The Unusual Role of Pro in Cu(II) Binding by His2-Cyclopentapeptide
Abstract
:1. Introduction
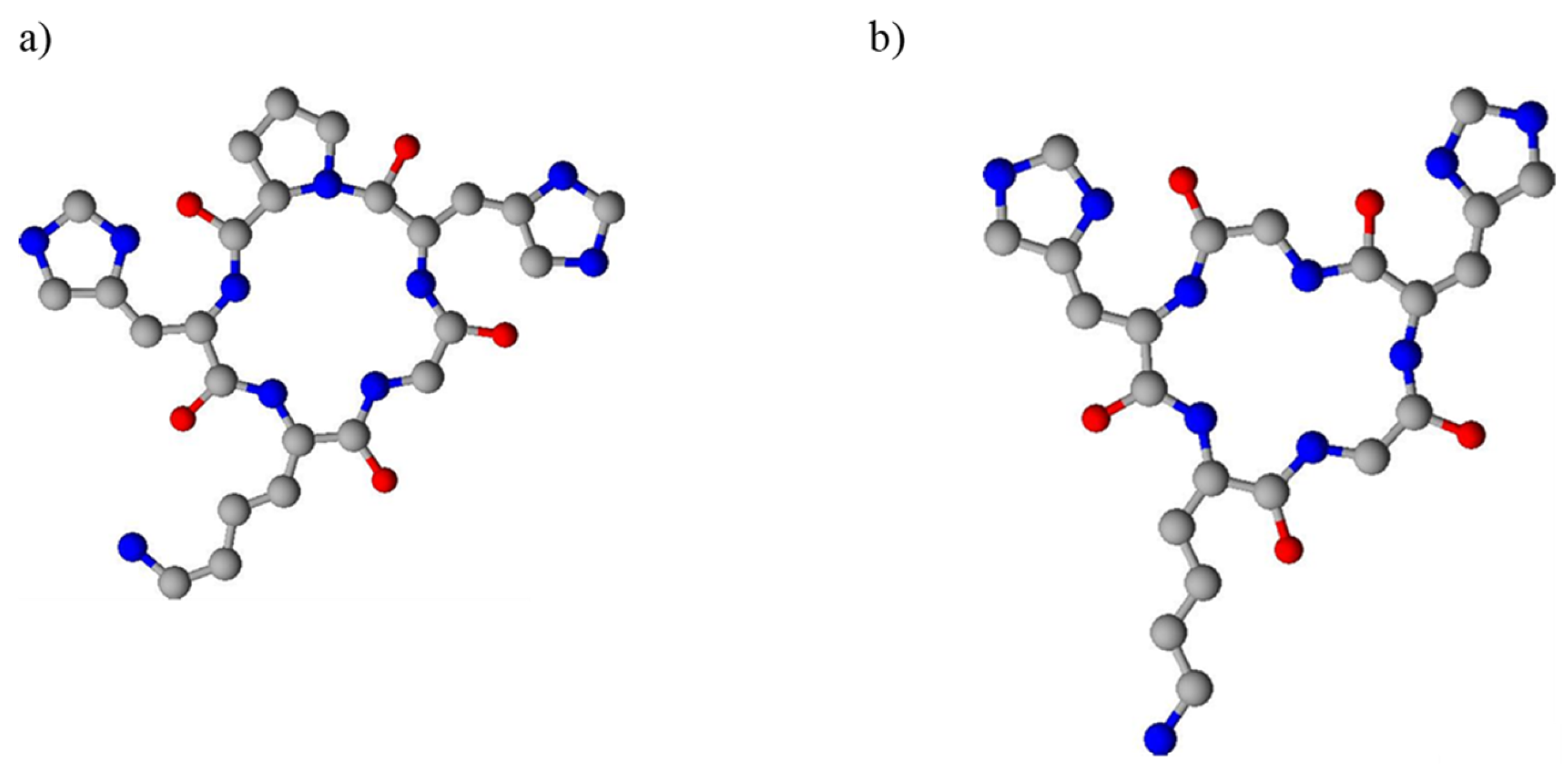
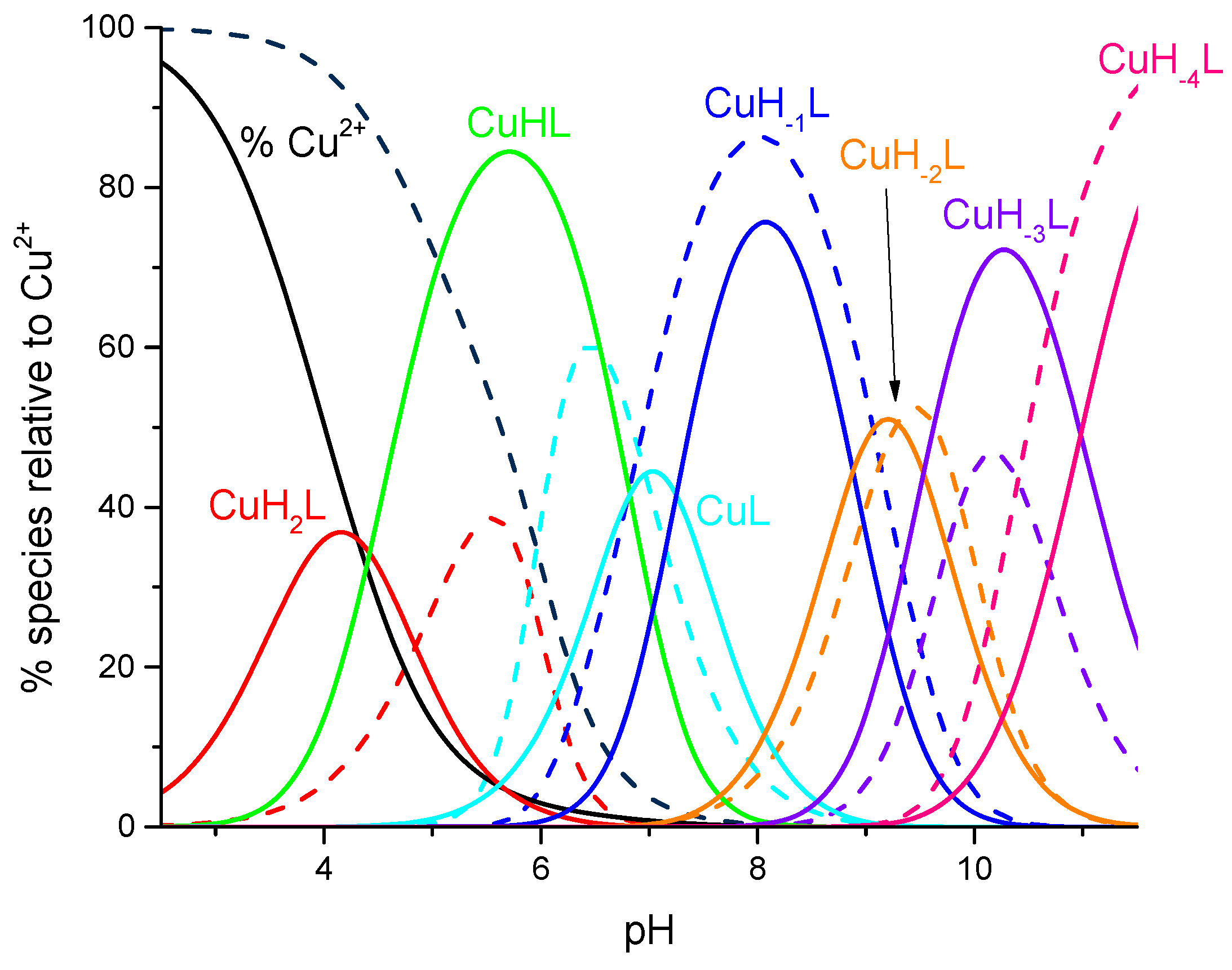
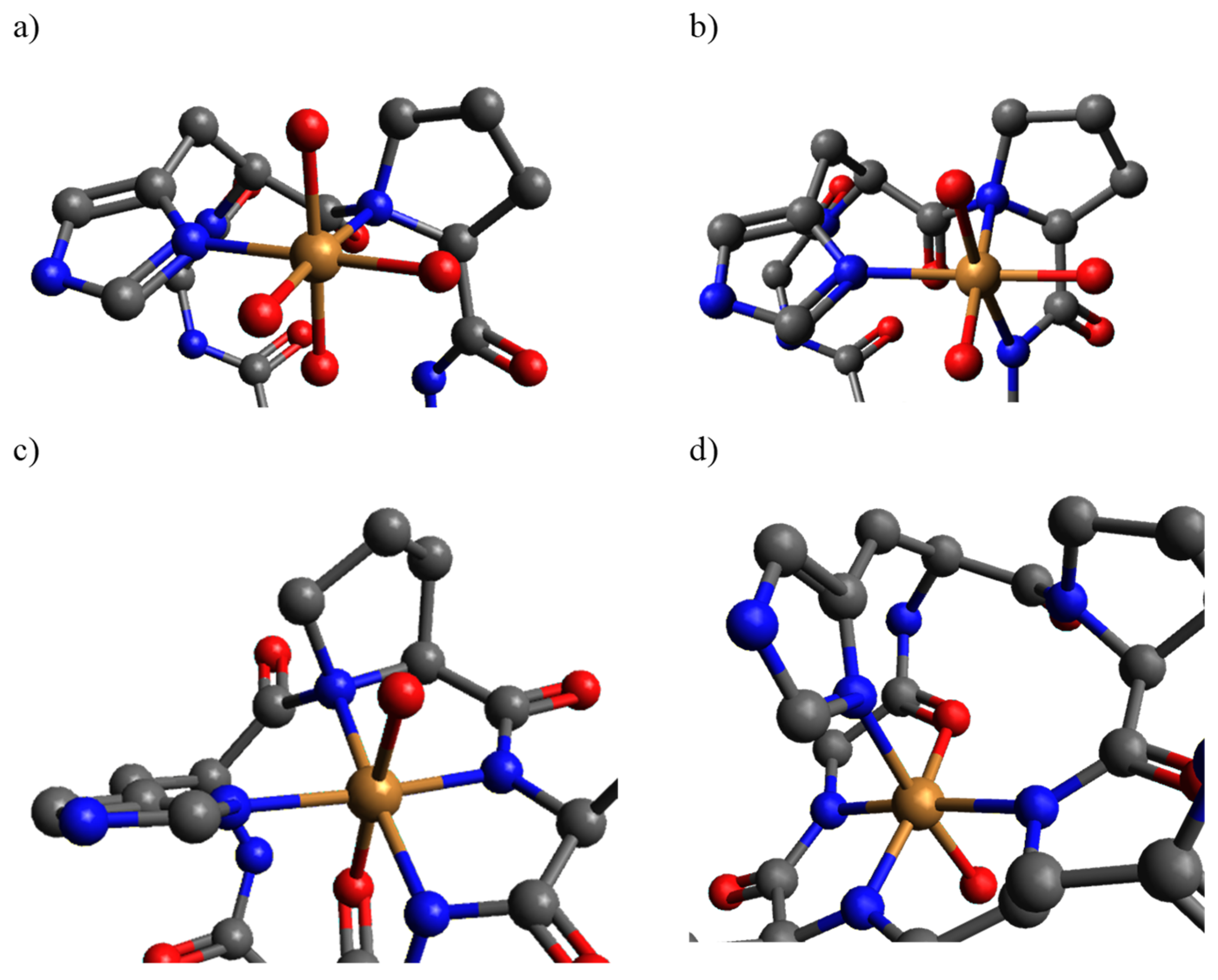
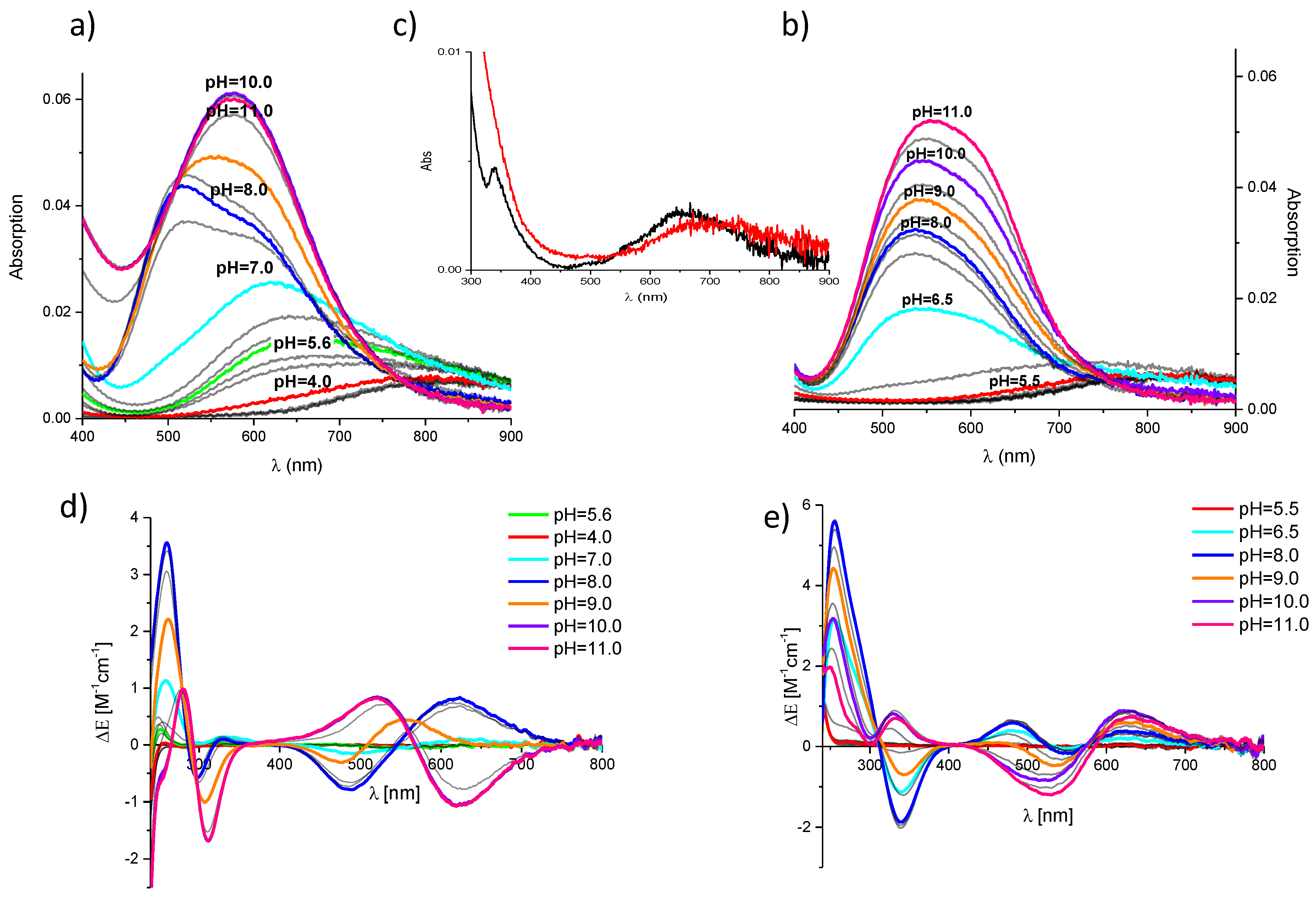
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Potentiometric Measurements
3.3. Spectroscopic Measurements
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bataille, M.; Formicka-Kozlowska, G.; Kozlowski, H.; Pettit, L.D.; Steel, I. The L-proline residue as a ‘break-point’ in the co-ordination of metal–peptide systems. J. Chem. Soc. Chem. Commun. 1984, 231–232. [Google Scholar] [CrossRef]
- Pettit, L.D.; Steel, I.; Formicka-Kozlowska, G.; Tatarowski, T.; Bataille, M. The L-proline residue as a ?break-point? in metal?peptide systems. J. Chem. Soc. Dalton Trans. 1985, 535–539. [Google Scholar] [CrossRef]
- Kozłowski, H.; Bal, W.; Dyba, M.; Kowalik-Jankowska, T. Specific structure–stability relations in metallopeptides. Co-ord. Chem. Rev. 1999, 184, 319–346. [Google Scholar] [CrossRef]
- Peluso, S.; Rückle, T.; Lehmann, C.; Mutter, M.; Peggion, C.; Crisma, M. Crystal structure of a synthetic cyclodecapeptide for template-assembled synthetic protein design. ChemBioChem 2001, 2, 432–437. [Google Scholar] [CrossRef]
