A Novel Regulator Participating in Nitrogen Removal Process of Bacillus subtilis JD-014
Abstract
:1. Introduction
2. Results and Discussion
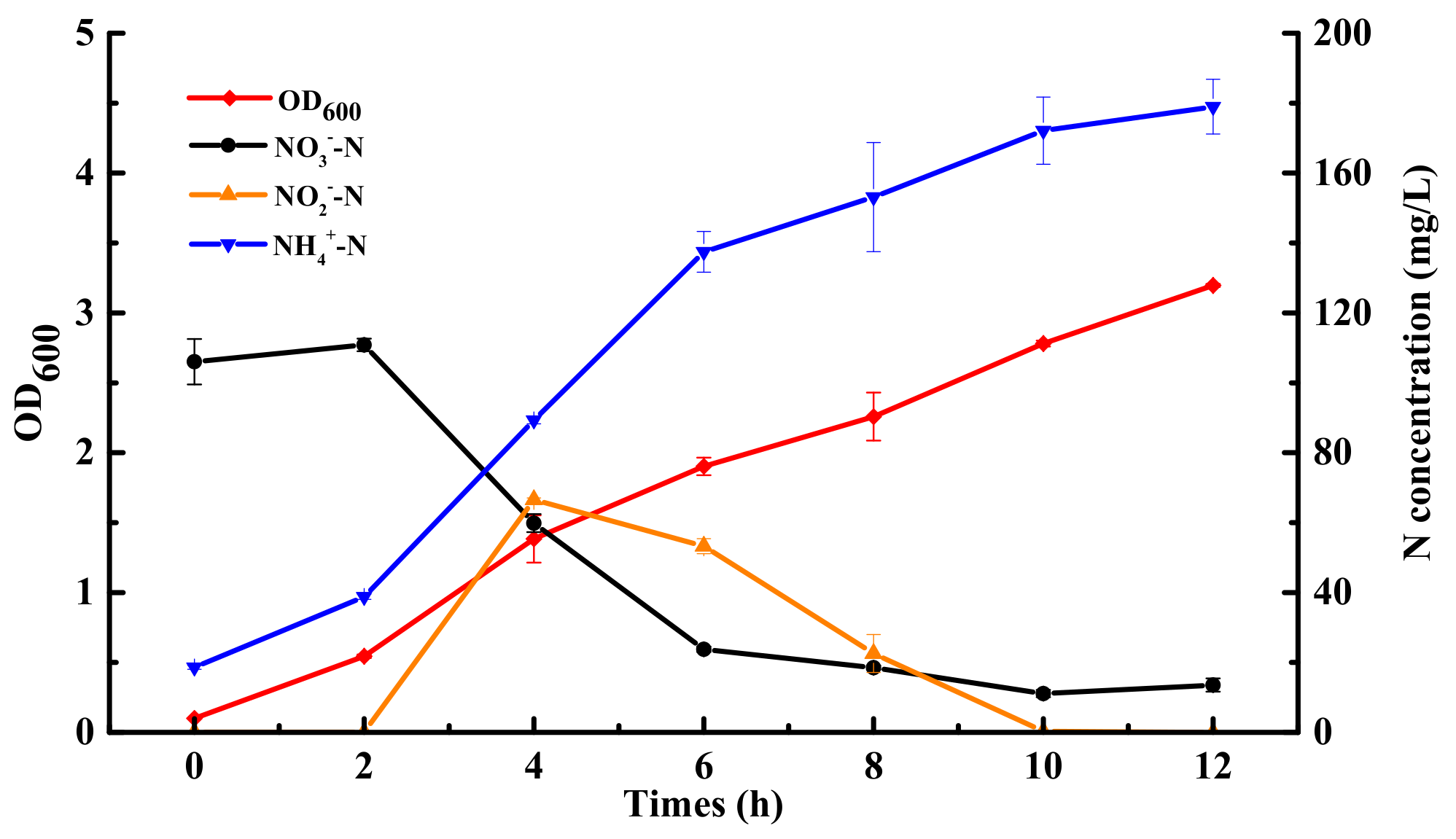
2.1. Effect of Carbon Sources and Nitrate Concentrations on Nitrogen Removal Performance of JD-014
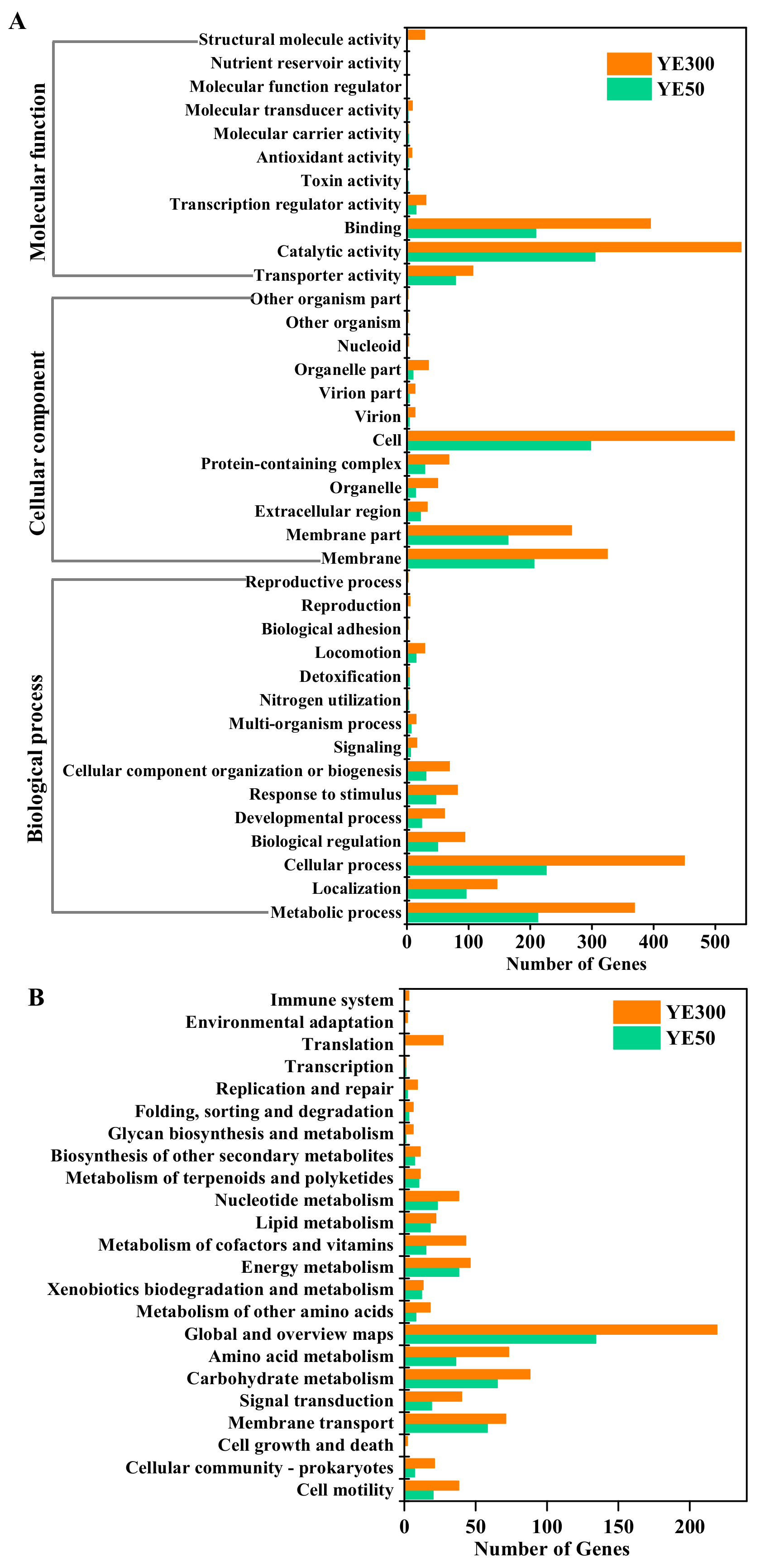
2.2. Transcriptomic Analysis of JD-014 during Denitrification Process
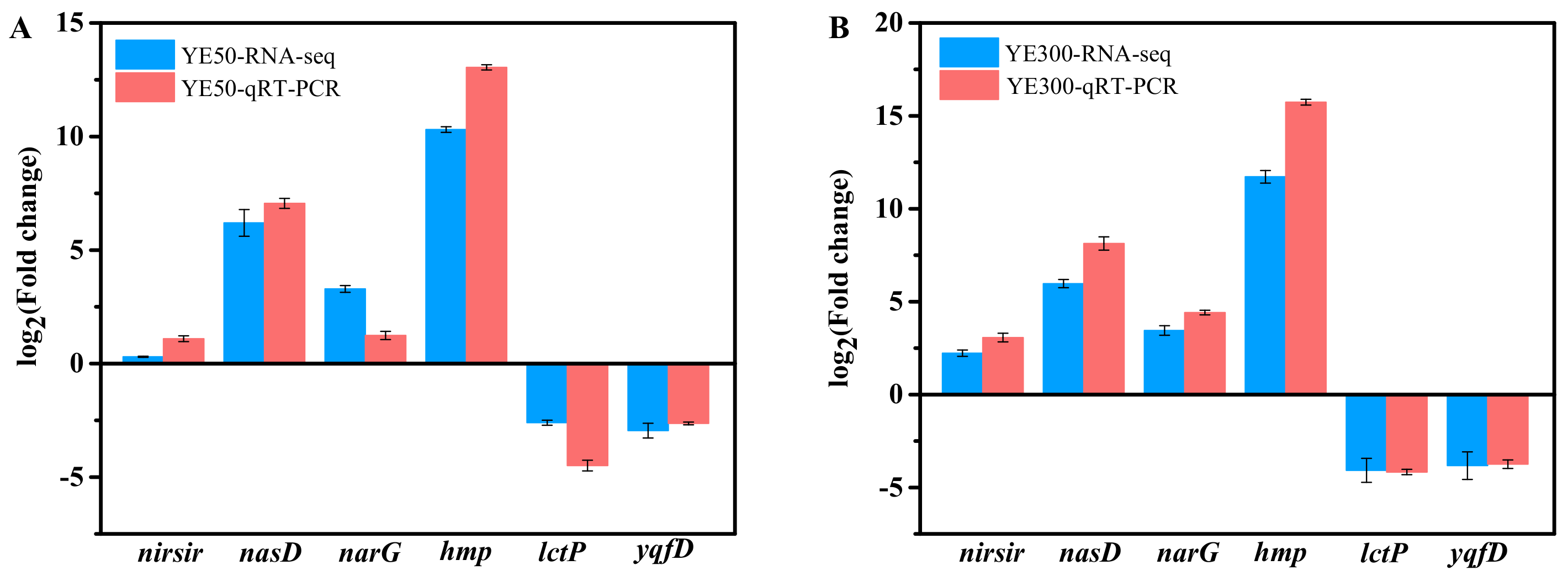
2.3. qRT-PCR Verification of Selected Genes
2.4. DEGs Related to Denitrification Process in JD-014
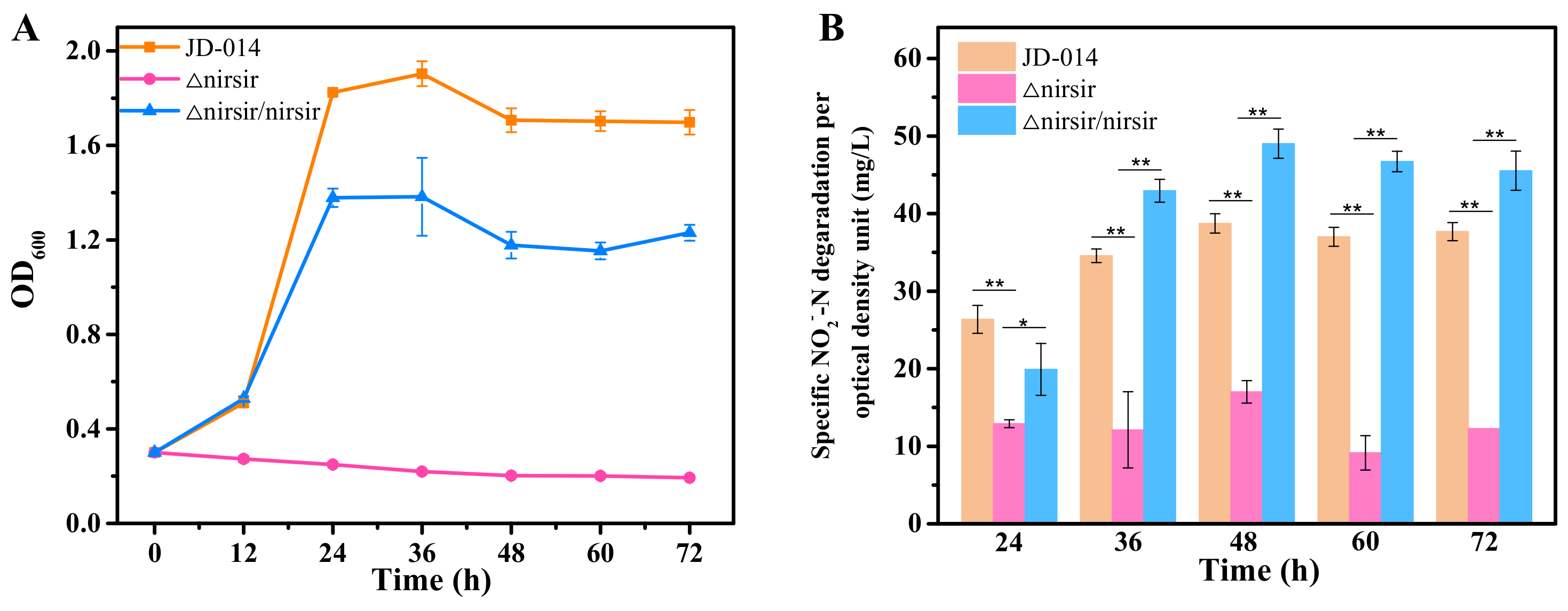
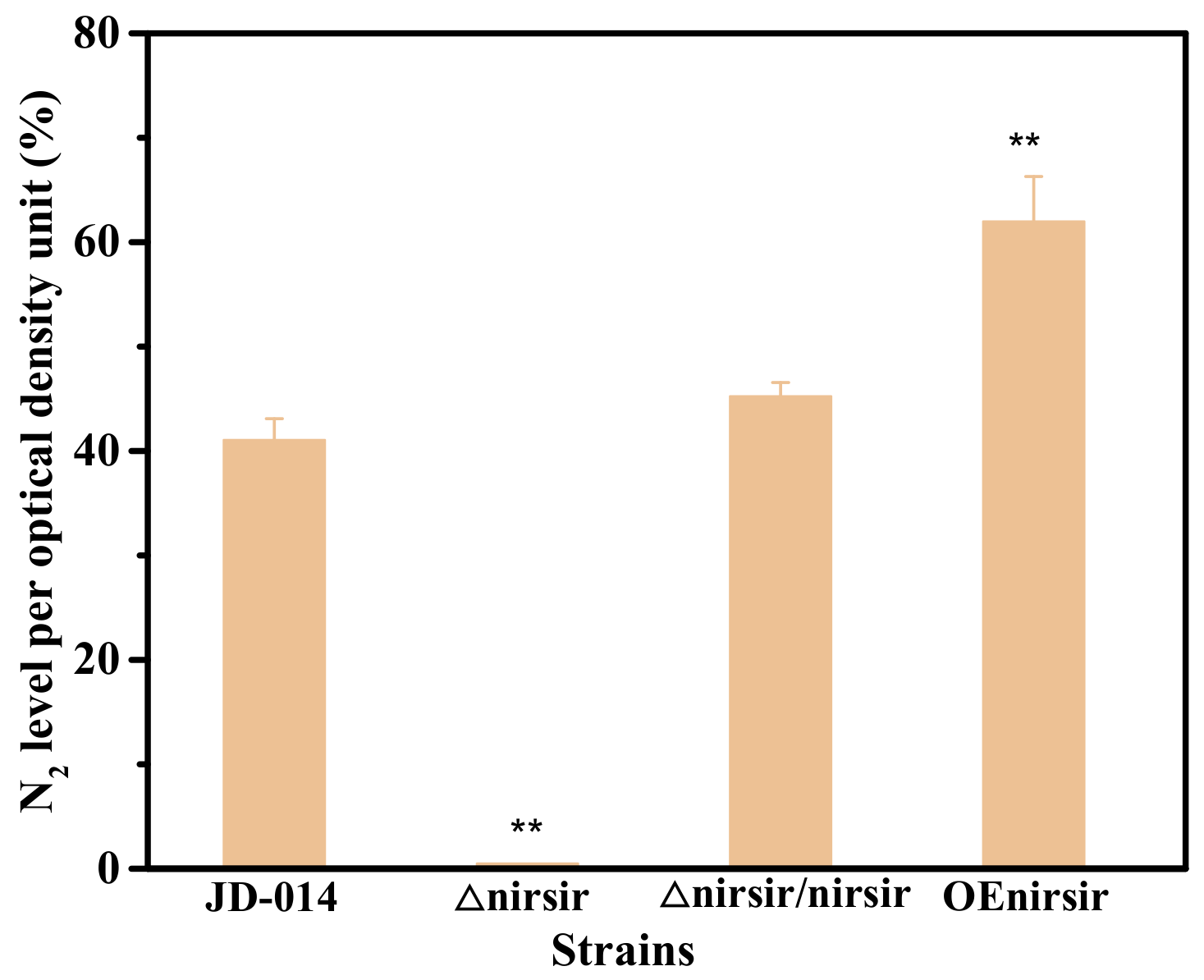
2.5. Functional Analysis of Nirsir under Aerobic Denitrification
3. Materials and Methods
3.1. Bacterial Strain and Culture Medium
3.2. Carbon Sources and Nitrate Concentrations Influence on Denitrification of JD-014
3.3. Transcriptome Analysis
3.4. Quantitative Real Time PCR
3.5. Construction of Nirsir Knockout Mutant Strain of JD-014
3.6. Construction of Nirsir Complementation and Overexpression Mutant Strain of JD-014
3.7. Analytical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Green, S.J.; Prakash, O.; Gihring, T.M.; Akob, D.M.; Jasrotia, P.; Jardine, P.M.; Watson, D.B.; Brown, S.D.; Palumbo, A.V.; Kostka, J.E. Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl. Environ. Microbiol. 2010, 76, 3244–3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezvani, F.; Sarrafzadeh, M.H.; Ebrahimi, S.; Oh, H.M. Nitrate removal from drinking water with a focus on biological methods: A review. Environ. Sci. Pollut. Res. 2019, 26, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Sun, R.; Wu, Y.; Hu, S.; Liu, X.; Chan, J.; Mi, X. Characteristics and metabolic pathway of the bacteria for heterotrophic nitrification and aerobic denitrification in aquatic ecosystems. Environ. Res. 2020, 191, 110069. [Google Scholar] [CrossRef]
