.E.; Sanyal, S.; Jiao, Z.; Allen, J.; Banach, K.; Fahrenbach, J.; Weiss, D.; Taylor, W.R.; Zafari, A.M.; et al. Human heart failure is associated with abnormal C-terminal splicing variants in the cardiac sodium channel. Circ. Res. 2007, 101, 1146–1154. [Google Scholar] [CrossRef]
- Minegishi, S.; Ishigami, T.; Kawamura, H.; Kino, T.; Chen, L.; Nakashima-Sasaki, R.; Doi, H.; Azushima, K.; Wakui, H.; Chiba, Y.; et al. An Isoform of Nedd4-2 Plays a Pivotal Role in Electrophysiological Cardiac Abnormalities. Int. J. Mol. Sci. 2017, 18, 1268. [Google Scholar] [CrossRef]
- Sanguinetti, M.C.; Tristani-Firouzi, M. hERG potassium channels and cardiac arrhythmia. Nature 2006, 440, 463–469. [Google Scholar] [CrossRef]
- Fishman, G.I.; Chugh, S.S.; Dimarco, J.P.; Albert, C.M.; Anderson, M.E.; Bonow, R.O.; Buxton, A.E.; Chen, P.S.; Estes, M.; Jouven, X.; et al. Sudden cardiac death prediction and prevention: Report from a National Heart, Lung, and Blood Institute and Heart Rhythm Society Workshop. Circulation 2010, 122, 2335–2348. [Google Scholar] [CrossRef] [PubMed]
- Kuriachan, V.P.; Sumner, G.L.; Mitchell, L.B. Sudden cardiac death. Curr. Probl. Cardiol. 2015, 40, 133–200. [Google Scholar] [CrossRef] [PubMed]
- Remo, B.F.; Giovannone, S.; Fishman, G.I. Connexin43 cardiac gap junction remodeling: Lessons from genetically engineered murine models. J. Membr. Biol. 2012, 245, 275–281. [Google Scholar] [CrossRef]
- Severs, N.J.; Bruce, A.F.; Dupont, E.; Rothery, S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc. Res. 2008, 80, 9–19. [Google Scholar] [CrossRef]
- Einsele, H. Bortezomib. Recent Results Cancer Res. 2014, 201, 325–345. [Google Scholar] [CrossRef]
- Hedhli, N.; Lizano, P.; Hong, C.; Fritzky, L.F.; Dhar, S.K.; Liu, H.; Tian, Y.; Gao, S.; Madura, K.; Vatner, S.F.; et al. Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H1385–H1393. [Google Scholar] [CrossRef]
- Hacihanefioglu, A.; Tarkun, P.; Gonullu, E. Acute severe cardiac failure in a myeloma patient due to proteasome inhibitor bortezomib. Int. J. Hematol. 2008, 88, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Nalepa, G.; Rolfe, M.; Harper, J.W. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 2006, 5, 596–613. [Google Scholar] [CrossRef] [PubMed]
- Petroski, M.D. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008, 9 (Suppl. 1), S7. [Google Scholar] [CrossRef]
- Zhang, W.; Sidhu, S.S. Development of inhibitors in the ubiquitination cascade. FEBS Lett. 2014, 588, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Sun, Y. SCF E3 ubiquitin ligases as anticancer targets. Curr. Cancer Drug Targets 2011, 11, 347–356. [Google Scholar] [CrossRef]
- Huang, X.; Dixit, V.M. Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Res. 2016, 26, 484–498. [Google Scholar] [CrossRef]
- Mund, T.; Lewis, M.J.; Maslen, S.; Pelham, H.R. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA 2014, 111, 16736–16741. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Rotblat, B.; Ansell, K.; Amelio, I.; Caraglia, M.; Misso, G.; Bernassola, F.; Cavasotto, C.N.; Knight, R.A.; Ciechanover, A.; et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis. 2014, 5, e1203. [Google Scholar] [CrossRef]
- Bongiorno-Borbone, L.; Giacobbe, A.; Compagnone, M.; Eramo, A.; De Maria, R.; Peschiaroli, A.; Melino, G. Anti-tumoral effect of desmethylclomipramine in lung cancer stem cells. Oncotarget 2015, 6, 16926–16938. [Google Scholar] [CrossRef]
- Azakir, B.A.; Angers, A. Reciprocal regulation of the ubiquitin ligase Itch and the epidermal growth factor receptor signaling. Cell Signal 2009, 21, 1326–1336. [Google Scholar] [CrossRef] [PubMed]
- Santini, S.; Stagni, V.; Giambruno, R.; Fianco, G.; Di Benedetto, A.; Mottolese, M.; Pellegrini, M.; Barila, D. ATM kinase activity modulates ITCH E3-ubiquitin ligase activity. Oncogene 2014, 33, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Malhi, G.S.; Gessler, D.; Outhred, T. The use of lithium for the treatment of bipolar disorder: Recommendations from clinical practice guidelines. J. Affect. Disord. 2017, 217, 266–280. [Google Scholar] [CrossRef]
- Wang, X.; Fang, Z.; Wang, A.; Luo, C.; Cheng, X.; Lu, M. Lithium Suppresses Hedgehog Signaling via Promoting ITCH E3 Ligase Activity and Gli1-SUFU Interaction in PDA Cells. Front. Pharmacol. 2017, 8, 820. [Google Scholar] [CrossRef] [PubMed]