- Dumy, P.; Eggleston, I.M.; Esposito, G.; Nicula, S.; Mutter, M. Solution structure of Regioselectively Addressable Functional-ized Templates: An NMR and restrained molecular dynamics investigation. Biopolymers 1996, 39, 297–308. [Google Scholar] [CrossRef]
- Fontanesi, F.; Soto, I.C.; Horn, D.; Barrientos, A. Assembly of mitochondrial cytochromec-oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Physiol. 2006, 291, C1129–C1147. [Google Scholar] [CrossRef] [Green Version]
- Kotynia, A.; Janek, T.; Czyżnikowska, Ż.; Bielińska, S.; Kamysz, W.; Brasuń, J. The Analysis of Cu(II)/Zn(II) Cyclopeptide System as Potential Cu, ZnSOD Mimic Center. Int. J. Pept. Res. Ther. 2017, 23, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decker, H.; Schweikardt, T.; Tuczek, F. The First Crystal Structure of Tyrosinase: All Questions Answered? Angew. Chem. Int. Ed. 2006, 45, 4546–4550. [Google Scholar] [CrossRef] [PubMed]
- Vorherr, T.; Lewis, I.; Berghausen, J.; Desrayaud, S.; Schaefer, M. Modulation of Oral Bioavailability and Metabolism for Closely Related Cyclic Hexapeptides. Int. J. Pept. Res. Ther. 2018, 24, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Bockus, A.T.; McEwen, C.M.; Lokey, R.S. Form and Function in Cyclic Peptide Natural Products: A Pharmacokinetic Perspective. Curr. Top. Med. Chem. 2013, 999, 29–35. [Google Scholar] [CrossRef]
