Therapeutic Effect of a Newly Isolated Lytic Bacteriophage against Multi-Drug-Resistant Cutibacterium acnes Infection in Mice
Abstract
1. Introduction
2. Results
2.1. Isolation and Characterization of Phage TCUCAP1
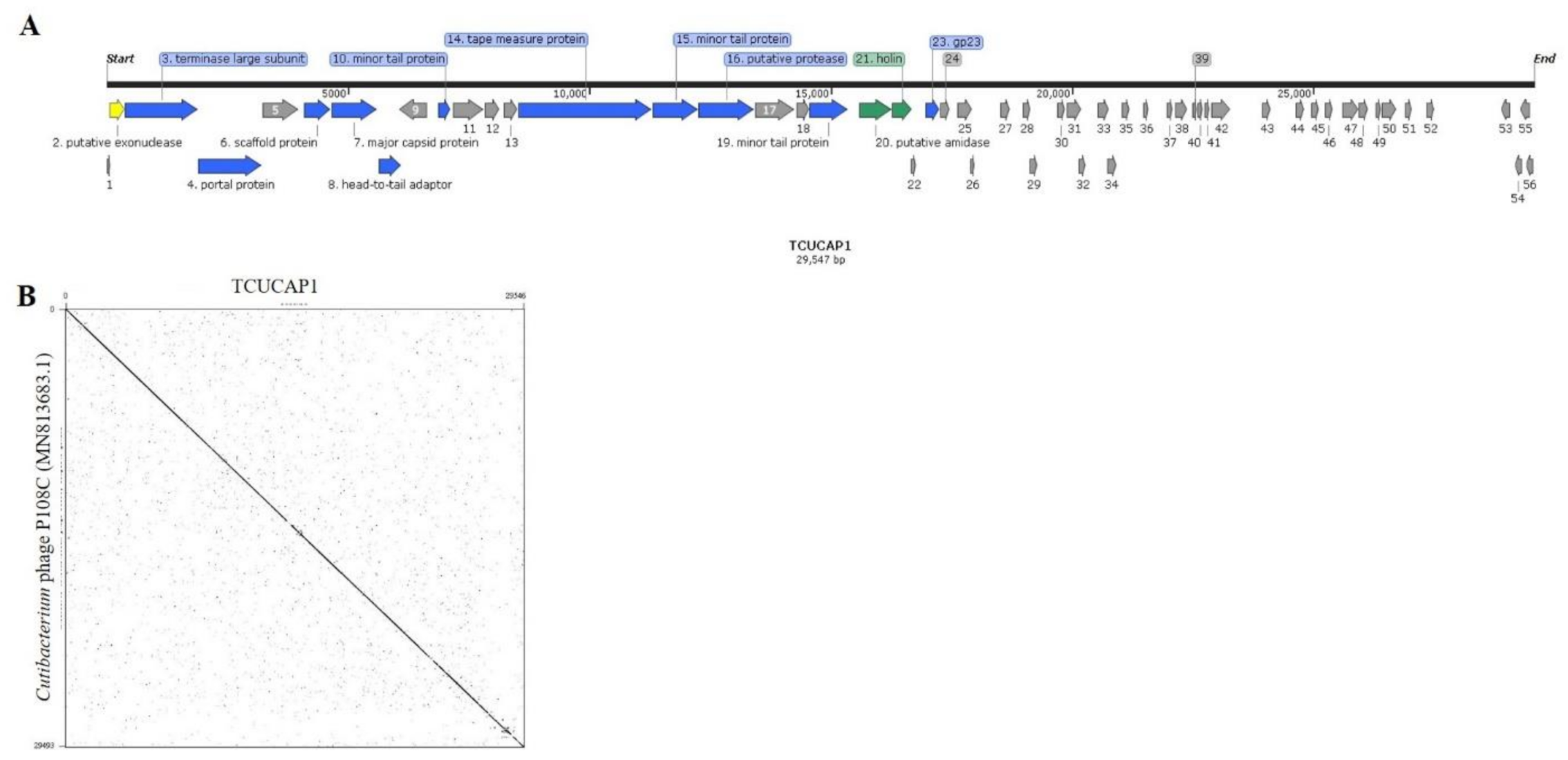
2.2. The Phage Genome
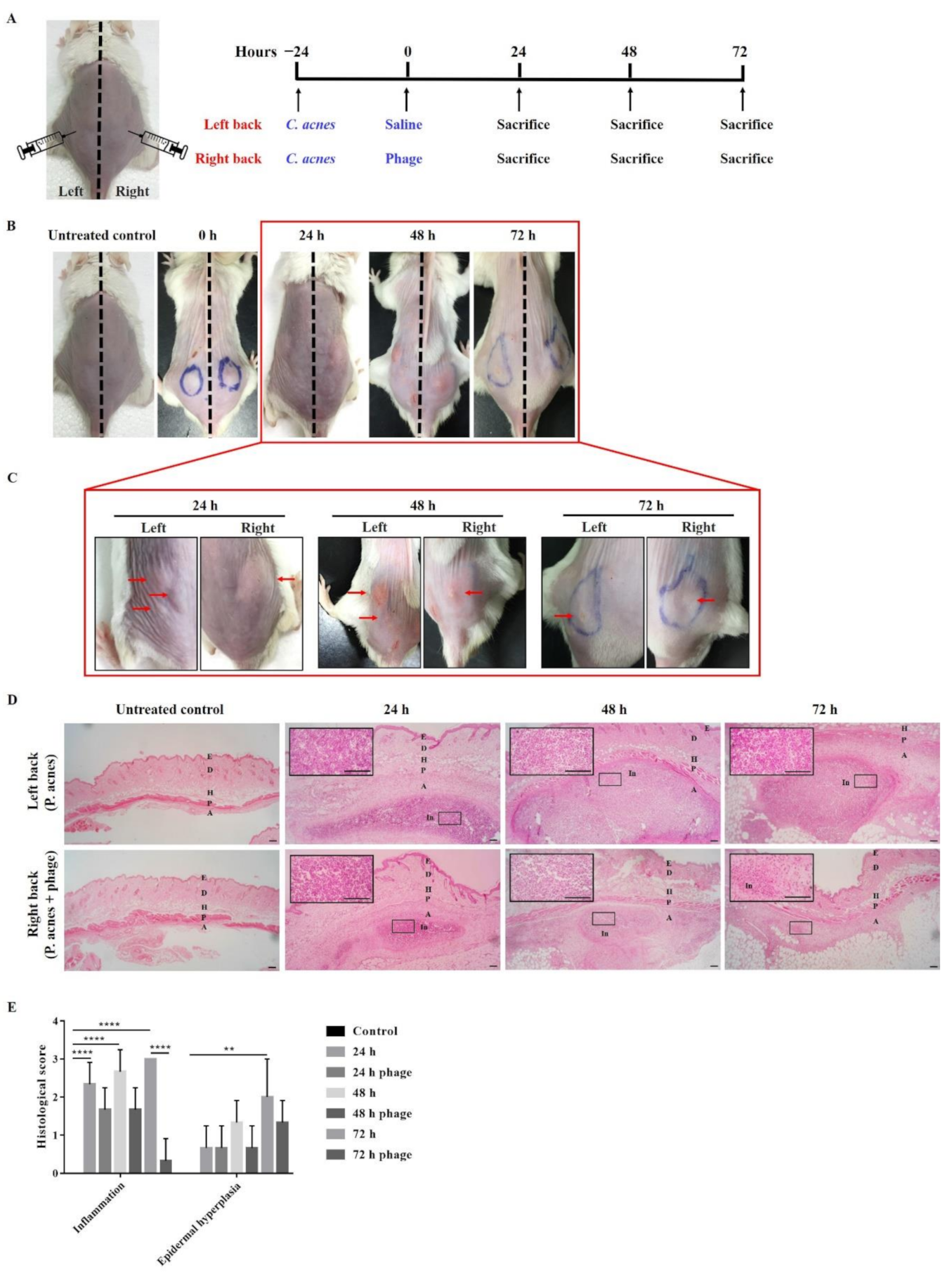
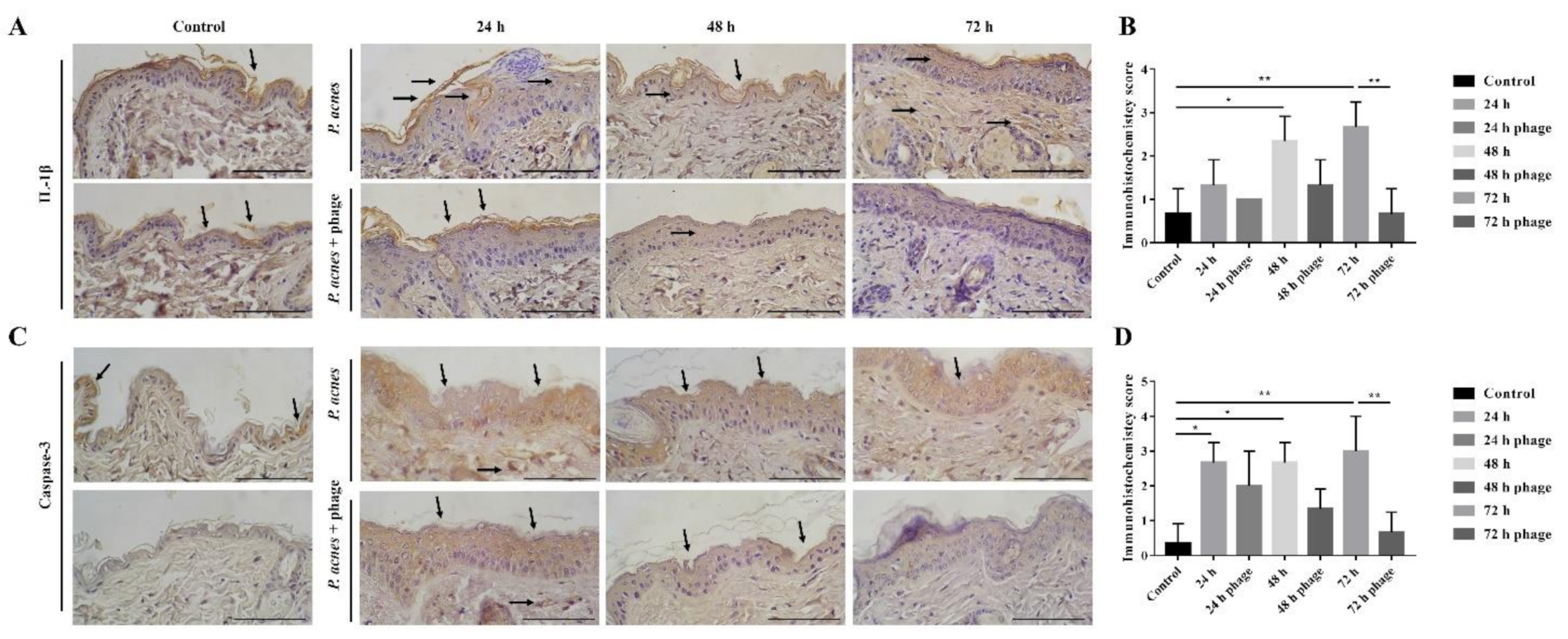
2.3. Phage Therapy in Mouse Intradermal Injection Model
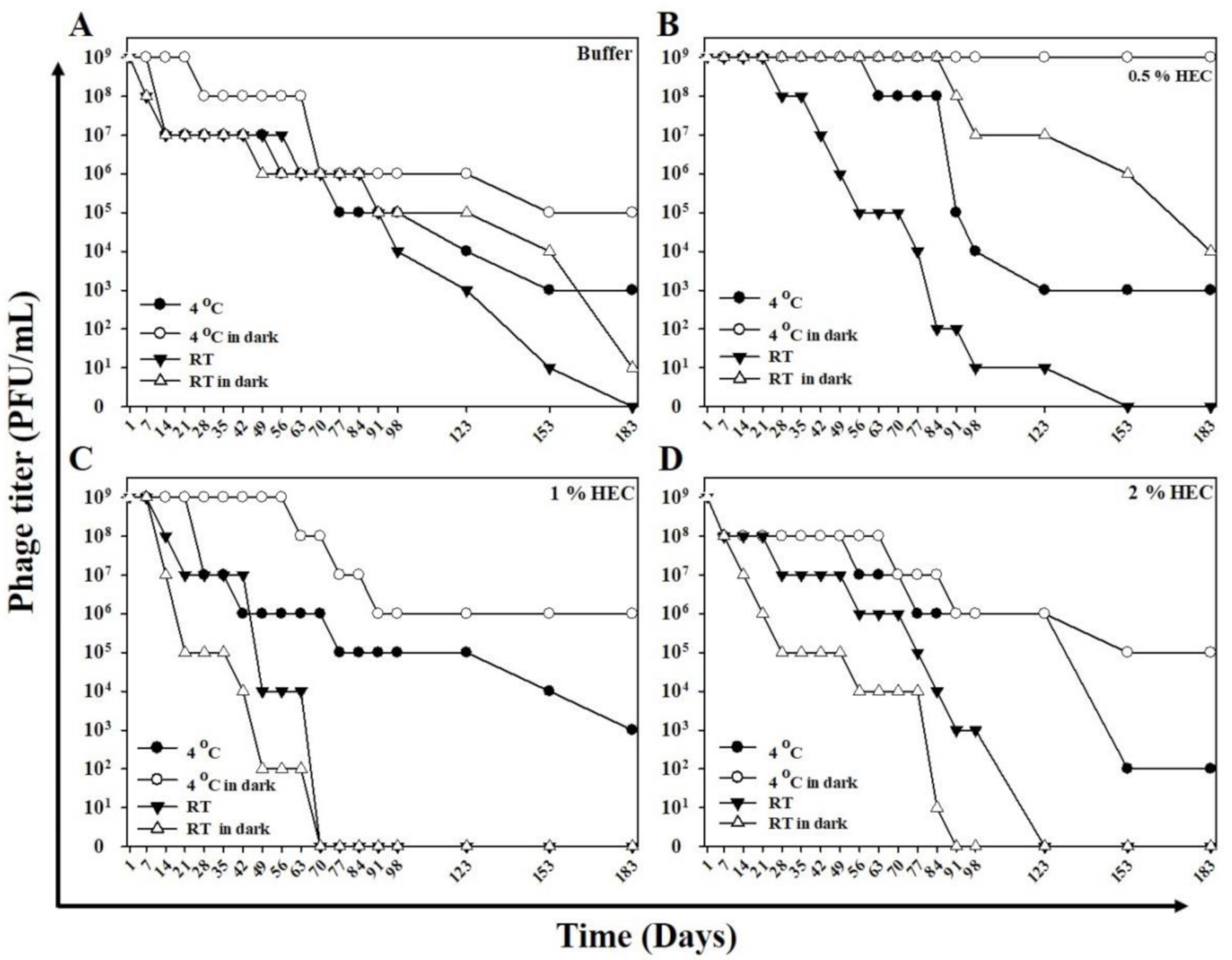
2.4. Lytic Capacity and Stability of the Phage HEC Cream
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Isolation and Propagation of Bacteriophage
4.3. Host Range Analysis
4.4. Caesium Chloride (CsCl) Gradient Purification
4.5. Transmission Electron Microscopy (TEM) Analysis
4.6. Extraction of Bacteriophage DNA
4.7. Genome Sequencing and Bioinformatic Analysis
4.8. Phage Therapy in Mouse Subcutaneous Injection Model
4.9. Examination of Pathological and Histological Changes
4.10. Immunohistochemisry Stain
4.11. Preparation of Hydroxyethyl Cellulose (HEC) Cream
4.12. Lytic Capacity of the Phage HEC Cream
4.13. Stability of the Phage HEC Cream
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elston, M.J.; Dupaix, J.P.; Opanova, M.I.; Atkinson, R.E. Cutibacterium acnes (formerly Proprionibacterium acnes) and Shoulder Surgery. Hawaii J. Health Soc. Welf. 2019, 78, 3–5. [Google Scholar] [PubMed]
- Perry, A.; Lambert, P. Propionibacterium acnes: Infection beyond the skin. Expert Rev. Anti. Infect. Ther. 2011, 9, 1149–1156. [Google Scholar] [CrossRef]
- Omer, H.; McDowell, A.; Alexeyev, O.A. Understanding the role of Propionibacterium acnes in acne vulgaris: The critical importance of skin sampling methodologies. Clin. Dermatol. 2017, 35, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.D.; Umari, T.; Dunnick, C.A.; Dellavalle, R.P. The epidemiology of acne vulgaris in late adolescence. Adolesc. Health Med. Ther. 2016, 7, 13–25. [Google Scholar] [CrossRef]
- Skroza, N.; Tolino, E.; Mambrin, A.; Zuber, S.; Balduzzi, V.; Marchesiello, A.; Bernardini, N.; Proietti, I.; Potenza, C. Adult Acne Versus Adolescent Acne: A Retrospective Study of 1,167 Patients. J. Clin. Aesthet. Dermatol. 2018, 11, 21–25. [Google Scholar]