- Koju, R.; Miao, S.; Liang, B.; Joshi, D.R.; Qu, J. Transcriptional and metabolic response against hydroxyethane-(1,1-bisphosphonic acid) on bacterial denitrification by a halophilic Pannonibacter sp. strain DN. Chemosphere 2020, 252, 126478. [Google Scholar] [CrossRef]
- Bai, H.; Liao, S.; Wang, A.; Huang, J.; Shu, W.; Ye, J. High-efficiency inorganic nitrogen removal by newly isolated Pannonibacter phragmitetus B1. Bioresour. Technol. 2019, 271, 91–99. [Google Scholar] [CrossRef]
- Rajta, A.; Bhatia, R.; Setia, H.; Pathania, P. Role of heterotrophic-aerobic denitrifying bacteria in nitrate removal from wastewater. J. Appl. Microbiol. 2019, 128, 1261–1278. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, J.; Li, Q.X.; Wang, Y.; Li, S.; Ren, T.; Wang, L. Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresour. Technol. 2012, 116, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Yang, J.; Ma, F.; Pi, S.; Xing, L.; Li, A. Characterisation of Pseudomonas stutzeri T13 for aerobic denitrification: Stoichiometry and reaction kinetics. Sci. Total Environ. 2020, 717, 135181. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.F.; An, J.; Fu, G.H.; Yang, X.L. Isolation and characterization of an aerobic denitrifying Bacillus sp. YX-6 from shrimp culture ponds. Aquaculture 2011, 319, 188–193. [Google Scholar] [CrossRef]
- Chen, J.; Gu, S.; Hao, H.; Chen, J. Characteristics and metabolic pathway of Alcaligenes sp. TB for simultaneous heterotrophic nitrification-aerobic denitrification. Appl. Microbiol. Biotechnol. 2016, 100, 9787–9794. [Google Scholar] [CrossRef] [PubMed]
- Padhi, S.K.; Tripathy, S.; Sen, R.; Mahapatra, A.S.; Mohanty, S.; Maiti, N.K. Characterisation of heterotrophic nitrifying and aerobic denitrifying Klebsiella pneumoniae CF-S9 strain for bioremediation of wastewater. Int. Biodeterior. Biodegrad. 2013, 78, 67–73. [Google Scholar] [CrossRef]