- Clark, R.; Fischer, H.; Dempster, L.; Daly, N.L.; Rosengren, K.J.; Nevin, S.T.; Meunier, F.A.; Adams, D.; Craik, D.J. Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the -conotoxin MII. Proc. Natl. Acad. Sci. USA 2005, 102, 13767–13772. [Google Scholar] [CrossRef] [Green Version]
- Akcan, M.; Stroud, M.R.; Hansen, S.J.; Clark, R.; Daly, N.L.; Craik, D.J.; Olson, J.M. Chemical Re-engineering of Chlorotoxin Improves Bioconjugation Properties for Tumor Imaging and Targeted Therapy. J. Med. Chem. 2011, 54, 782–787. [Google Scholar] [CrossRef] [Green Version]
- Namjoshi, S.; Benson, H.A.E. Cyclic peptides as potential therapeutic agents for skin disorders. Biopolymer 2010, 94, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old’ class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Chakraborty, S.; Liu, S. Radiolabeled Cyclic RGD Peptides as Radiotracers for Imaging Tumors and Thrombosis by SPECT. Theranostics 2011, 1, 58–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S. Radiolabeled Cyclic RGD Peptide Bioconjugates as Radiotracers Targeting Multiple Integrins. Bioconjugate Chem. 2015, 26, 1413–1438. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Wang, F.; Liu, S. Radiolabeled cyclic RGD peptides as radiotracers for tumor imaging. Biophys. Rep. 2016, 2, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, G.; Gurrath, M.; Kessler, H.; Timpl, R. Dynamic Forcing, a Method for Evaluating Activity and Selectivity Profiles of RGD(Arg-Gly-Asp) Peptides. Angew. Chem. Int. Ed. 1992, 31, 326–328. [Google Scholar] [CrossRef]
