Exploring New Potential Anticancer Activities of the G-Quadruplexes Formed by [(GTG2T(G3T)3] and Its Derivatives with an Abasic Site Replacing Single Thymidine
Abstract
:1. Introduction
2. Results and Discussion
2.1. NMR Spectroscopy
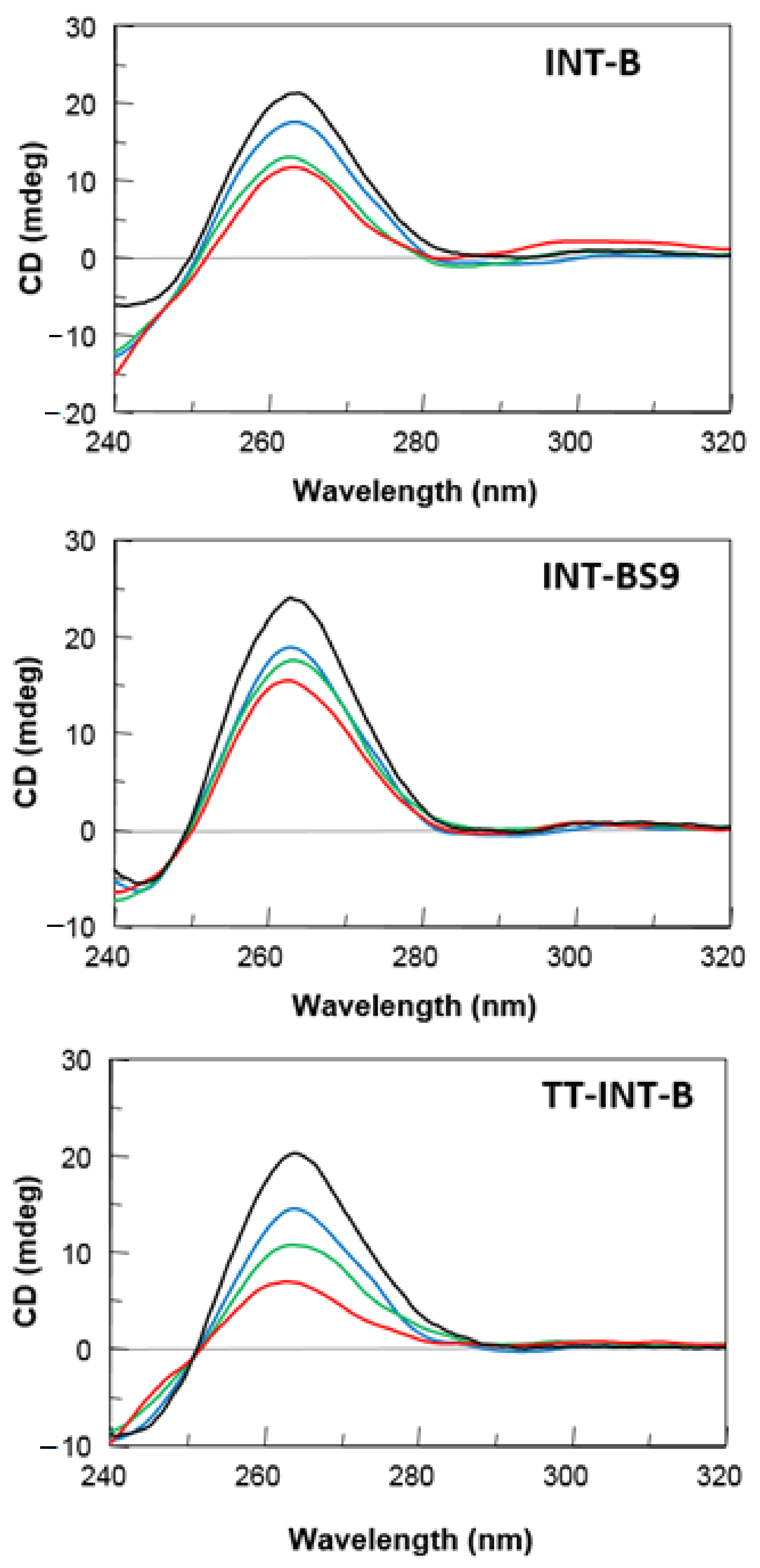
2.2. CD Spectroscopy
2.3. Polyacrylamide Gel Electrophoresis (PAGE)
2.4. Nuclease Stability Assay
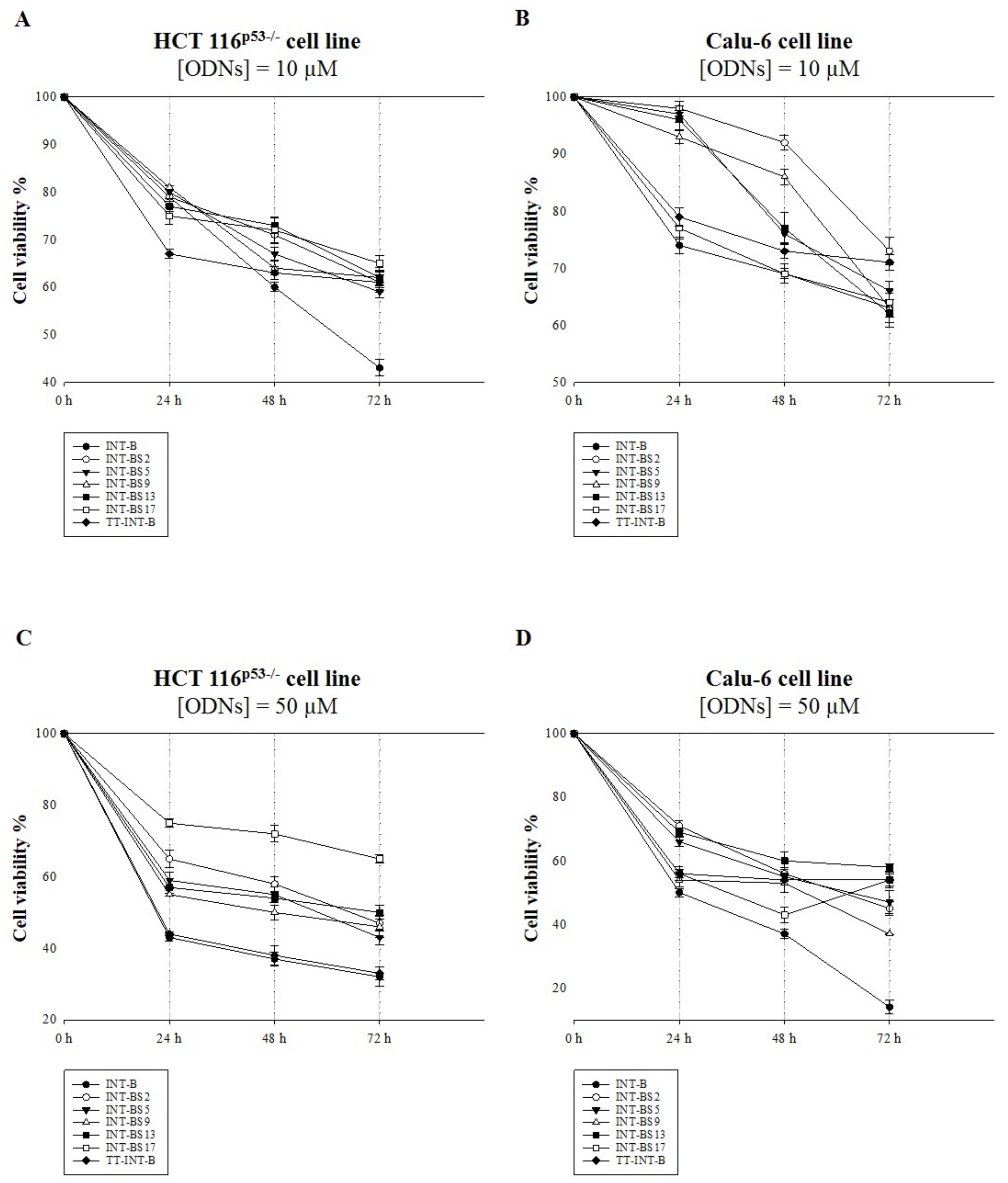
2.5. Antiproliferative Activity
2.6. Anti-Motility Property
3. Materials and Methods
3.1. Oligonucleotide Synthesis and Purification
3.2. NMR Spectroscopy
3.3. CD Spectroscopy
3.4. Gel Electrophoresis
3.5. Nuclease Stability Assay
3.6. Cell Cultures and Treatments with the ODNs
3.7. MTT Assay
3.8. Wound Healing Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-Quadruplex-Forming Aptamers-Characteristics, Applications, and Perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisz, K.; Jana, J. Thermodynamic Stability of G-Quadruplexes: Impact of Sequence and Environment. ChemBioChem 2021, 1–10. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef]
- Masai, H.; Tanaka, T. G-quadruplex DNA and RNA: Their roles in regulation of DNA replication and other biological functions. Biochem. Biophys. Res. Commun. 2020, 531, 25–38. [Google Scholar] [CrossRef]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin Binding Aptamer, More than a Simple Aptamer: Chemically Modified Derivatives and Biomedical Applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef] [Green Version]
- Russo Krauss, I.; Napolitano, V.; Petraccone, L.; Troisi, R.; Spiridonova, V.; Mattia, C.A.; Sica, F. Duplex/quadruplex oligonucleotides: Role of the duplex domain in the stabilization of a new generation of highly effective anti-thrombin aptamers. Int. J. Biol. Macromol. 2017, 107, 1697–1705. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Do, N.Q.; Phan, A.T. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011, 39, 8984–8991. [Google Scholar] [CrossRef]
