Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae: 2. Role of Filamentous Extracellular Network and Cell-to-Cell Connections at the Biofilm Surface
Abstract
:1. Introduction
2. Results
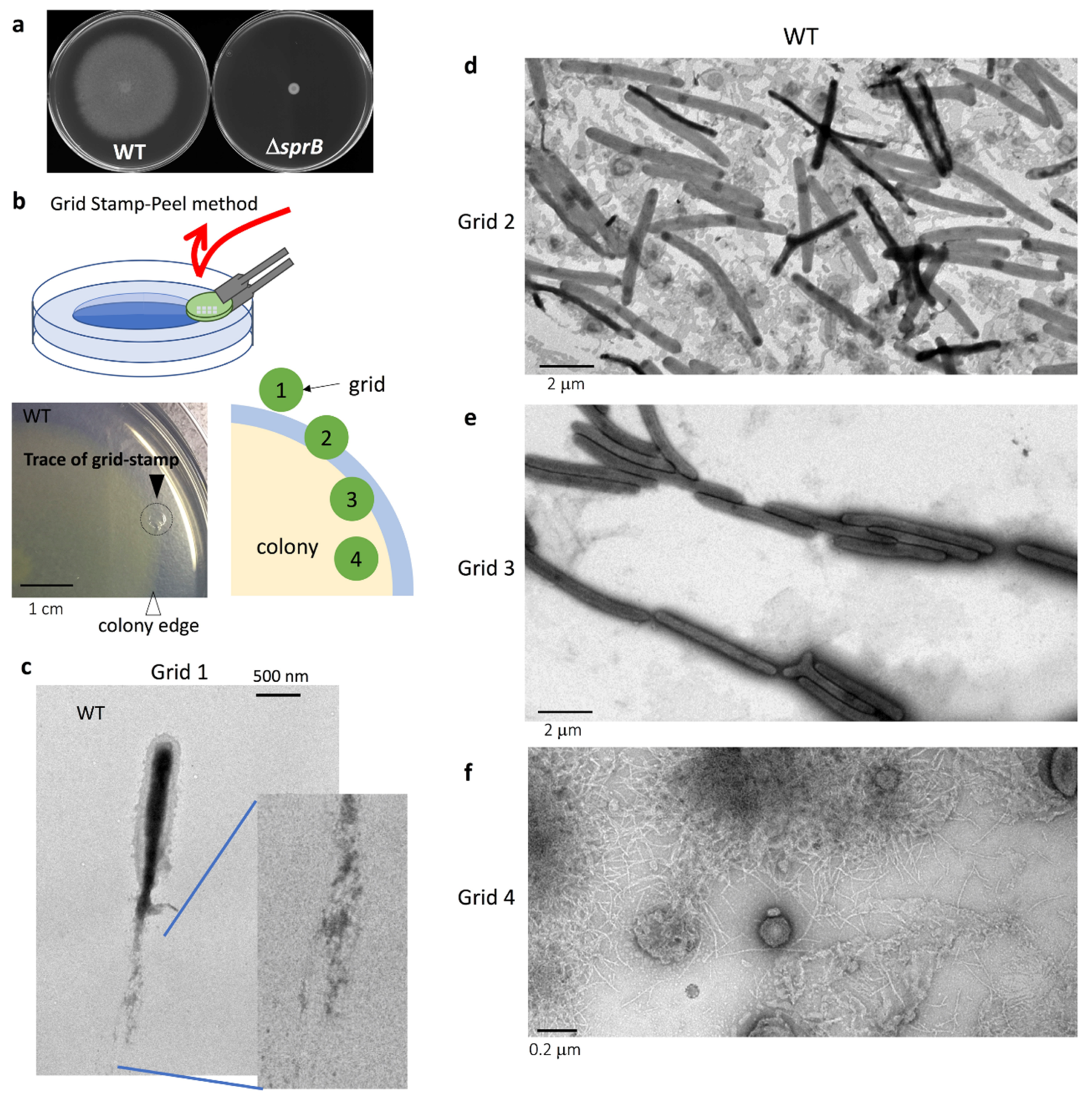
2.1. Grid Stamp-Peel Method
2.2. The Surface Structure at the Leading Edge of Spreading WT Colonies
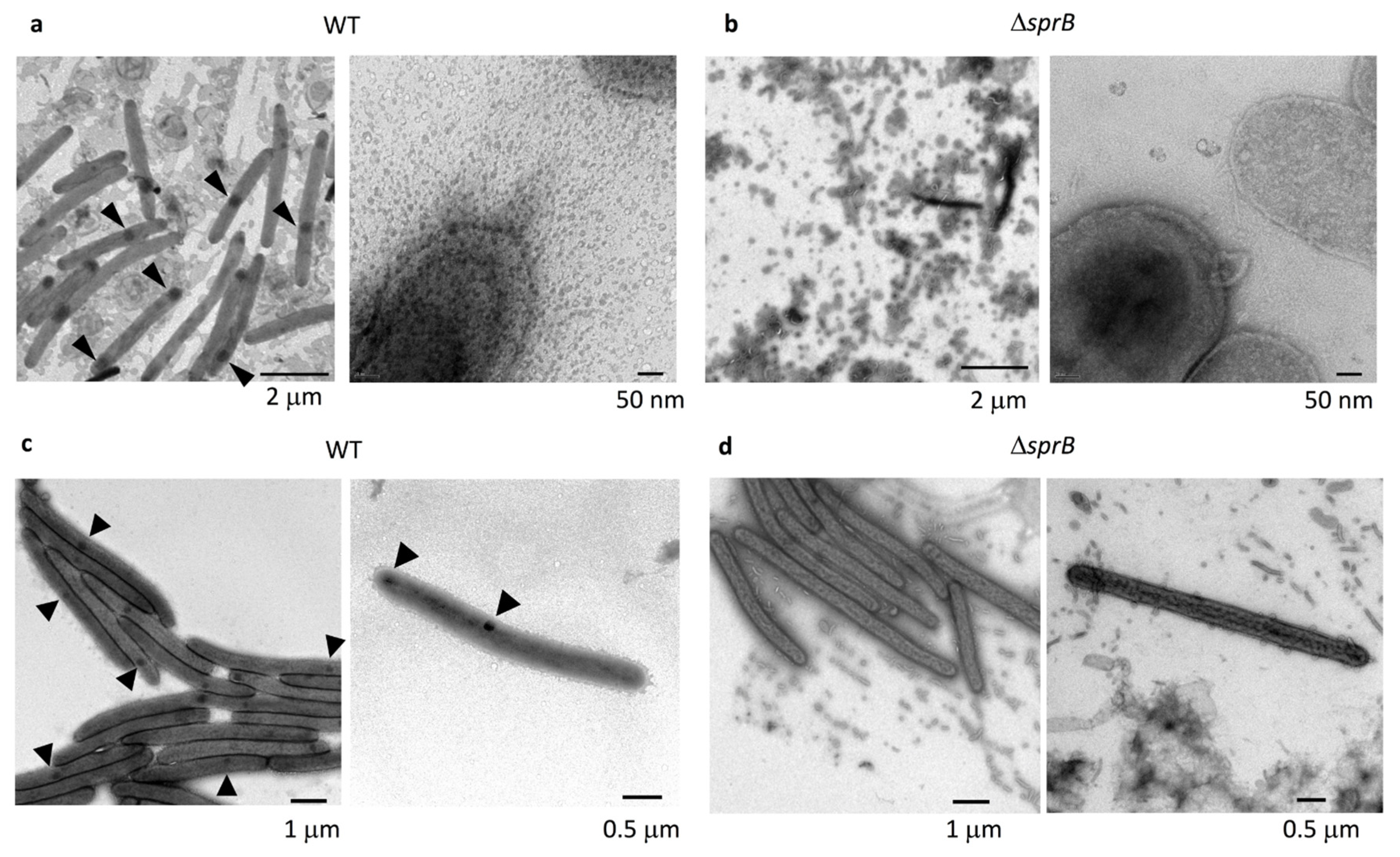
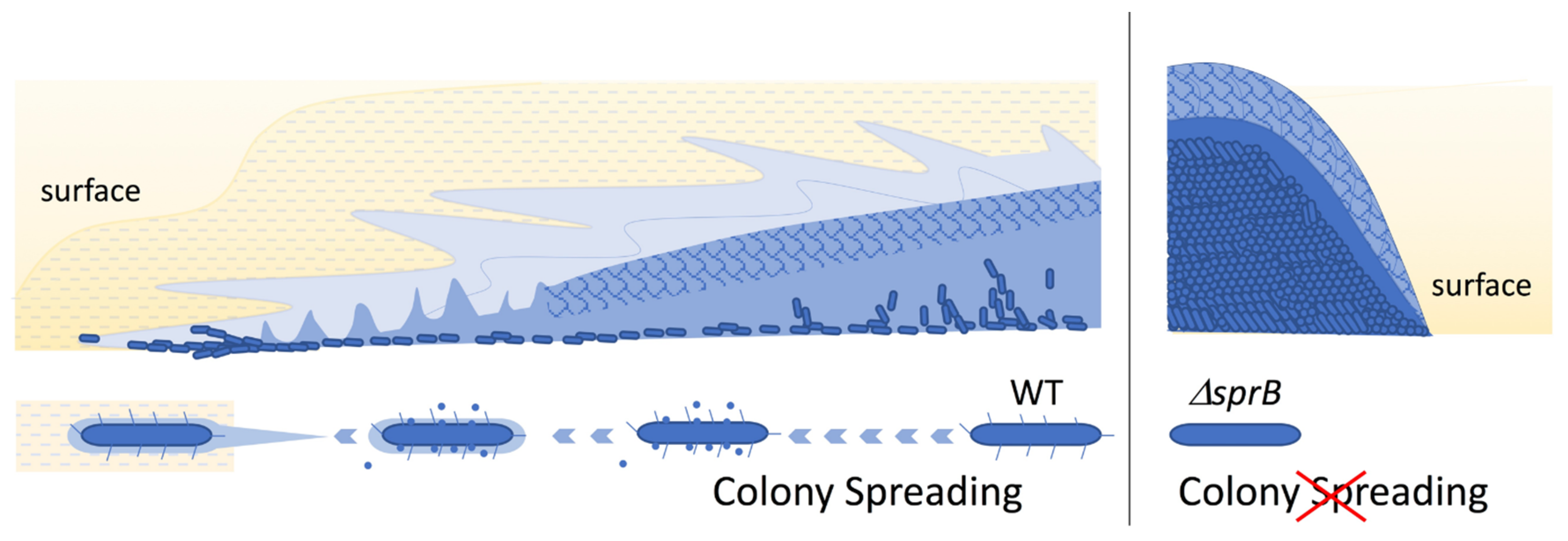
2.3. Comparison of the Surface Structure at the Edge of WT and Adhesin sprB-Deficient Colonies
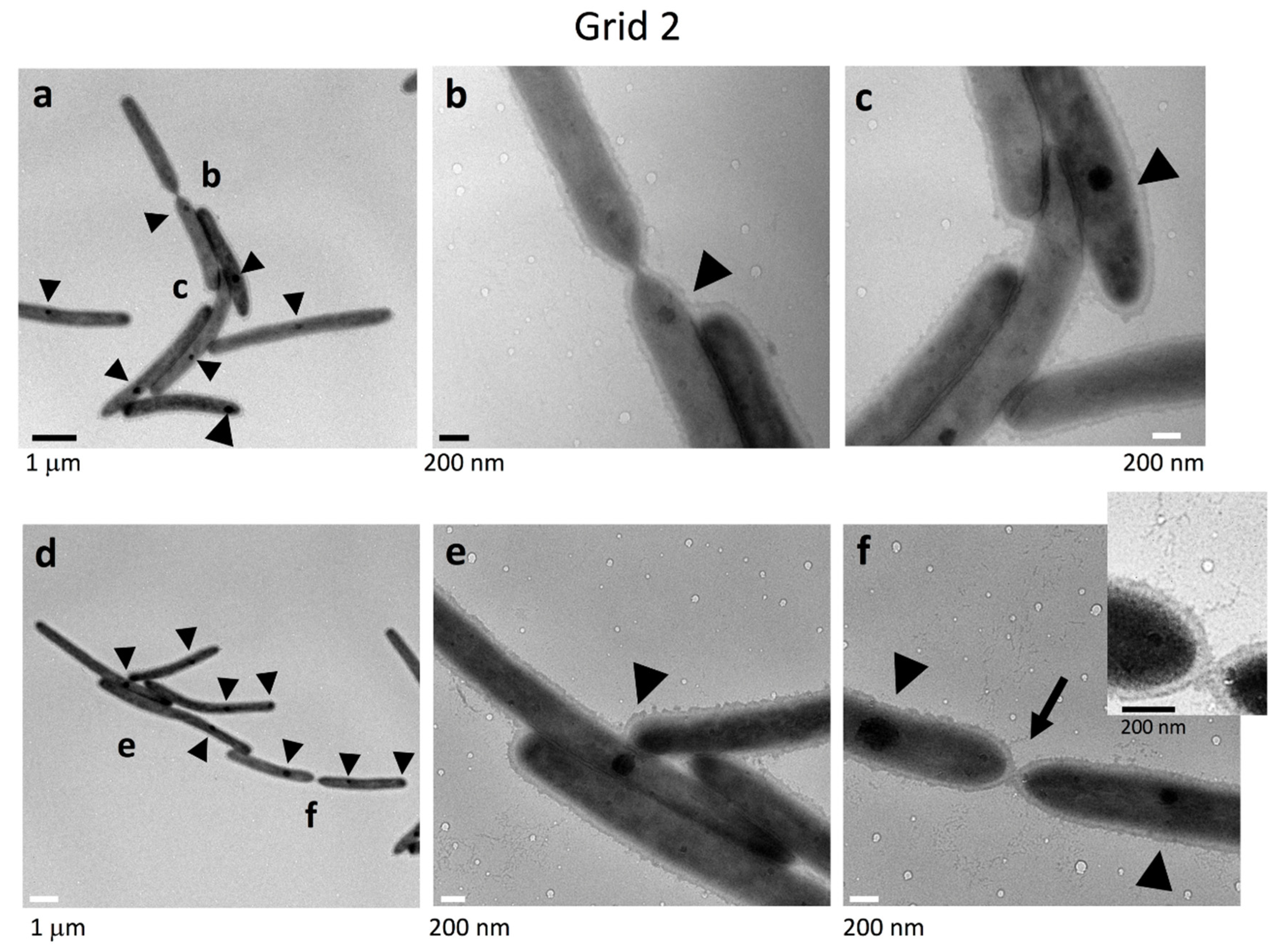
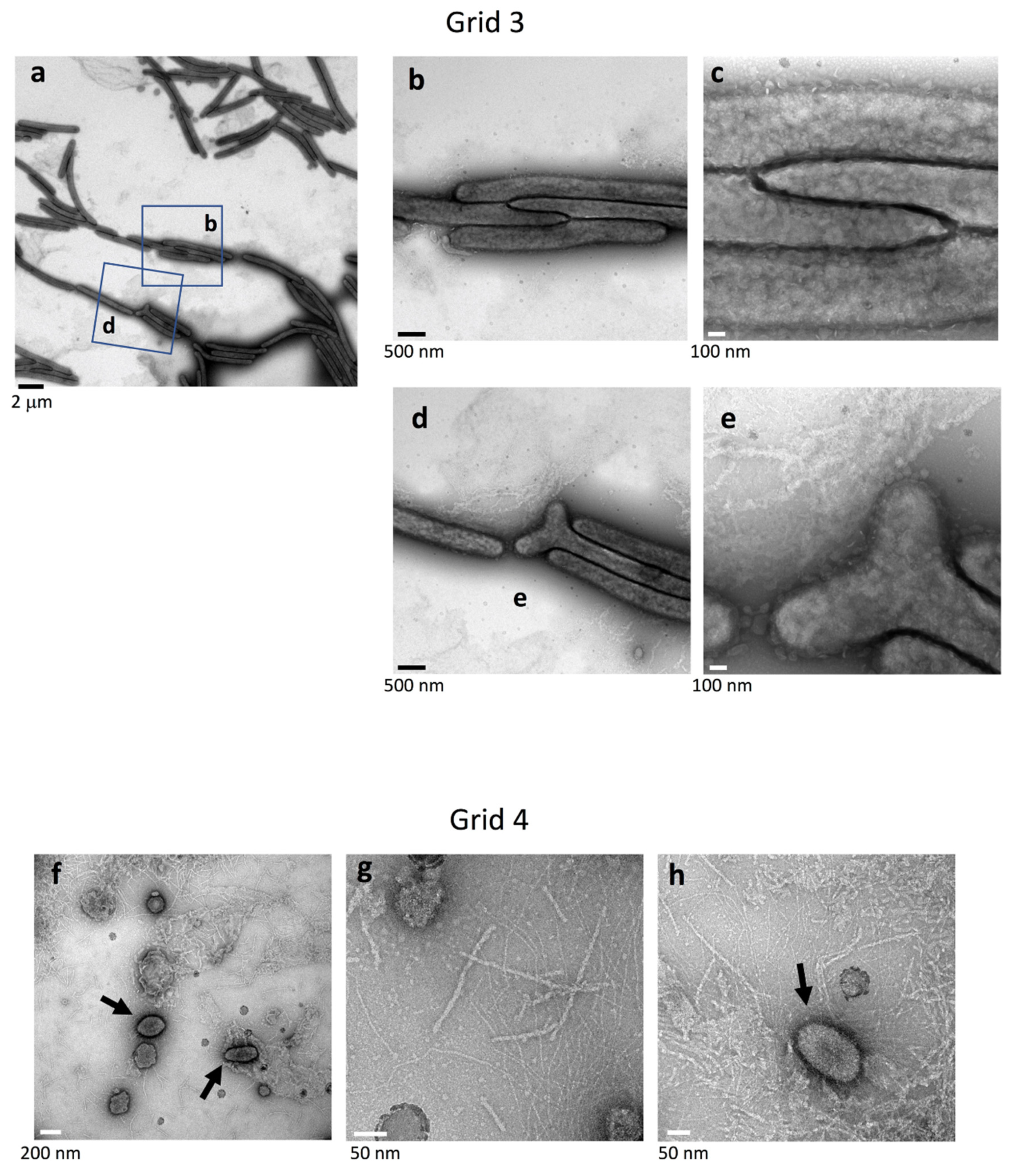
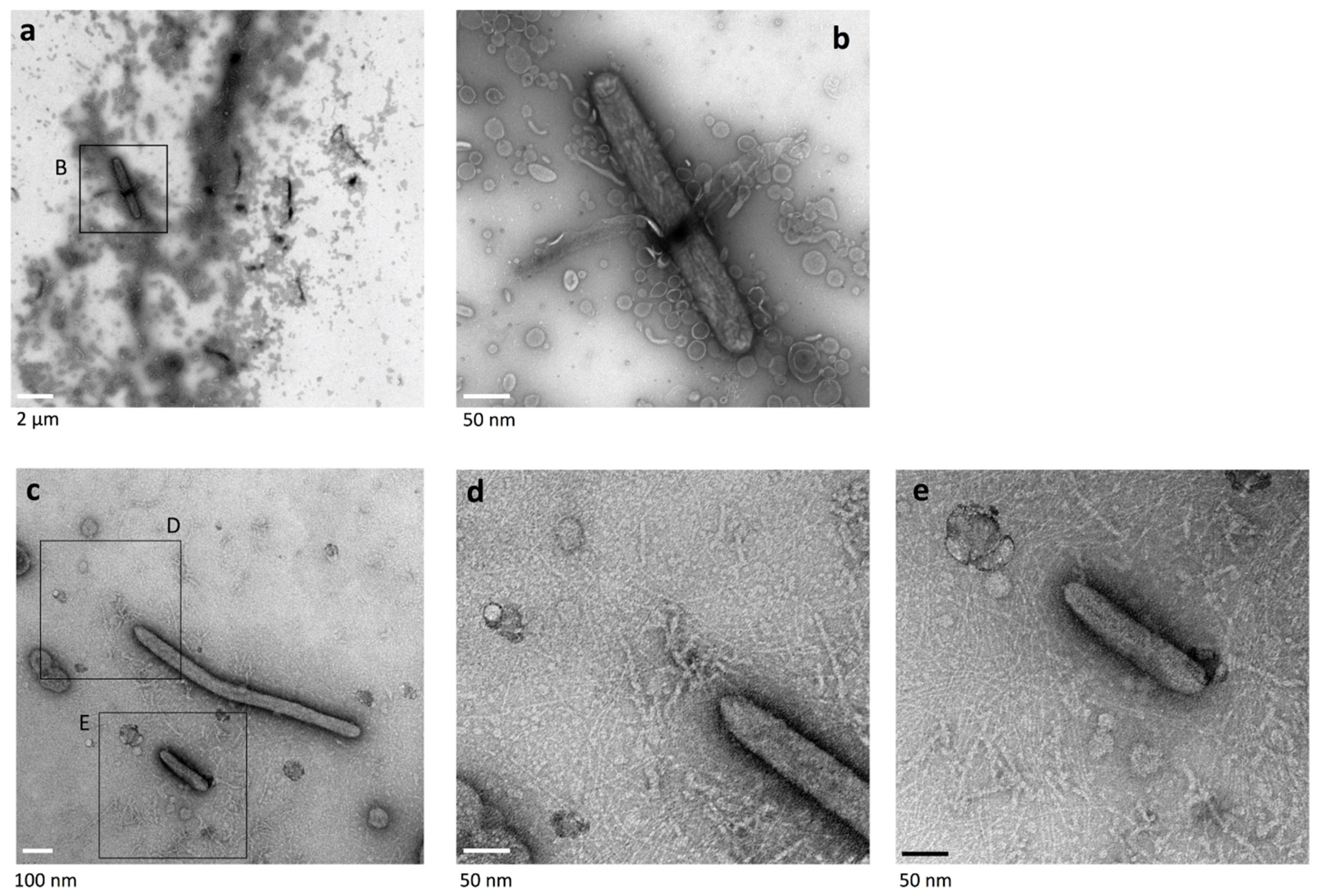
2.4. Cell Connections at the Translucent Edges of Spreading WT Colonies
2.5. Surface Structure Inside the Edge of WT Colony Bodies
2.6. Top Surface Inside the Edge of Nonspreading sprB Colonies
2.7. Surface Structure of Nonspreading gldK Mutant Colonies Deficient in T9SS
2.8. Surface Structure of Nonspreading gldG Mutant Colonies Deficient in Gliding Motility Protein
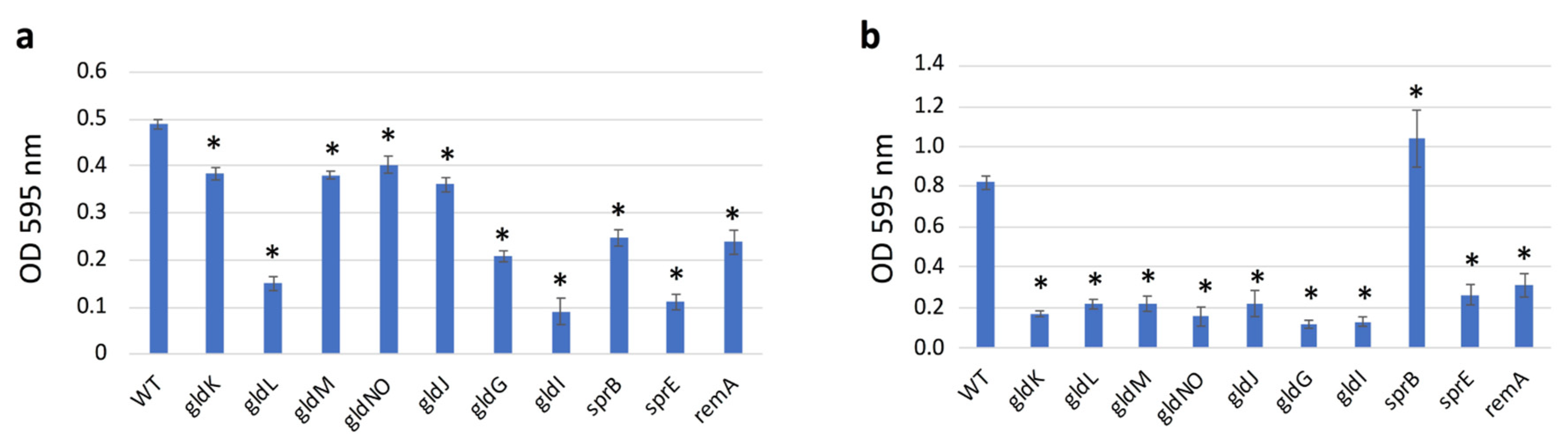
2.9. Quantification of Biofilms Formed
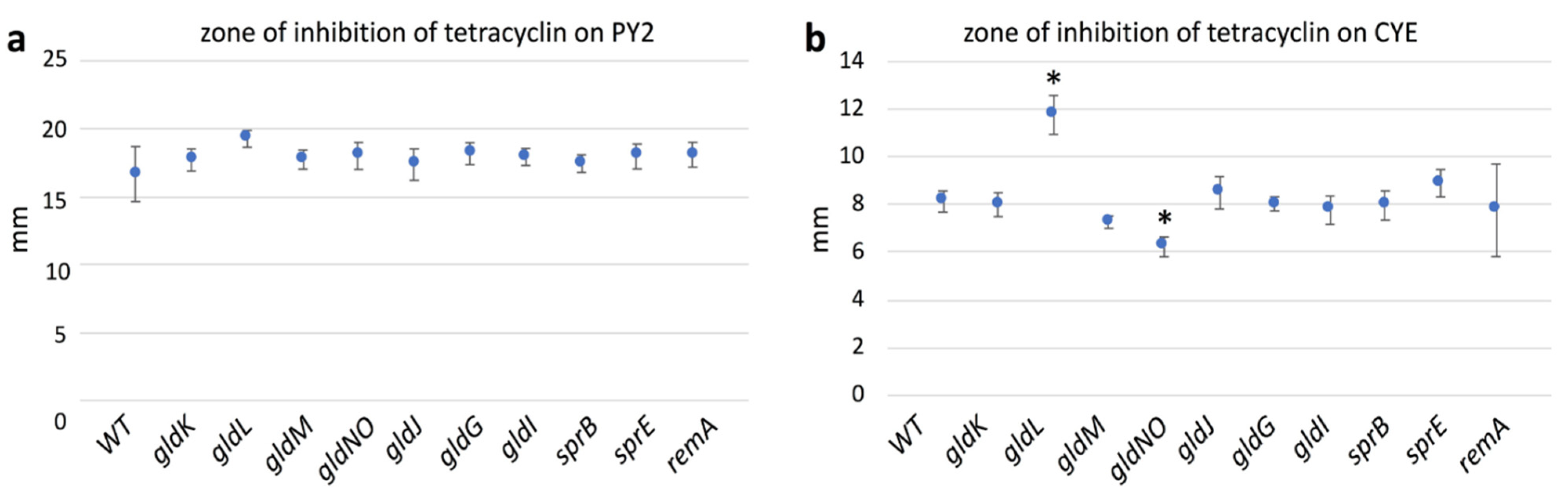
2.10. Antibiotic Susceptibility
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Biofilm Cultivation
4.2. Carbon-Grid Stamp-Peel Method
4.3. TEM Imaging
4.4. Crystal Violet Biofilm Assay
4.5. Disk Diffusion Susceptibility Test
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| WT | Wild-type |
| sprB | sprB deletion mutant CJ1922 |
| gldK | Spontaneous gldK mutant |
| gldL | gldL::HimarEm1 |
| gldM | Spontaneous gldM mutant |
| gldNO | gldNO::HimarEm1 |
| gldJ | Spontaneous gldJ mutant |
| gldG | Spontaneous gldG mutant |
| gldI | Spontaneous gldI mutant |
| sprE | Spontaneous sprE mutant |
| remA | remA deletion mutant |
| T9SS | Type IX secretion system |
| EPM | Extracellular polymeric matrix |