- Haubner, R.; Gratias, R.; Diefenbach, B.; Goodman, S.L.; Jonczyk, A.; Kessler, H. Structural and Functional Aspects of RGD-Containing Cyclic Pentapeptides as Highly Potent and Selective Integrin αVβ3Antagonists. J. Am. Chem. Soc. 1996, 118, 7461–7472. [Google Scholar] [CrossRef]
- Ngu-Schwemlein, M.; Gilbert, W.; Askew, K.; Schwemlein, S. Thermodynamics and fluorescence studies of the interactions of cyclooctapeptides with Hg2+, Pb2+, and Cd2+. Bioorganic Med. Chem. 2008, 16, 5778–5787. [Google Scholar] [CrossRef]
- Ngu-Schwemlein, M.; Butko, P.; Cook, B.; Whigham, T. Interactions of an acidic cyclooctapeptide with metal ions: Microcalorimetric and fluorescence analyses. J. Pept. Res. 2008, 66, 72–81. [Google Scholar] [CrossRef]
- Bonomo, R.P.; Impellizzeri, G.; Pappalardo, G.; Purrello, R.; Rizzarelli, E.; Tabbì, G. Co-ordinating properties of cyclopeptides. Thermodynamic and spectroscopic study on the formation of copper(II) complexes with cyclo(Gly-His)4 and cyclo(Gly-His-Gly)2 and their superoxide dismutase-like activity. J. Chem. Soc. Dalton Trans. 1998, 3851–3858. [Google Scholar] [CrossRef]