- Rocha, M.A.; Bagatin, E. Adult-onset acne: Prevalence, impact, and management challenges. Clin. Cosmet. Investig. Dermatol. 2018, 11, 59–69. [Google Scholar] [CrossRef]
- Kraft, J.; Freiman, A. Management of acne. Can. Med. Assoc. J. 2011, 183, 430–435. [Google Scholar] [CrossRef]
- Zhu, T.; Zhu, W.; Wang, Q.; He, L.; Wu, W.; Liu, J.; Li, Y.; Sun, D. Antibiotic susceptibility of Propionibacterium acnes isolated from patients with acne in a public hospital in Southwest China: Prospective cross-sectional study. BMJ Open 2019, 9, e022938. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, N.; Hernandez, P.O.; Tyring, S.K.; Haitz, K.A.; Motta, A. Antimicrobial susceptibility of Propionibacterium acnes isolates from acne patients in Colombia. Int. J. Dermatol. 2013, 52, 688–692. [Google Scholar] [CrossRef]
- Luk, N.M.; Hui, M.; Lee, H.C.; Fu, L.H.; Liu, Z.H.; Lam, L.Y.; Eastel, M.; Chan, Y.K.; Tang, L.S.; Cheng, T.S.; et al. Antibiotic-resistant Propionibacterium acnes among acne patients in a regional skin centre in Hong Kong. J. Eur. Acad. Dermatol. Venereol. 2013, 27, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Abdel Fattah, N.S.; Darwish, Y.W. In vitro antibiotic susceptibility patterns of Propionibacterium acnes isolated from acne patients: An Egyptian university hospital-based study. J. Eur. Acad. Dermatol. Venereol. 2013, 27, 1546–1551. [Google Scholar] [CrossRef] [PubMed]
- Jahns, A.C.; Lundskog, B.; Ganceviciene, R.; Palmer, R.H.; Golovleva, I.; Zouboulis, C.C.; McDowell, A.; Patrick, S.; Alexeyev, O.A. An increased incidence of Propionibacterium acnes biofilms in acne vulgaris: A case-control study. Br. J. Dermatol. 2012, 167, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Peeters, E.; Nelis, H.J. Biofilm formation by Propionibacterium acnes is associated with increased resistance to antimicrobial agents and increased production of putative virulence factors. Res. Microbiol. 2007, 158, 386–392. [Google Scholar] [CrossRef]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G., Jr. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Nikolich, M.P.; Filippov, A.A. Bacteriophage Therapy: Developments and Directions. Antibiotics 2020, 9, 135. [Google Scholar] [CrossRef]