- Zhao, B.; He, Y.L.; Zhang, X.F. Nitrogen removal capability through simultaneous heterotrophic nitrification and aerobic denitrification by Bacillus sp. LY. Environ. Technol. 2010, 31, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, M. Potential of Aerobic denitrification by Pseudomonas stutzeri TR2 has the potential to reduce nitrous oxide emission from wastewater treatment plants. Appl. Environ. Microbiol. 2010, 76, 4619–4625. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Yang, J.; Wang, X.; Wang, E.; Li, B.; He, R.; Yuan, H. Removal of nitrogen by heterotrophic nitrification-aerobic denitrification of a phosphate accumulating bacterium Pseudomonas stutzeri YG-24. Bioresour. Technol. 2015, 182, 18–25. [Google Scholar] [CrossRef]
- Felix, K.A.K.; Abarike, D.E.; Lu, Y. A review on the application of Bacillus as probiotics in aquaculture. Fish Shellfish Immunol. 2019, 87, 820–828. [Google Scholar] [CrossRef]
- Ziaei-Nejad, S.; Rezaei, M.H.; Takami, G.A.; Lovett, D.L.; Mirvaghefi, A.R.; Shakouri, M. The effect of Bacillus spp. bacteria used as probiotics on digestive enzyme activity, survival and growth in the Indian white shrimp Fenneropenaeus indicus. Aquaculture 2006, 252, 516–524. [Google Scholar] [CrossRef]
- Hong, H.A.; Duc, L.H.; Cutting, S.M. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 2005, 29, 813–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Xin, Y.; Zhang, L.; Gu, Z.; Li, Y.; Ding, Z.; Shi, G. Characterization on the aerobic denitrification process of Bacillus strains. Biomass Bioenergy 2020, 140, 105677. [Google Scholar] [CrossRef]