- Bonomo, R.P.; Conte, E.; Impellizzeri, G.; Pappalardo, G.; Purrello, R.; Rizzarelli, E. Copper(II) complexes with cyclo(L-aspartyl-L-aspartyl) and cyclo(L-glutamyl-L-glutamyl) derivatives and their antioxidant properties. J. Chem. Soc. Dalton Trans. 1996, 3093–3099. [Google Scholar] [CrossRef]
- Kotynia, A.; Pap, J.S.; Brasun, J. The binding abilities of homodetic cyclic His-peptides toward copper ions. Inorg. Chim. Acta 2018, 472, 3–11. [Google Scholar] [CrossRef]
- Kotynia, A.; Bielinska, S.; Kamysz, W.; Brasun, J. The coordination abilities of the multiHis-cyclopeptide with two metal-binding centers—Potentiometric and spectroscopic investigation. Dalton Trans. 2012, 41, 12114. [Google Scholar] [CrossRef]
- Kotynia, A.; Czyżnikowska, Ż.; Bielinska, S.; Szyrwiel, Ł.; Kamysz, W.; Malinka, W.; Brasun, J. The impact of two–GlyProGly–motifs on formation of di-copper complexes by His 4-cyclopeptides. New J. Chem. 2014, 38, 5198–5206. [Google Scholar] [CrossRef]
- Kállay, C.; Várnagy, K.; Malandrinos, G.; Hadjiliadis, N.; Sanna, D.; Sóvágó, I. Copper(ii) complexes of terminally protected pentapeptides containing three histidyl residues in alternating positions, Ac-His-Xaa-His-Yaa-His-NH2. Dalton Trans. 2006, 4545–4552. [Google Scholar] [CrossRef] [PubMed]