| TEM | Transmission electron microscopy |
References
- Agarwal, S.; Hunnicutt, D.W.; McBride, M.J. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 1997, 94, 12139–12144. [Google Scholar] [CrossRef] [Green Version]
- McBride, M.J. Cytophaga-flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 2004, 7, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Nelson, S.S.; Bollampalli, S.; McBride, M.J. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 2008, 190, 2851–2857. [Google Scholar] [CrossRef] [Green Version]
- Nakane, D.; Sato, K.; Wada, H.; McBride, M.J.; Nakayama, K. Helical flow of surface protein required for bacterial gliding motility. Proc. Natl. Acad. Sci. USA 2013, 110, 11145–11150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarrell, K.F.; McBride, M.J. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 2008, 6, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Berg, H.C. Towards a model for Flavobacterium gliding. Curr. Opin. Microbiol. 2015, 28, 93–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrivastava, A.; Rhodes, R.G.; Pochiraju, S.; Nakane, D.; McBride, M.J. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J. Bacteriol. 2012, 194, 3678–3688. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Naito, M.; Yukitake, H.; Hirakawa, H.; Shoji, M.; McBride, M.J.; Rhodes, R.G.; Nakayama, K. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 276–281. [Google Scholar] [CrossRef] [Green Version]
- McBride, M.J.; Zhu, Y. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J. Bacteriol. 2013, 195, 270–278. [Google Scholar] [CrossRef] [Green Version]
- James, R.H.; Deme, J.C.; Kjӕr, A.; Alcock, F.; Silale, A.; Lauber, F.; Johnson, S.; Berks, B.C.; Lea, S.M. Structure and mechanism of the proton-driven motor that powers type 9 secretion and gliding motility. Nat. Microbiol. 2021, 6, 221–233. [Google Scholar] [CrossRef]