- Liu, G.; Vijayaraman, S.B.; Dong, Y.; Li, X.; Andongmaa, B.T.; Zhao, L.; Tu, J.; He, J.; Lin, L. Bacillus velezensis LG37: Transcriptome profiling and functional verification of GlnK and MnrA in ammonia assimilation. BMC Genom. 2020, 21, 215. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Yang, Q.; Shi, Y.; Xin, Y.; Zhang, L.; Gu, Z.; Shi, G. Insight into the denitrification mechanism of Bacillus subtilis JD-014 and its application potential in bioremediation of nitrogen wastewater. Process Biochem. 2021, 103, 78–86. [Google Scholar] [CrossRef]
- Kraft, B.; Strous, M.; Tegetmeyer, H.E. Microbial nitrate respiration—Genes, enzymes and environmental distribution. J. Biotechnol. 2011, 155, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Camargo, J.A.; Alonso, A. Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: A global assessment. Environ. Int. 2006, 32, 831–849. [Google Scholar] [CrossRef]
- Hong, Y.H.; Deng, M.C.; Xu, X.M.; Wu, C.F.; Xiao, X.; Zhu, Q.; Sun, X.X.; Zhou, Q.Z.; Peng, J.; Yuan, J.P.; et al. Characterization of the transcriptome of Achromobacter sp. HZ01 with the outstanding hydrocarbon-degrading ability. Gene 2016, 584, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Wu, Q.; Zhang, J.; Guo, W.; Ding, Y.; Wang, J.; Wu, H.; Sun, M.; Hou, L.; Wei, X.; et al. Isolation and transcriptome analysis of phenol-degrading bacterium from carbon-sand filters in a full-scale drinking water treatment plant. Front. Microbiol. 2018, 9, 2162. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.; Zhao, Y.G.; Guo, L.; Gao, M.; Jin, C.; She, Z. Transcriptomics of Planococcus kocurii O516 reveals the degrading metabolism of sulfamethoxazole in marine aquaculture wastewater. Environ. Pollut. 2020, 265, 114939. [Google Scholar] [CrossRef]