- Do, N.Q.; Chung, W.J.; Truong, T.H.A.; Heddi, B.; Phan, A.T. G-quadruplex structure of an anti-proliferative DNA sequence. Nucleic Acids Res. 2017, 45, 7487–7493. [Google Scholar] [CrossRef] [Green Version]
- Esposito, V.; Galeone, A.; Mayol, L.; Oliviero, G.; Virgilio, A.; Randazzo, L. A Topological Classification of G-Quadruplex Structures. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1155–1159. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Napolitano, E.; Platella, C.; Musumeci, D.; Montesarchio, D. G-quadruplex-based aptamers targeting human thrombin: Discovery, chemical modifications and antithrombotic effects. Pharmacol. Ther. 2021, 217, 107649. [Google Scholar] [CrossRef]
- Kelly, J.A.; Feigon, J.; Yeates, T.O. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1996, 256, 417–422. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Tulinsky, A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 272–282. [Google Scholar] [CrossRef] [Green Version]
- Krauss, I.R.; Merlino, A.; Giancola, C.; Randazzo, A.; Mazzarella, L.; Sica, F. Thrombin-aptamer recognition: A revealed ambiguity. Nucleic Acids Res. 2011, 39, 7858–7867. [Google Scholar] [CrossRef] [Green Version]
- Virgilio, A.; Petraccone, L.; Vellecco, V.; Bucci, M.; Varra, M.; Irace, C.; Santamaria, R.; Pepe, A.; Mayol, L.; Esposito, V.; et al. Site-specific replacement of the thymine methyl group by fluorine in thrombin binding aptamer significantly improves structural stability and anticoagulant activity. Nucleic Acids Res. 2015, 43, 10602–10611. [Google Scholar] [CrossRef] [Green Version]