- Shrivastava, A.; Johnston, J.J.; van Baaren, J.M.; McBride, M.J. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J. Bacteriol. 2013, 195, 3201–3212. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, S.S.; Johnston, J.J.; Zhu, Y.; Hying, Z.T.; McBride, M.J. The Carboxy-Terminal Region of Flavobacterium johnsoniae SprB Facilitates Its Secretion by the Type IX Secretion System and Propulsion by the Gliding Motility Machinery. J. Bacteriol. 2019, 201, e00218–e00219. [Google Scholar] [CrossRef] [Green Version]
- Veith, P.D.; Muhammad, N.A.N.; Dashper, S.G.; Likić, V.A.; Gorasia, D.G.; Chen, D.; Byrne, S.J.; Catmull, D.V.; Reynolds, E.C. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J. Proteome Res. 2013, 12, 4449–4461. [Google Scholar] [CrossRef] [PubMed]
- Hunnicutt, D.W.; Kempf, M.J.; Mark, J.; McBride, M.J. Mutations in Flavobacterium johnsoniae gldF and gldG Disrupt Gliding Motility and Interfere with Membrane Localization of GldA. J. Bacteriol. 2002, 184, 2370–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Pascual, D.; Rochat, T.; Kerouault, B.; Gómez, E.; Neulat-Ripoll, F.; Henry, C.; Quillet, E.; Guijarro, J.A.; Bernardet, J.F.; Duchaud, E. More Than Gliding: Involvement of GldD and GldG in the Virulence of Flavobacterium psychrophilum. Front. Microbiol. 2017, 8, 2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, S.; Okuda, K.; Miyakawa, R.; Sato, M.; Arita-Morioka, K.; Chiba, A.; Yamanaka, K.; Ogura, T.; Mizunoe, Y.; Sato, C. Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy. Sci. Rep. 2016, 6, 25889. [Google Scholar] [CrossRef] [Green Version]
- Okuda, K.I.; Nagahori, R.; Yamada, S.; Sugimoto, S.; Sato, C.; Sato, M.; Iwase, T.; Hashimoto, K.; Mizunoe, Y. The composition and structure of biofilms developed by Propionibacterium acnes isolated from cardiac pacemaker devices. Front. Microbiol. 2018, 9, 182. [Google Scholar] [CrossRef] [Green Version]