- Hamstra, S.I.; Kurgan, N.; Baranowski, R.W.; Qiu, L.; Watson, C.J.F.; Messner, H.N.; MacPherson, R.E.K.; MacNeil, A.J.; Roy, B.D.; Fajardo, V.A. Low-dose lithium feeding increases the SERCA2a-to-phospholamban ratio, improving SERCA function in murine left ventricles. Exp. Physiol. 2020, 105, 666–675. [Google Scholar] [CrossRef]
- Heitmeier, T.; Sydykov, A.; Lukas, C.; Vroom, C.; Korfei, M.; Petrovic, A.; Klingel, K.; Gunther, A.; Eickelberg, O.; Weissmann, N.; et al. Altered proteasome function in right ventricular hypertrophy. Cardiovasc. Res. 2020, 116, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Segura, A.M.; Frazier, O.H.; Buja, L.M. Fibrosis and heart failure. Heart Fail. Rev. 2014, 19, 173–185. [Google Scholar] [CrossRef] [PubMed]

| HECT-Type E3 Ligase | Substrate/Target | Main Findings | Reference |
|---|---|---|---|
| Cardiac hypertrophy | |||
| ITCH | Dishevelled | Cardiac-specific ITCH transgenic mice inhibited maladaptive hypertrophy via Wnt/β catenin signal inhibition. | [80] |
| NEDD4-2 | ENaC in kidney | Cardiac hypertrophy was observed in NEDD4-2 null mice on chronic high-salt diet. | [81,82] |
| Circular RNA WWP1 | ANF and miR-23a | Circular RNA WWP1 was dysregulated in the heart treated with isoproterenol. | [83] |
| WWP2 | PARP1 | WWP2 conditional knockout mice (MycCre+;WWP2Fl/Fl) exacerbated isoproterenol-induced cardiac hypertrophy. | [84] |
| E6AP | Increased myocardium E6AP expression after pressure overload. | [16] | |
| HUWE1 | c-myc | HUWE1 conditional knockout mice spontaneously developed cardiac hypertrophy. | [85] |
| HACE1 | Unknown | HACE1 conditional knockout mice spontaneously developed cardiac hypertrophy. | [86] |
| HECTD3 | SUMO2/STAT1 | AAV9-medited overexpression of HECTD3 inhibited pathological hypertrophy in mice. | [87] |
| Cardiac fibrosis | |||
| WWP2 | SMAD2 | WWP2mut/mut mice attenuated cardiac fibrosis after angiotensin II infusion and myocardial infarction. | [88] |
| SMURF1 | SMURF1 was involved in BMP-2 antagonization for TGF-β1 signal. SMURF1 was a target of miR-10b-5p, which inhibits cardiac fibroblast activation. | [89,90] | |
| SMURF2 | SMAD7 | Mediator of TGF-β signal. SMURF2 mediated SMAD7 degradation was inhibited by SMAD3 inhibitor. | [91,92] |
| HFpEF | |||
| WWP1 | Not described | Cardiac-specific overexpression of WWP1 developed cardiac hypertrophy with diastolic dysfunction. | [93] |
| HECT-Type E3 Ligase | Substrate/Target | Main Findings | Reference |
|---|---|---|---|
| I/R injury | |||
| NEDD4-1 | p-Akt, PTEN | Overexpression of NEDD4-1 ameliorated myocardial apoptosis after I/R injury in rat injected with NEDD4-1 lentivirus vector. | [180] |
| SMURF2 | EZH2 | miR-322/503 ameliorated I/R injury via inhibition of SMURF2 translation. | [181] |
| Doxorubicin cardiotoxicity | |||
| ITCH | TXNIP Unknown | Cardiac specific ITCH transgenic mice attenuated doxorubicin cardiotoxicity and myocardial infarction. miR-34b/c inhibited myocardial injury through ITCH. | [182,183] |
| Circular RNA ITCH | miR-17-5p miR-330-5p | Inhibition of apoptosis caused by H2O2. Inhibition of doxorubicin cardiotoxicity. | [184] [185] |
| Arrhythmia | |||
| NEDD4-2 | Nav1.5 | Contribution to Nav1.5 downregulation in HF. | [186] |
| WWP1 | Connexin 43 | Cardiac-specific overexpression of WWP1 die due to ventricular arrhythmia. | [187] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goto, J.; Otaki, Y.; Watanabe, T.; Watanabe, M. The Role of HECT-Type E3 Ligase in the Development of Cardiac Disease. Int. J. Mol. Sci. 2021, 22, 6065. https://doi.org/10.3390/ijms22116065
Goto J, Otaki Y, Watanabe T, Watanabe M. The Role of HECT-Type E3 Ligase in the Development of Cardiac Disease. International Journal of Molecular Sciences. 2021; 22(11):6065. https://doi.org/10.3390/ijms22116065
Chicago/Turabian StyleGoto, Jun, Yoichiro Otaki, Tetsu Watanabe, and Masafumi Watanabe. 2021. "The Role of HECT-Type E3 Ligase in the Development of Cardiac Disease" International Journal of Molecular Sciences 22, no. 11: 6065. https://doi.org/10.3390/ijms22116065
APA StyleGoto, J., Otaki, Y., Watanabe, T., & Watanabe, M. (2021). The Role of HECT-Type E3 Ligase in the Development of Cardiac Disease. International Journal of Molecular Sciences, 22(11), 6065. https://doi.org/10.3390/ijms22116065