- Kállay, C.; Várnagy, K.; Malandrinos, G.; Hadjiliadis, N.; Sanna, D.; Sóvágó, I. Thermodynamic and structural characterization of the macrochelates formed in the reactions of copper(II) and zinc(II) ions with peptides of histidine. Inorg. Chim. Acta 2009, 362, 935–945. [Google Scholar] [CrossRef]
- Brasun, J.; Gabbiani, C.; Ginanneschi, M.; Messori, L.; Orfei, M.; Swiatek-Kozlowska, J. The copper(II) binding properties of the cyclic peptide c(HGHK). J. Inorg. Biochem. 2004, 98, 2016–2021. [Google Scholar] [CrossRef]
- Brasuń, J.; Matera-Witkiewicz, A.; Ołdziej, S.; Pratesi, A.; Ginanneschi, M.; Messori, L. Impact of ring size on the copper(II) coordination abilities of cyclic tetrapeptides. J. Inorg. Biochem. 2009, 103, 813–817. [Google Scholar] [CrossRef]
- Kállay, C.; Nagy, Z.; Várnagy, K.; Malandrinos, G.; Hadjiliadis, N.; Sóvágó, I. Thermodynamic and Structural Characterization of the Copper(II) Complexes of Peptides Containing Both Histidyl and Aspartyl Residues. Bioinorg. Chem. Appl. 2007, 2007, 030394. [Google Scholar] [CrossRef] [Green Version]
- Csire, G.; Timári, S.; Asztalos, J.; Király, J.M.; Kiss, M.; Várnagy, K. Coordination, redox properties and SOD activity of Cu(II) complexes of multihistidine peptides. J. Inorg. Biochem. 2017, 177, 198–210. [Google Scholar] [CrossRef]