- Decostere, A.; Haesebrouck, F.; Charlier, G.; Ducatelle, R. The association of Flavobacterium columnare strains of high and low virulence with gill tissue of black mollies (Poecilia sphenops). Vet. Microbiol. 1999, 67, 287–298. [Google Scholar] [CrossRef]
- Levipan, H.A.; Avendaño-Herrera, R. Different Phenotypes of Mature Biofilm in Flavobacterium psychrophilum Share a Potential for Virulence That Differs from Planktonic State. Front. Cell Infect. Microbiol. 2017, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Kawai, K.; Kurohara, K.; Oshima, S. Adherence of Flavobacterium psychrophilum on the body surface of the ayu Plecoglossus altivelis. Microbes Infect. 2002, 4, 279–283. [Google Scholar] [CrossRef]
- Gavriilidou, A.; Gutleben, J.; Versluis, D.; Forgiarini, F.; Passel, M.W.J.; Ingham, C.J.; Smidt, H.; Sipkema, D. Comparative genomic analysis of Flavobacteriaceae: Insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis. BMC Genom. 2020, 21, 569. [Google Scholar] [CrossRef]
- Li, N.; Zhu, Y.; LaFrentz, B.R.; Evenhuis, J.P.; Hunnicutt, D.W.; Conrad, R.A.; Barbier, P.; Gullstrand, C.W.; Roets, J.E.; Powers, J.L.; et al. The Type IX Secretion System Is Required for Virulence of the Fish Pathogen Flavobacterium columnare. Appl. Environ. Microbiol. 2017, 83, e01769-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbier, P.; Rochat, T.; Mohammed, H.H.; Wiens, G.D.; Bernardet, J.F.; Halpern, D.; Duchaud, E.; McBride, M.J. The Type IX Secretion System Is Required for Virulence of the Fish Pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2020, 86, e00799-20. [Google Scholar] [CrossRef] [PubMed]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa Biofilms: Host Response and Clinical Implications in Lung Infections. Am. J. Respir. Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Live, L.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumru, S.; Tekedar, H.C.; Gulsoy, N.; Waldbieser, G.C.; Lawrence, M.L.; Karsi, A. Comparative Analysis of the Flavobacterium columnare Genomovar I and II Genomes. Front. Microbiol. 2017, 8, 1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrivastava, A.; Patel, V.K.; Tang, Y.; Yost, S.C.; Dewhirst, F.E.; Berg, H.C. Cargo transport shapes the spatial organization of a microbial community. Proc. Natl. Acad. Sci. USA 2018, 115, 8633–8638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loch, T.P.; Faisal, M. Emerging flavobacterial infections in fish: A review. J. Adv. Res. 2015, 6, 283–300. [Google Scholar] [CrossRef]
- Brenner, D.J.; Hollis, D.G.; Fanning, G.R.; Weaver, R.E. Capnocytophaga canimorsus sp. nov. (formerly CDC group DF-2), a cause of septicemia following dog bite, and C. cynodegmi sp. nov., a cause of localized wound infection following dog bite. J. Clin. Microbiol. 1989, 27, 231–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laanto, E.; Penttinen, R.K.; Bamford, J.K.; Sundberg, L.R. Comparing the different morphotypes of a fish pathogen--implications for key virulence factors in Flavobacterium columnare. BMC Microbiol. 2014, 14, 170. [Google Scholar] [CrossRef] [Green Version]
- Kayansamruaj, P.; Dong, H.T.; Hirono, I.; Kondo, H.; Senapin, S.; Rodkhum, C. Comparative genome analysis of fish pathogen Flavobacterium columnare reveals extensive sequence diversity within the species. Infect. Genet. Evol. 2017, 54, 7–17. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Fuente-Núñez, C.D.l.; Reffuveille, F.; Fernández, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef]