- Huang, X.; Li, W.; Zhang, D.; Qin, W. Ammonium removal by a novel oligotrophic Acinetobacter sp. Y16 capable of heterotrophic nitrification-aerobic denitrification at low temperature. Bioresour. Technol. 2013, 146, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lv, R.; Xiao, Y.; Hu, W.; Mai, Y.; Zhang, J.; Lin, L.; Hu, X. A novel nitrite-base aerobic denitrifying bacterium Acinetobacter sp. YT03 and its transcriptome analysis. Front. Microbiol. 2019, 10, 2580. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Lv, Y.; Liu, Y.; Ren, R. Removal of nitrogen by heterotrophic nitrification-aerobic denitrification of a novel metal resistant bacterium Cupriavidus sp. S1. Bioresour. Technol. 2016, 220, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, J.; Zhang, S.; Liu, F.; Peng, J.; Peng, Y.; Wu, J. Nitrogen removal characteristics of a novel heterotrophic nitrification and aerobic denitrification bacteria, Alcaligenes faecalis strain WT14. J. Environ. Manag. 2021, 282, 111961. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, T.; Song, Y.; Chen, H.; Lv, Y. Heterotrophic nitrogen removal by Acinetobacter sp. Y1 isolated from coke plant wastewater. J. Biosci. and Bioeng. 2015, 120, 549–554. [Google Scholar] [CrossRef]
- Elefsiniotis, P.; Wareham, D.G.; Smith, M.O. Use of volatile fatty acids from an acid-phase digester for denitrification. J. Biotechnol. 2004, 114, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ni, J. Heterotrophic nitrification-aerobic denitrification by novel isolated bacteria. J. Ind. Microbiol. Biotechnol. 2011, 38, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, S.; Zhou, L. Effect of carbon source, C/N ratio, nitrate and dissolved oxygen concentration on nitrite and ammonium production from denitrification process by Pseudomonas stutzeri D6. Bioresour. Technol. 2012, 104, 65–72. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Jiang, X.; Zhang, L.; Hou, C.; Su, G.; Wang, L.; Mu, Y.; Shen, J. Nitrate stimulation of N-Methylpyrrolidone biodegradation by Paracoccus pantotrophus: Metabolite mechanism and genomic characterization. Bioresour. Technol. 2019, 294, 122185. [Google Scholar] [CrossRef]