- Esposito, V.; Pirone, L.; Mayol, L.; Pedone, E.; Virgilio, A.; Galeone, A. Exploring the binding of d(GGGT)4 to the HIV-1 integrase: An approach to investigate G-quadruplex aptamer/target protein interactions. Biochimie 2016, 127, 19–22. [Google Scholar] [CrossRef] [Green Version]
- Virgilio, A.; Amato, T.; Petraccone, L.; Esposito, F.; Grandi, N.; Tramontano, E.; Romero, R.; Haider, S.; Gomez-Monterrey, I.; Novellino, E.; et al. Improvement of the activity of the anti-HIV-1 integrase aptamer T30175 by introducing a modified thymidine into the loops. Sci. Rep. 2018, 8, 7447. [Google Scholar] [CrossRef]
- Dapić, V.; Abdomerović, V.; Marrington, R.; Peberdy, J.; Rodger, A.; Trent, J.O.; Bates, P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 2097–2107. [Google Scholar] [CrossRef] [Green Version]
- Jing, N.; Li, Y.; Xu, X.; Sha, W.; Li, P.; Feng, L.; Tweardy, D.J. Targeting Stat3 with G-Quartet Oligodeoxynucleotides in Human Cancer Cells. DNA Cell Biol. 2003, 22, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Ogloblina, A.M.; Khristich, A.N.; Karpechenko, N.Y.; Semina, S.E.; Belitsky, G.A.; Dolinnaya, N.G.; Yakubovskaya, M.G. Multi-targeted effects of G4-aptamers and their antiproliferative activity against cancer cells. Biochimie 2018, 145, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Qi, C.; Meng, J.; Zhang, N.; Bing, T.; Yang, X.; Cao, Z.; Shangguan, D. General Cell-Binding Activity of Intramolecular G-Quadruplexes with Parallel Structure. PLoS ONE 2013, 8, e62348. [Google Scholar]