- Rajković, S.; Kállay, C.; Serényi, R.; Malandrinos, G.; Hadjiliadis, N.; Sanna, D.; Sóvágó, I. Complex formation processes of terminally protected peptides containing two or three histidyl residues. Characterization of the mixed metal complexes of peptides. Dalton Trans. 2008, 5059–5071. [Google Scholar] [CrossRef] [PubMed]
- Irving, H.; Miles, M.; Pettit, L. A study of some problems in determining the stoicheiometric proton dissociation constants of complexes by potentiometric titrations using a glass electrode. Anal. Chim. Acta 1967, 38, 475–488. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. SUPERQUAD: An improved general program for computation of formation constants from potentiometric data. J. Chem. Soc. Dalton Trans. 1985, 1195–1200. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
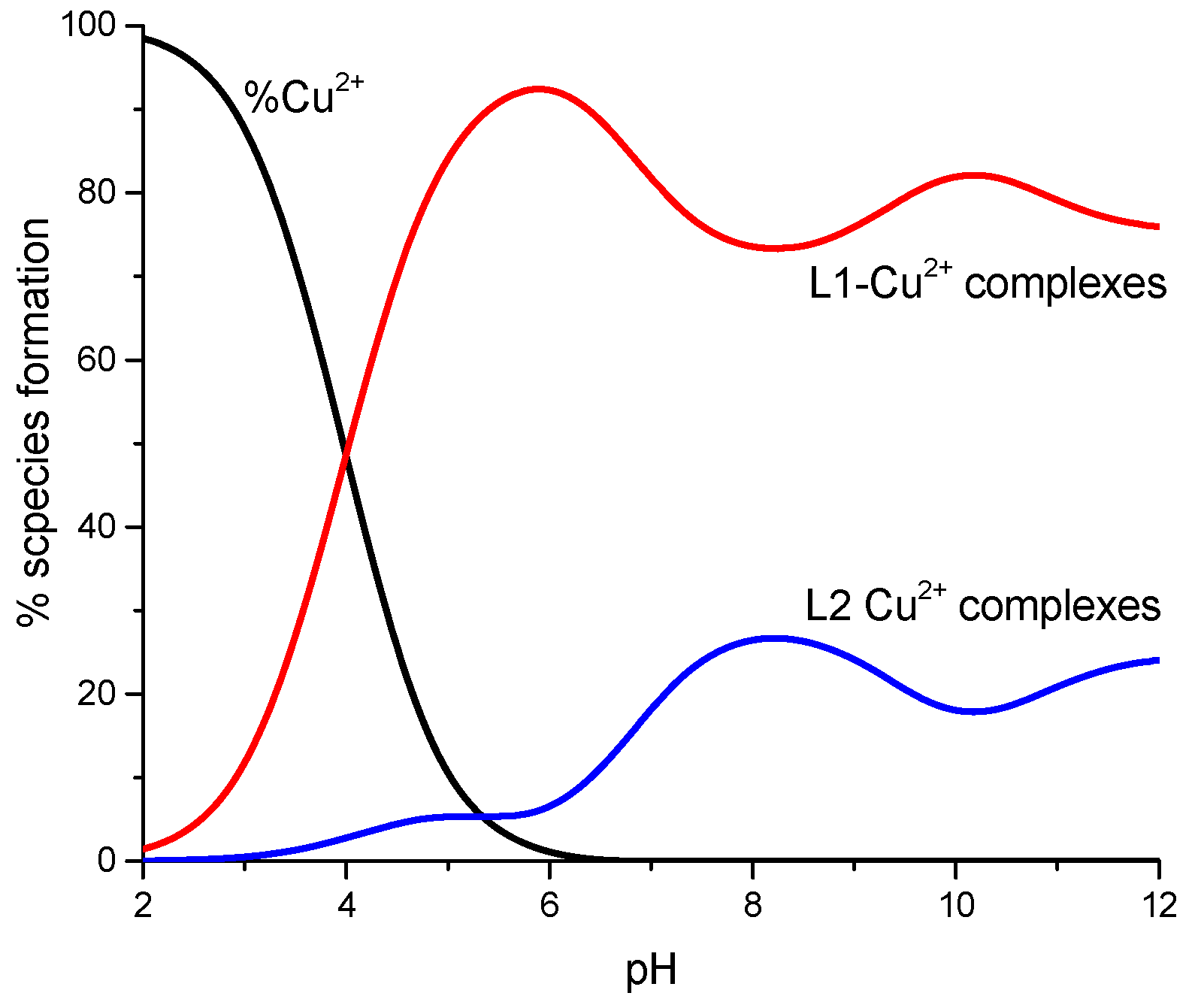
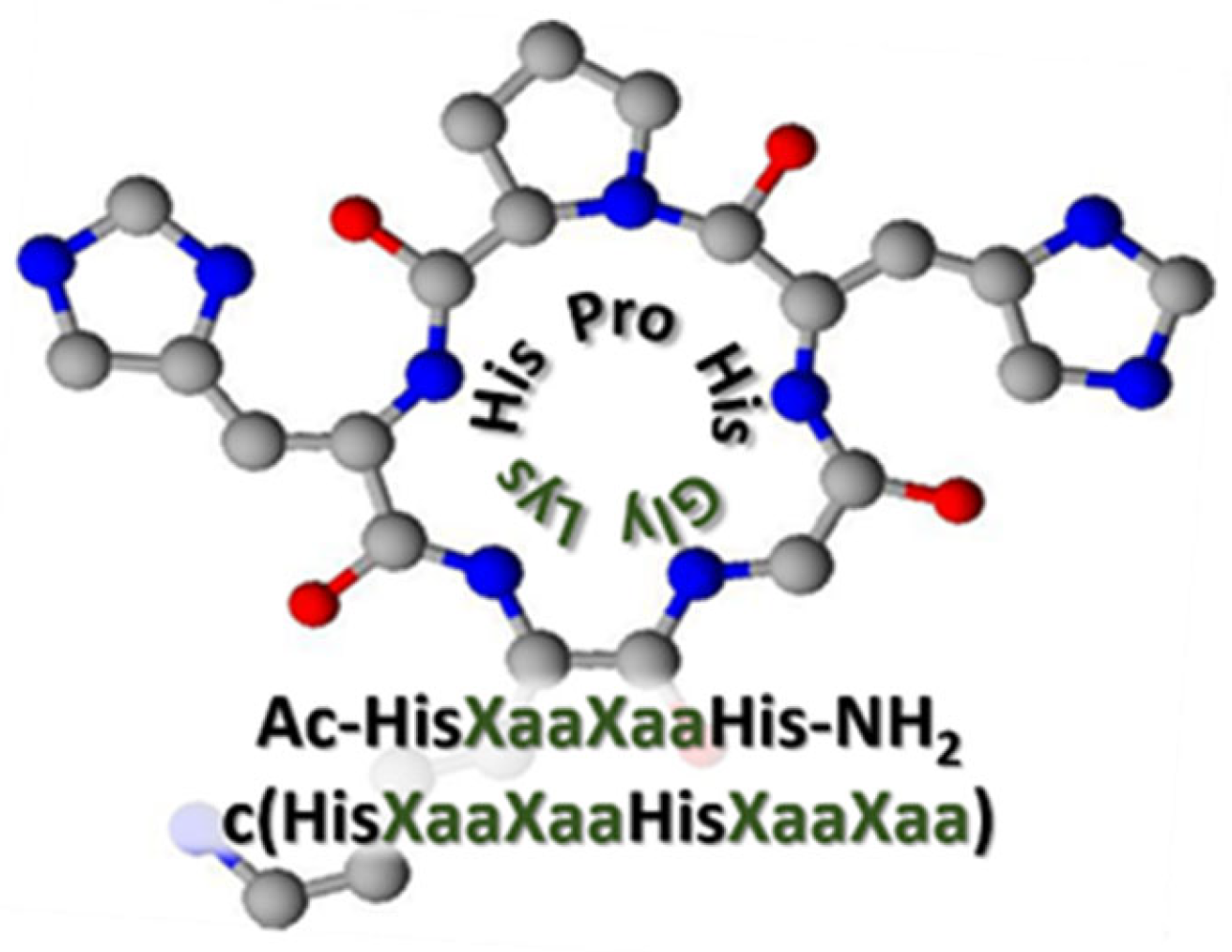
| L1 | L2 | |||
|---|---|---|---|---|
| logβ | logK | logβ | logK | |
| HL | 9.79 ± 0.04 | 9.79 | 9.63 ± 0.05 | 9.63 |
| H2L | 16.53 ± 0.06 | 6.74 | 16.71 ± 0.08 | 7.08 |
| H3L | 21.89 ± 0.06 | 5.36 | 22.53 ± 0.08 | 5.82 |
| CuH2L | 21.06 ± 0.05 | 4.42 | 20.31 ± 0.07 | − |
| CuHL | 16.64 ± 0.03 | 6.82 | − | − |
| CuL | 9.82 ± 0.05 | 7.25 | 8.51 ± 0.06 | 6.90 |
| CuH-1L | 2.57 ± 0.04 | 8.88 | 1.61 ± 0.05 | 9.13 |
| CuH-2L | −6.31 ± 0.04 | 9.53 | −7.52 ± 0.07 | 9.87 |
| CuH-3L | −15.84 ± 0.05 | 10.98 | −17.39 ± 0.07 | 10.40 |
| CuH-4L | −26.82 ± 0.05 | − | −27.79 ± 0.07 | − |