- Wu, S.; Zachary, E.; Wells, K.; Loc-Carrillo, C. Phage Therapy: Future Inquiries. Postdoc J. 2013, 1, 24–35. [Google Scholar] [CrossRef][Green Version]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef]
- Melo, L.D.R.; Pires, D.P.; Monteiro, R.; Azeredo, J. Phage Therapy of Infectious Biofilms: Challenges and Strategies. In Phage Therapy: A Practical Approach; Górski, A., Międzybrodzki, R., Borysowski, J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 295–313. [Google Scholar] [CrossRef]
- Hansen, M.F.; Svenningsen, S.L.; Røder, H.L.; Middelboe, M.; Burmølle, M. Big Impact of the Tiny: Bacteriophage-Bacteria Interactions in Biofilms. Trends Microbiols 2019, 27, 739–752. [Google Scholar] [CrossRef]
- Górski, A.; Targońska, M.; Borysowski, J.; Weber-Dabrowska, B. The potential of phage therapy in bacterial infections of the eye. Ophthalmologica 2009, 223, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, L.D. Bacteriophages for managing Shigella in various clinical and non-clinical settings. Bacteriophage 2013, 3, e25098. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Anggård, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- Rhoads, D.D.; Wolcott, R.D.; Kuskowski, M.A.; Wolcott, B.M.; Ward, L.S.; Sulakvelidze, A. Bacteriophage therapy of venous leg ulcers in humans: Results of a phase I safety trial. J. Wound Care 2009, 18, 237–243. [Google Scholar] [CrossRef]
- Rashel, M.; Uchiyama, J.; Ujihara, T.; Uehara, Y.; Kuramoto, S.; Sugihara, S.; Yagyu, K.; Muraoka, A.; Sugai, M.; Hiramatsu, K.; et al. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phi MR11. J. Infect. Dis. 2007, 196, 1237–1247. [Google Scholar] [CrossRef]
- Latz, S.; Krüttgen, A.; Häfner, H.; Buhl, E.M.; Ritter, K.; Horz, H.P. Differential Effect of Newly Isolated Phages Belonging to PB1-Like, phiKZ-Like and LUZ24-Like Viruses against Multi-Drug Resistant Pseudomonas aeruginosa under Varying Growth Conditions. Viruses 2017, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, B.; Xu, M.; Yan, Q.; Liu, S.; Zhu, X.; Sun, Z.; Reed, E.; Ding, L.; Gong, J.; et al. Use of bacteriophage in the treatment of experimental animal bacteremia from imipenem-resistant Pseudomonas aeruginosa. Int. J. Mol. Med. 2006, 17, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.W.; Shin, T.H.; Kim, J.H.; Shin, S.P.; Han, J.E.; Heo, G.J.; De Zoysa, M.; Shin, G.W.; Chai, J.Y.; Park, S.C. Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic-resistant O3:K6 pandemic clinical strain. J. Infect. Dis. 2014, 210, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.H.; Wang, J.L.; Wen, F.S.; Chang, K.M.; Kuo, C.F.; Lin, C.H.; Luo, H.R.; Hung, C.H. Isolation and characterization of φkm18p, a novel lytic phage with therapeutic potential against extensively drug resistant Acinetobacter baumannii. PLoS ONE 2012, 7, e46537. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.Y.; Yang, Z.C.; Gong, Y.L.; Huang, G.T.; Yin, S.P.; Jiang, B.; Peng, Y.Z. Therapeutic effect of phages on extensively drug-resistant Acinetobacter baumannii-induced sepsis in mice. Chin. J. Burns 2016, 32, 523–528. [Google Scholar] [CrossRef]
- Jeon, J.; Yong, D. Two Novel Bacteriophages Improve Survival in Galleria mellonella Infection and Mouse Acute Pneumonia Models Infected with Extensively Drug-Resistant Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2019, 85, e02900–e02918. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Huang, G.; Zhang, Y.; Jiang, B.; Yang, Z.; Dong, Z.; You, B.; Yuan, Z.; Hu, F.; Zhao, Y.; et al. Phage Abp1 Rescues Human Cells and Mice from Infection by Pan-Drug Resistant Acinetobacter Baumannii. Cell. Physiol. Biochem. 2017, 44, 2337–2345. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mi, Z.; Tong, Y. Potential of the Phage Depolymerase from a Myoviridae Bacteriophage vB_AbaM_IME200 Against Pandrug-Resistant Acinetobacter Baumannii. Am. J. Respir. Crit. Care Med. 2018, 197, A5488. [Google Scholar]
- Farrar, M.D.; Howson, K.M.; Bojar, R.A.; West, D.; Towler, J.C.; Parry, J.; Pelton, K.; Holland, K.T. Genome sequence and analysis of a Propionibacterium acnes bacteriophage. J. Bacteriol. 2007, 189, 4161–4167. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.L.; Petrovski, S.; Dyson, Z.A.; Seviour, R.; Tucci, J. The Formulation of Bacteriophage in a Semi Solid Preparation for Control of Propionibacterium acnes Growth. PLoS ONE 2016, 11, e0151184. [Google Scholar] [CrossRef] [PubMed]