- Hu, J.; Zhao, Z.; Liu, Q.; Ye, M.; Hu, B.; Wang, J.; Tan, W. Study of the Function of G-Rich Aptamers Selected for Lung Adenocarcinoma. Chem. An. Asian J. 2015, 10, 1519–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legatova, V.; Samoylenkova, N.; Arutyunyan, A.; Tashlitsky, V.; Zavyalova, E.; Usachev, D.; Pavlova, G.; Kopylov, A. Covalent bi-modular parallel and antiparallel G-Quadruplex DNA nanocostructs reduce viability of patient glioma primary cell cultures. Int. J. Mol. Sci. 2021, 22, 3372. [Google Scholar] [CrossRef]
- Bates, P.J.; Reyes-Reyes, E.M.; Malik, M.T.; Murphy, E.M.; O’Toole, M.G.; Trent, J.O. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1414–1428. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef]
- Fan, X.; Sun, L.; Wu, Y.; Zhang, L.; Yang, Z. Bioactivity of 2′-deoxyinosine-incorporated aptamer AS1411. Sci. Rep. 2016, 6, 25799. [Google Scholar] [CrossRef] [Green Version]
- Ogloblina, A.M.; Iaccarino, N.; Capasso, D.; Di Gaetano, S.; Garzarella, E.U.; Dolinnaya, N.G.; Yakubovskaya, M.G.; Pagano, B.; Amato, J.; Randazzo, A. Toward G-Quadruplex-Based Anticancer Agents: Biophysical and Biological Studies of Novel AS1411 Derivatives. Int. J. Mol. Sci. 2020, 21, 7781. [Google Scholar] [CrossRef]
- Scuotto, M.; Rivieccio, E.; Varone, A.; Corda, D.; Bucci, M.; Vellecco, V.; Cirino, G.; Virgilio, A.; Esposito, V.; Galeone, A.; et al. Site specific replacements of a single loop nucleoside with a dibenzyl linker may switch the activity of TBA from anticoagulant to antiproliferative. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zhu, Y.; Wang, C.; Guan, Z.; Zhang, L.; Yang, Z. Alkylation of phosphorothioated thrombin binding aptamers improves the selectivity of inhibition of tumor cell proliferation upon anticoagulation. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1864–1869. [Google Scholar] [CrossRef]