- Olsen, I. Biofilm-specific antibiotic tolerance and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Sato, K.; Naya, M.; Hatano, Y.; Kondo, Y.; Sato, M.; Narita, Y.; Nagano, K.; Naito, M.; Sato, C. Biofilm spreading by the adhesin-dependent gliding motility of Flavobacterium johnsoniae. 1. Internal structure of the biofilm. Int. J. Mol. Sci. 2021, 22, 1894. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Naya, M.; Hatano, Y.; Kondo, Y.; Sato, M.; Narita, Y.; Nagano, K.; Naito, M.; Nakayama, K.; Sato, C. Colony spreading of the gliding bacterium Flavobacterium johnsoniae in the absence of the motility adhesin SprB. Sci. Rep. 2021, 11, 697. [Google Scholar] [CrossRef]
- Cai, W.; Fuenteb, L.D.L.; Arias, C.R. Biofilm Formation by the Fish Pathogen Flavobacterium columnare: Development and Parameters Affecting Surface Attachment. Appl. Environ. Microbiol. 2013, 79, 5633–5642. [Google Scholar] [CrossRef] [Green Version]
- Faure, L.M.; Fiche, J.B.; Espinosa, L.; Ducret, A.; Anantharaman, V.; Luciano, J.; Lhospice, S.; Islam, S.T.; Tréguier, J.; Sotes, M.; et al. The mechanism of force transmission at bacterial focal adhesion complexes. Nature 2016, 539, 530–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arciola, C.R.; Campoccia, D.; Montanaro, L. Implant infections: Adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 2018, 16, 397–409. [Google Scholar] [CrossRef]
- Veerachamy, S.; Yarlagadda, T.; Manivasagam, G.; Yarlagadda, P.K. Bacterial adherence and biofilm formation on medical implants. Proc. Inst. Mech. Eng. H 2014, 228, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilm formation: A clinically relevant microbiological process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef] [Green Version]
- Hajishengallis, G. Complement and periodontitis. Biochem. Pharmacol. 2010, 80, 1992–2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosaddad, S.A.; Tahmasebi, E.; Yazdanian, A.; Rezvani, M.B.; Seifalian, A.; Yazdanian, M.; Tebyanian, H. Oral microbial biofilms: An update. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2005–2019. [Google Scholar] [CrossRef]