- Lood, R.; Mörgelin, M.; Holmberg, A.; Rasmussen, M.; Collin, M. Inducible Siphoviruses in superficial and deep tissue isolates of Propionibacterium acnes. BMC Microbiol. 2008, 8, 139. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Fitz-Gibbon, S.; Hayes, C.; Bowman, C.; Inkeles, M.; Loncaric, A.; Russell, D.A.; Jacobs-Sera, D.; Cokus, S.; Pellegrini, M.; et al. Propionibacterium acnes Bacteriophages Display Limited Genetic Diversity and Broad Killing Activity against Bacterial Skin Isolates. mBio 2012, 3, e00279-12. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yan, R.; Zhong, Q.; Ngo, S.; Bangayan, N.J.; Nguyen, L.; Lui, T.; Liu, M.; Erfe, M.C.; Craft, N.; et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 2015, 9, 2078–2093. [Google Scholar] [CrossRef]
- Zierdt, C.H. Properties of Corynebacterium acnes bacteriophage and description of an interference phenomenon. J. Virol. 1974, 14, 1268–1273. [Google Scholar] [CrossRef]
- Comeau, A.M.; Chan, A.M.; Suttle, C.A. Genetic richness of vibriophages isolated in a coastal environment. Environ. Microbiol. 2006, 8, 1164–1176. [Google Scholar] [CrossRef]
- Holmfeldt, K.; Middelboe, M.; Nybroe, O.; Riemann, L. Large Variabilities in Host Strain Susceptibility and Phage Host Range Govern Interactions between Lytic Marine Phages and Their Flavobacterium Hosts. Appl. Environ. Microbiol. 2007, 73, 6730–6739. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.S.; Kong, B.J.; Park, S.N. Physicochemical properties of pH-sensitive hydrogels based on hydroxyethyl cellulose–hyaluronic acid and for applications as transdermal delivery systems for skin lesions. Eur. J. Pharm. Biopharm. 2015, 92, 146–154. [Google Scholar] [CrossRef]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Frank, A.C.; Lobry, J.R. Oriloc: Prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics 2000, 16, 560–561. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.H.; Lee, K.C.; Lee, S.J.; Kim, D.W.; Lee, W.J. HR-1 Mice: A New Inflammatory Acne Mouse Model. Ann. Dermatol. 2015, 27, 257–264. [Google Scholar] [CrossRef]