- Esposito, V.; Russo, A.; Amato, T.; Varra, M.; Vellecco, V.; Bucci, M.; Russo, G.; Virgilio, A.; Galeone, A. Backbone modified TBA analogues endowed with antiproliferative activity. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1213–1221. [Google Scholar] [CrossRef] [Green Version]
- Esposito, V.; Russo, A.; Vellecco, V.; Bucci, M.; Russo, G.; Mayol, L.; Virgilio, A.; Galeone, A. Thrombin binding aptamer analogues containing inversion of polarity sites endowed with antiproliferative and anti-motility properties against Calu-6 cells. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2645–2650. [Google Scholar] [CrossRef]
- Esposito, V.; Russo, A.; Amato, T.; Vellecco, V.; Bucci, M.; Mayol, L.; Russo, G.; Virgilio, A.; Galeone, A. The “Janus face” of the thrombin binding aptamer: Investigating the anticoagulant and antiproliferative properties through straightforward chemical modifications. Bioorg. Chem. 2018, 76, 202–209. [Google Scholar] [CrossRef]
- Kotkowiak, W.; Lisowiec-Wachnicka, J.; Grynda, J.; Kierzek, R.; Wengel, J.; Pasternak, A. Thermodynamic, Anticoagulant, and Antiproliferative Properties of Thrombin Binding Aptamer Containing Novel UNA Derivative. Mol. Ther. Nucleic Acids 2018, 10, 304–316. [Google Scholar] [CrossRef] [Green Version]
- Antipova, O.; Samoylenkova, N.; Savchenko, E.; Zavyalova, E.; Revishchin, A.; Pavlova, G.; Kopylov, A. Bimodular antiparallel G-quadruplex nanoconstruct with antiproliferative activity. Molecules 2019, 24, 3625. [Google Scholar] [CrossRef] [Green Version]
- Pecoraro, A.; Virgilio, A.; Esposito, V.; Galeone, A.; Russo, G.; Russo, A. UL3 mediated nucleolar stress pathway as a new mechanism of action of antiproliferative G-quadruplex TBA derivatives in colon cancer cells. Biomolecules 2020, 10, 583. [Google Scholar] [CrossRef] [Green Version]