- Wolff, D.; Frese, C.; Maier-Kraus, T.; Krueger, T.; Wolff, B. Bacterial biofilm composition in caries and caries-free subjects. Caries Res. 2013, 47, 69–77. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Dental biofilms: Difficult therapeutic targets. Periodontol 2000 2002, 28, 12–55. [Google Scholar] [CrossRef]
- Sato, C.; Yamazaki, D.; Sato, M.; Takeshima, H.; Memtily, N.; Hatano, Y.; Tsukuba, T.; Sakai, E. Calcium phosphate mineralization in bone tissues directly observed in aqueous liquid by atmospheric SEM (ASEM) without staining: Microfluidics crystallization chamber and immuno-EM. Sci. Rep. 2019, 9, 7352. [Google Scholar] [CrossRef]
- Naya, M.; Sato, C. Pyrene Excimer-Based Fluorescent Labeling of Neighboring Cysteines by Protein Dynamics: ASEM-Induced Thiol-Ene Click Reaction for High Spatial Resolution CLEM. Int. J. Mol. Sci. 2020, 21, 7550. [Google Scholar] [CrossRef]
- Rhodes, R.G.; Pucker, H.G.; McBride, M.J. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant gliding motility genes remF, remG, remH, and remI. J. Bacteriol. 2011, 193, 2418–2428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, L.E.; Pate, J.L.; Betzig, R.J. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsoniae. J. Bacteriol. 1984, 159, 26–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, T.F.; Khubbar, M.K.; Saffarini, D.A.; McBride, M.J. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J. Bacteriol. 2005, 187, 6943–6952. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Strain | Description | Reference |
|---|---|---|
| Cj1827 | Wild-type rpsL2 | [52] |
| UW102-57 | Spontaneous gldK mutant | [53] |
| CJ1300 | gldL::HimarEm1 | [54] |