- Cisło, M.; Dabrowski, M.; Weber-Dabrowska, B.; Woytoń, A. Bacteriophage treatment of suppurative skin infections. Arch. Immunol. Ther. 1987, 35, 175–183. [Google Scholar]
- Porras, A.M.; Brito, I.L. The internationalization of human microbiome research. Curr. Opin. Microbiol. 2019, 50, 50–55. [Google Scholar] [CrossRef]
- Leeming, J.P.; Holland, K.T.; Cunliffe, W.J. The microbial ecology of pilosebaceous units isolated from human skin. J. Gen. Microbiol. 1984, 130, 803–807. [Google Scholar] [CrossRef]
- Tomida, S.; Nguyen, L.; Chiu, B.H.; Liu, J.; Sodergren, E.; Weinstock, G.M.; Li, H. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio 2013, 4, e00003–e00013. [Google Scholar] [CrossRef]
- Loessner, M.J. Bacteriophage endolysins—Current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef]
- Rajesh, T.; Anthony, T.; Saranya, S.; Pushpam, P.L.; Gunasekaran, P. Functional characterization of a new holin-like antibacterial protein coding gene tmp1 from goat skin surface metagenome. Appl. Microbiol. Biotechnol. 2011, 89, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, N.; Yan, Y.; Wang, H.; Li, Y.; Lu, C.; Sun, J. Combined antibacterial activity of phage lytic proteins holin and lysin from Streptococcus suis bacteriophage SMP. Curr. Microbiol. 2012, 65, 28–34. [Google Scholar] [CrossRef]
- Kistowska, M.; Gehrke, S.; Jankovic, D.; Kerl, K.; Fettelschoss, A.; Feldmeyer, L.; Fenini, G.; Kolios, A.; Navarini, A.; Ganceviciene, R.; et al. IL-1β Drives Inflammatory Responses to Propionibacterium acnes In Vitro and In Vivo. J. Investig. Dermatol. 2014, 134, 677–685. [Google Scholar] [CrossRef]
- Qin, M.; Pirouz, A.; Kim, M.H.; Krutzik, S.R.; Garbán, H.J.; Kim, J. Propionibacterium acnes Induces IL-1β secretion via the NLRP3 inflammasome in human monocytes. J. Investig. Dermatol. 2014, 134, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Jiao, Y.; Yuan, Y.; Zhou, Z.; Zheng, Y.; Xiao, J.; Li, C.; Chen, Z.; Cao, P. Propionibacterium acnes induces intervertebral disc degeneration by promoting nucleus pulposus cell apoptosis via the TLR2/JNK/mitochondrial-mediated pathway. Emerg. Microbes Infect. 2018, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-R.; Kim, K.-H.; An, H.-J.; Kim, J.-Y.; Han, S.-M.; Lee, K.