- Yang, J.; Feng, L.; Pi, S.; Cui, D.; Ma, F.; Zhao, H.P.; Li, A. A critical review of aerobic denitrification: Insights into the intracellular electron transfer. Sci. Total Environ. 2020, 731, 139080. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Strous, M. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys. Acta 2013, 1827, 136–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, M.; Zhao, Y.; Zhu, L.; Song, X.; Wei, Z. Denitrification during composting: Biochemistry, implication and perspective. Int. Biodeterior. Biodegrad. 2020, 153, 105043. [Google Scholar] [CrossRef]
- Onley, J.R.; Ahsan, S.; Sanford, R.A.; Loffler, F.E. Denitrification by Anaeromyxobacter dehalogenans, a common soil bacterium lacking nitrite reductase genes (nirS/nirK). Appl. Environ. Microbiol. 2017, 84, e01985-17. [Google Scholar] [CrossRef] [Green Version]
- Jin, P.; Chen, Y.; Yao, R.; Zheng, Z.; Du, Q. New insight into the nitrogen metabolism of simultaneous heterotrophic nitrification-aerobic denitrification bacterium in mRNA expression. J. Hazard. Mater. 2019, 371, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, H.; Tan, J.; Wei, P.; Yu, S.; Liu, R.; Gao, J. Transcriptome profiling analysis of the intoxication response in midgut tissue of Agrotis ipsilon larvae to Bacillus thuringiensis Vip3Aa protoxin. Pestic. Biochem. Physiol. 2019, 160, 20–29. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, D.; Mawlong, G.T.; Sarki, Y.N.; Singh, A.K.; Chikkaputtaiah, C.; Boruah, H.P.D. Transcriptome analysis of crude oil degrading Pseudomonas aeruginosa strains for identification of potential genes involved in crude oil degradation. Gene 2020, 755, 144909. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Zakataeva, N.P.; Nikitina, O.V.; Gronskiy, S.V.; Romanenkov, D.V.; Livshits, V.A. A simple method to introduce marker-free genetic modifications into the chromosome of naturally nontransformable Bacillus amyloliquefaciens strains. Appl. Microbiol. Biotechnol. 2010, 85, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association; American Water Works Association; Water Environment Federation. Standard Methods for the Examination of Water and Wastewater, 20th ed.; American Public Health Association: Washington, DC, USA, 1998; pp. 1171–1175. [Google Scholar]



| Genes | Description | Strains | Size (aa) | Identity (%) | Cover (%) | Accession Number |
|---|---|---|---|---|---|---|
| nirsir | sulfite reductase subunit beta | Bacillus subtilis JD-014 | 571 | - | - | in CP045478 |
| cysI | sulfite reductase [NADPH] hemoprotein beta-component | Bacillus sp. URHB0009 | 572 | 84.09 | 99 | WP027319807 |
| Mesorhizobium sp. Root172 | 573 | 50.95 | 97 | WP056566970 | ||
| Geobacillus sp. BCO2 | 349 | 68.77 | 52 | KPD00931 | ||
| Paenibacillus sp. P1XP2 | 517 | 58.99 | 86 | KHF35799 | ||
| Verrucomicrobiales | 394 | 54.06 | 67 | MAJ16745 | ||
| nirS (partial) | cytochrome cd1 nitrite reductase | Bacillus cereus GS-5 | 136 | 13.73 | 8 | APM87481 |
| Pseudomonas aeruginosa CCUG 241 | 259 | 10.99 | 6 | AAD26540 | ||
| Uncultured bacterium wA20 | 265 | 11.48 | 17 | CAB76794 | ||
| Pseudomonas sp. R125 | 154 | 17.82 | 10 | CAF25139 | ||
| nirK | copper-type nitrite reductase | Bacillus azotoformans LMG 9581 | 353 | 13.41 | 13 | EKN68572 |
| Bacillus firmus GY-49 | 353 | 10.66 | 30 | AMQ34899 | ||
| Nitrosococcus oceani NS58 | 372 | 13.74 | 20 | CCA61349 | ||
| Anammox organism KSU-1 | 337 | 11.56 | 21 | GAB64238 |
| Primer | Primer Sequences (5′-3′) |
|---|---|
| hmp-F1-RT | CAAACAGCCTGAACGGCAAA |
| hmp-R1-RT | CGGCTCGCGATACACAAATG |
| nasD-F1-RT | AAAGAAGCCATTTGCGGCTG |