| FJ113 | Spontaneous gldM mutant | [54] |
| CJ1631A | gldNO::HimarEm1 | [54] |
| UW102-55 | Spontaneous gldJ mutant | [53] |
| CJ1922 | sprB deletion mutant | [52] |
| FJ149 | Spontaneous sprE mutant | [54] |
| UW102-34 | Spontaneous gldG mutant | [53] |
| UW102-41 | Spontaneous gldI mutant | [53] |
| CJ1984 | remA deletion mutant | [7] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, K.; Naya, M.; Hatano, Y.; Kasahata, N.; Kondo, Y.; Sato, M.; Takebe, K.; Naito, M.; Sato, C. Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae: 2. Role of Filamentous Extracellular Network and Cell-to-Cell Connections at the Biofilm Surface. Int. J. Mol. Sci. 2021, 22, 6911. https://doi.org/10.3390/ijms22136911
Sato K, Naya M, Hatano Y, Kasahata N, Kondo Y, Sato M, Takebe K, Naito M, Sato C. Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae: 2. Role of Filamentous Extracellular Network and Cell-to-Cell Connections at the Biofilm Surface. International Journal of Molecular Sciences. 2021; 22(13):6911. https://doi.org/10.3390/ijms22136911
Chicago/Turabian StyleSato, Keiko, Masami Naya, Yuri Hatano, Naoki Kasahata, Yoshio Kondo, Mari Sato, Katsuki Takebe, Mariko Naito, and Chikara Sato. 2021. "Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae: 2. Role of Filamentous Extracellular Network and Cell-to-Cell Connections at the Biofilm Surface" International Journal of Molecular Sciences 22, no. 13: 6911. https://doi.org/10.3390/ijms22136911
APA StyleSato, K., Naya, M., Hatano, Y., Kasahata, N., Kondo, Y., Sato, M., Takebe, K., Naito, M., & Sato, C. (2021). Biofilm Spreading by the Adhesin-Dependent Gliding Motility of Flavobacterium johnsoniae: 2. Role of Filamentous Extracellular Network and Cell-to-Cell Connections at the Biofilm Surface. International Journal of Molecular Sciences, 22(13), 6911. https://doi.org/10.3390/ijms22136911