-G.; Park, K.-K. Protective effect of melittin against inflammation and apoptosis on Propionibacterium acnes-induced human THP-1 monocytic cell. Eur. J. Pharmacol. 2014, 740, 218–226. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.D.T.; de la Fuente-Nunez, C.; Lu, T.K. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019, 179, 459–469. [Google Scholar] [CrossRef]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: An ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; He, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a Bacteriophage Cocktail to Constrain the Emergence of Phage-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Chen, Y.; Batra, H.; Dong, J.; Chen, C.; Rao, V.B.; Tao, P. Genetic Engineering of Bacteriophages Against Infectious Diseases. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ryan, E.M.; Alkawareek, M.Y.; Donnelly, R.F.; Gilmore, B.F. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. 2012, 65, 395–398. [Google Scholar] [CrossRef]
- Liu, C.G.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, e01462–e01520. [Google Scholar]
- Roach, D.R.; Donovan, D.M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef]
- Lemire, S.; Yehl, K.M.; Lu, T.K. Phage-Based Applications in Synthetic Biology. Annu. Rev. Virol. 2018, 5, 453–476. [Google Scholar] [CrossRef] [PubMed]
- Zaenglein, A.L.; Pathy, A.L.; Schlosser, B.J.; Alikhan, A.; Baldwin, H.E.; Berson, D.S.; Bowe, W.P.; Graber, E.M.; Harper, J.C.; Kang, S.; et al. Guidelines of care for the management of acne vulgaris. J. Am. Acad. Dermatol. 2016, 74, 945–973. [Google Scholar] [CrossRef] [PubMed]
- Dobler, D.; Schmidts, T.; Zinecker, C.; Schlupp, P.; Schäfer, J.; Runkel, F. Hydrophilic Ionic Liquids as Ingredients of Gel-Based Dermal Formulations. AAPS PharmSciTech 2016, 17, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.L.; Petrovski, S.; Chan, H.T.; Angove, M.J.; Tucci, J. Semi-Solid and Solid Dosage Forms for the Delivery of Phage Therapy to Epithelia. Pharmaceuticals 2018, 11, 26. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Lee, K.C.; Kim, M.J.; Jang, Y.H.; Lee, S.J.; Kim, D.W. Efficacy of Red or Infrared Light-Emitting Diodes in a Mouse Model of Propionibacterium acnes-Induced Inflammation. Ann. Dermatol. 