| logβ Independent of the Protonation State of the Ligand | |||||
|---|---|---|---|---|---|
| Ligand | 1NIm | 2NIm | 1Nam | 2Nam | 3Nam |
| Ac-HGGH-NH2 [32] | − | 5.97 | − | −7.88 | −16.10 |
| Ac-HAAH-NH2 [32] | − | 6.08 | −0.93 | −8.05 | −16.39 |
| Ac-HVVH-NH2 [33] | − | 5.79 | −1.05 | −9.29 | −17.60 |
| c(HGKHP)—L1 | 4.54 | 6.85 | 0.03 | −7.22 | −15.84 |
| c(HGGHGG) [22] | 3.63 | 5.45 | −1.40 | −8.00 | − |
| logβ* = logβCuHxL − logβHxL | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pieniężna, A.; Kotynia, A.; Brasuń, J. The Unusual Role of Pro in Cu(II) Binding by His2-Cyclopentapeptide. Int. J. Mol. Sci. 2021, 22, 6628. https://doi.org/10.3390/ijms22126628
Pieniężna A, Kotynia A, Brasuń J. The Unusual Role of Pro in Cu(II) Binding by His2-Cyclopentapeptide. International Journal of Molecular Sciences. 2021; 22(12):6628. https://doi.org/10.3390/ijms22126628
Chicago/Turabian StylePieniężna, Aleksandra, Aleksandra Kotynia, and Justyna Brasuń. 2021. "The Unusual Role of Pro in Cu(II) Binding by His2-Cyclopentapeptide" International Journal of Molecular Sciences 22, no. 12: 6628. https://doi.org/10.3390/ijms22126628
APA StylePieniężna, A., Kotynia, A., & Brasuń, J. (2021). The Unusual Role of Pro in Cu(II) Binding by His2-Cyclopentapeptide. International Journal of Molecular Sciences, 22(12), 6628. https://doi.org/10.3390/ijms22126628