| nasD-R1-RT | TTCCAGCCGAGCACATTCAT |
| narG-F1-RT | CTGGTTCAACTCCGACACGA |
| narG-R1-RT | AATCGTCTGCCACCCTTCAG |
| nirsir-F1-RT | AGCACTTTTGGATACGATCGCAGC |
| nirsir-R1-RT | TTCATGATATGCTCTCGTCCGCG |
| lctP-F1-RT | GATTGGCGTGTTCATCACCG |
| lctP-R1-RT | CAGCAAATCTGAACCCGCAC |
| yqfD-F1-RT | TGACAGTCCCGCTTGAAACA |
| yqfD-R1-RT | CCCAGATCGGGATTGCCAAA |
| gyrB-F1-RT | AAGCTGGGCAACTCAGAAGCACGG |
| gyrB-R1-RT | AGCCATTCTTGCTCTTGCCGCC |
| nirsir-KpnI-F | CGGGGTACCATGGTGACCAAAATTCTAAAAGCACCG |
| nirsir-XhoI-R | CCGCTCGAGTCGTACCGTCAGTTGTTGCTT |
| FX-nirsir-NheI-F | CTAGCTAGCGGCTGACAGCCAATCAGAACTT |
| FX-nirsir-BamHI-R | CGCGGATCCACCACTCGTATACCTCTGAGTGGA |
| FRT-BamHI-Tet-F | ACGGGATCCGAAGTTCCTATTCCGAAGTTCCTATTCTCTAG |
| AAAGTATAGGAACTTCGGATCAATTTTGAACTCTCTCC | |
| FRT-PstI-Tet-R | AACTGCAGGAAGTTCCTATACTTTCTAGAGAATAGGAACTT |
| CGGAATAGGAACTTCGGGCCATATTGTTGTATAAG | |
| yz-nirsir-F | GCATGACGTCCATAACACATTGCT |
| yz-nirsir-R | AGGAGTTCTCTCCACACTTGTCT |
| pMA5-a-F | GGAGCGATTTACATATGAGTTATGCAG |
| pMA5-a-R | ATCAGCTTGCTTTCGAGGTGAATTTCGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, T.; Shi, Y.; Yang, Q.; Xin, Y.; Gu, Z.; Zhang, L. A Novel Regulator Participating in Nitrogen Removal Process of Bacillus subtilis JD-014. Int. J. Mol. Sci. 2021, 22, 6543. https://doi.org/10.3390/ijms22126543
Yang T, Shi Y, Yang Q, Xin Y, Gu Z, Zhang L. A Novel Regulator Participating in Nitrogen Removal Process of Bacillus subtilis JD-014. International Journal of Molecular Sciences. 2021; 22(12):6543. https://doi.org/10.3390/ijms22126543
Chicago/Turabian StyleYang, Ting, Yi Shi, Qian Yang, Yu Xin, Zhenghua Gu, and Liang Zhang. 2021. "A Novel Regulator Participating in Nitrogen Removal Process of Bacillus subtilis JD-014" International Journal of Molecular Sciences 22, no. 12: 6543. https://doi.org/10.3390/ijms22126543