2016, 28, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.Y.P.; Liang, T.-R.; Jiang, S.-J.; Peng, S.-Y. Albendazole-Schisandrin B Co-Therapy on Angiostrongylus cantonensis-Induced Meningoencephalitis in Mice. Biomolecules 2020, 10, 1001. [Google Scholar] [CrossRef] [PubMed]

| Bacteriophage | Host | CsCl Density (g/cm3) | Morphology | Head Diameter (nm) | Head Length (nm) | Tail Diameter (nm) | Tail Length (nm) | Family |
|---|---|---|---|---|---|---|---|---|
| TCUCAP1 | C. acnes | 1.4–1.5 | Isometric head with non-contractile tail | 50.4 ± 1.1 | 48.5 ± 2.8 | 9.2 ± 0.5 | 155.9 ± 31.0 | Siphoviridae |
| Characteristic | TCUCAP1 |
|---|---|
| Length (bp) | 29,547 |
| Overall G+C content (%) | 53.83% |
| No. of annotated genes | 56 |
| Avg gene length (bp) | 398.1 |
| Gene density (no. of genes/kb) | 1.9 |
| Gene GC content (%) | 54.5 |
| No. of tRNAs | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, H.Y.P.; Lai, M.-J.; Chen, T.-Y.; Wu, W.-J.; Peng, S.-Y.; Chang, K.-C. Therapeutic Effect of a Newly Isolated Lytic Bacteriophage against Multi-Drug-Resistant Cutibacterium acnes Infection in Mice. Int. J. Mol. Sci. 2021, 22, 7031. https://doi.org/10.3390/ijms22137031
Lam HYP, Lai M-J, Chen T-Y, Wu W-J, Peng S-Y, Chang K-C. Therapeutic Effect of a Newly Isolated Lytic Bacteriophage against Multi-Drug-Resistant Cutibacterium acnes Infection in Mice. International Journal of Molecular Sciences. 2021; 22(13):7031. https://doi.org/10.3390/ijms22137031
Chicago/Turabian StyleLam, Ho Yin Pekkle, Meng-Jiun Lai, Ting-Yu Chen, Wen-Jui Wu, Shih-Yi Peng, and Kai-Chih Chang. 2021. "Therapeutic Effect of a Newly Isolated Lytic Bacteriophage against Multi-Drug-Resistant Cutibacterium acnes Infection in Mice" International Journal of Molecular Sciences 22, no. 13: 7031. https://doi.org/10.3390/ijms22137031
APA StyleLam, H. Y. P., Lai, M.-J., Chen, T.-Y., Wu, W.-J., Peng, S.-Y., & Chang, K.-C. (2021). Therapeutic Effect of a Newly Isolated Lytic Bacteriophage against Multi-Drug-Resistant Cutibacterium acnes Infection in Mice. International Journal of Molecular Sciences, 22(13), 7031. https://doi.org/10.3390/ijms22137031