- Virgilio, A.; Esposito, V.; Pecoraro, A.; Russo, A.; Vellecco, V.; Pepe, A.; Bucci, M.; Russo, G.; Galeone, A. Structural properties and anticoagulant/cytotoxic activities of heterochiral enantiomeric thrombin binding aptamer (TBA) derivatives. Nucleic Acids Res. 2020, 48, 12556–12565. [Google Scholar] [CrossRef]
- Do, N.Q.; Phan, A.T. Monomer-dimer equilibrium for the 5’-5’ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chemistry 2012, 18, 14752–14759. [Google Scholar] [CrossRef]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G4 matters—The influence of g-quadruplex structural elements on the antiproliferative properties of g-rich oligonucleotides. Int. J. Mol. Sci. 2021, 22, 4941. [Google Scholar] [CrossRef]
- Dalvit, C. Efficient multiple-solvent suppression for the study of the interactions of organic solvents with biomolecules. J. Biomol. NMR 1998, 11, 437–444. [Google Scholar] [CrossRef]
- Russo, A.; Saide, A.; Smaldone, S.; Faraonio, R.; Russo, G. Role of uL3 in Multidrug Resistance in p53-Mutated Lung Cancer Cells. Int. J. Mol. Sci. Artic. 2017, 18, 547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosca, L.; Pagano, M.; Pecoraro, A.; Borzacchiello, L.; Mele, L.; Cacciapuoti, G.; Porcelli, M.; Russo, G.; Russo, A. S-adenosyl-l-methionine overcomes ul3-mediated drug resistance in p53 deleted colon cancer cells. Int. J. Mol. Sci. 2021, 22, 103. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, A.; Carotenuto, P.; Russo, G.; Russo, A. Ribosomal protein uL3 targets E2F1 and Cyclin D1 in cancer cell response to nucleolar stress. Sci. Rep. 2019, 9, 15431. [Google Scholar] [CrossRef] [Green Version]
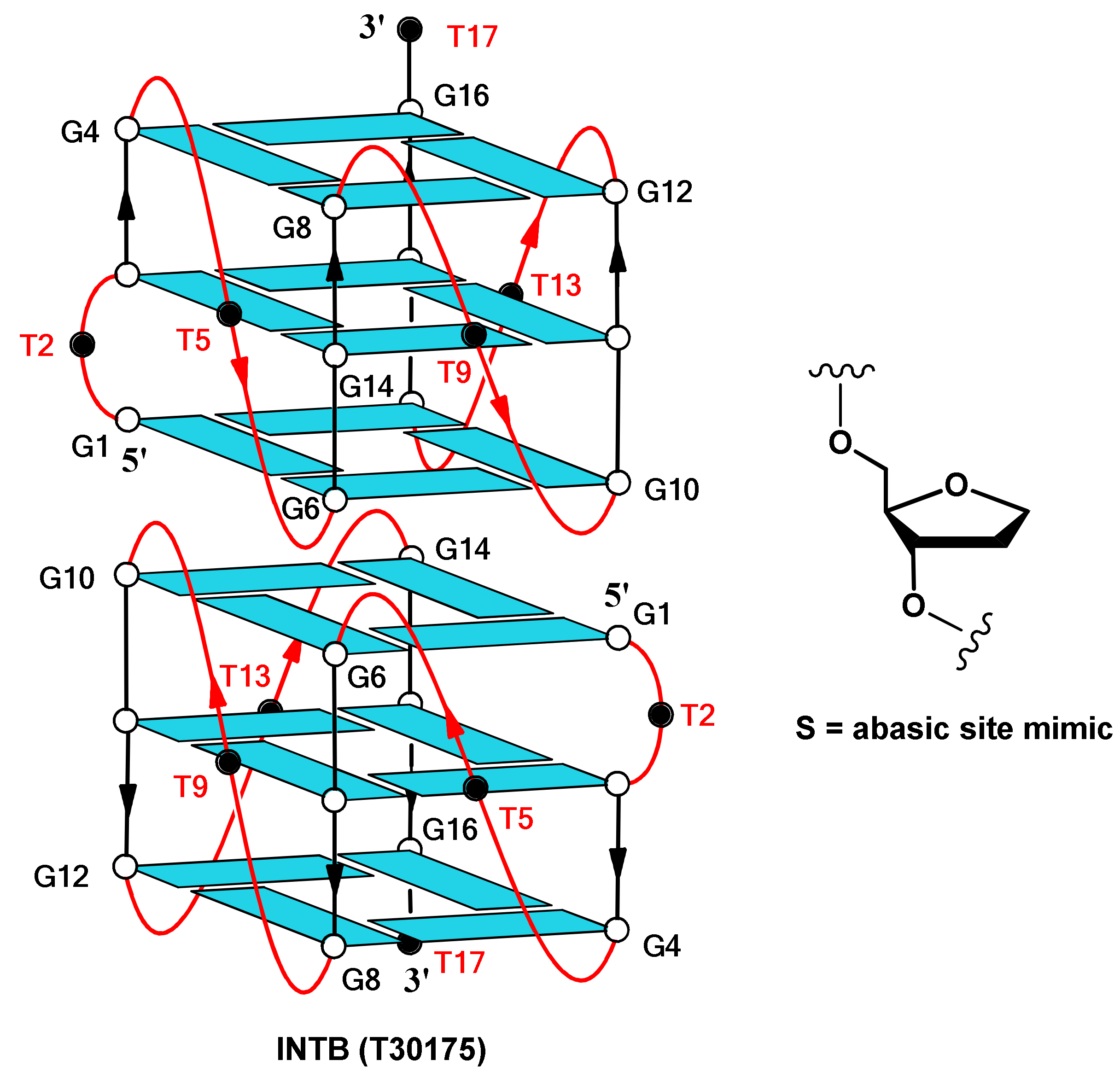
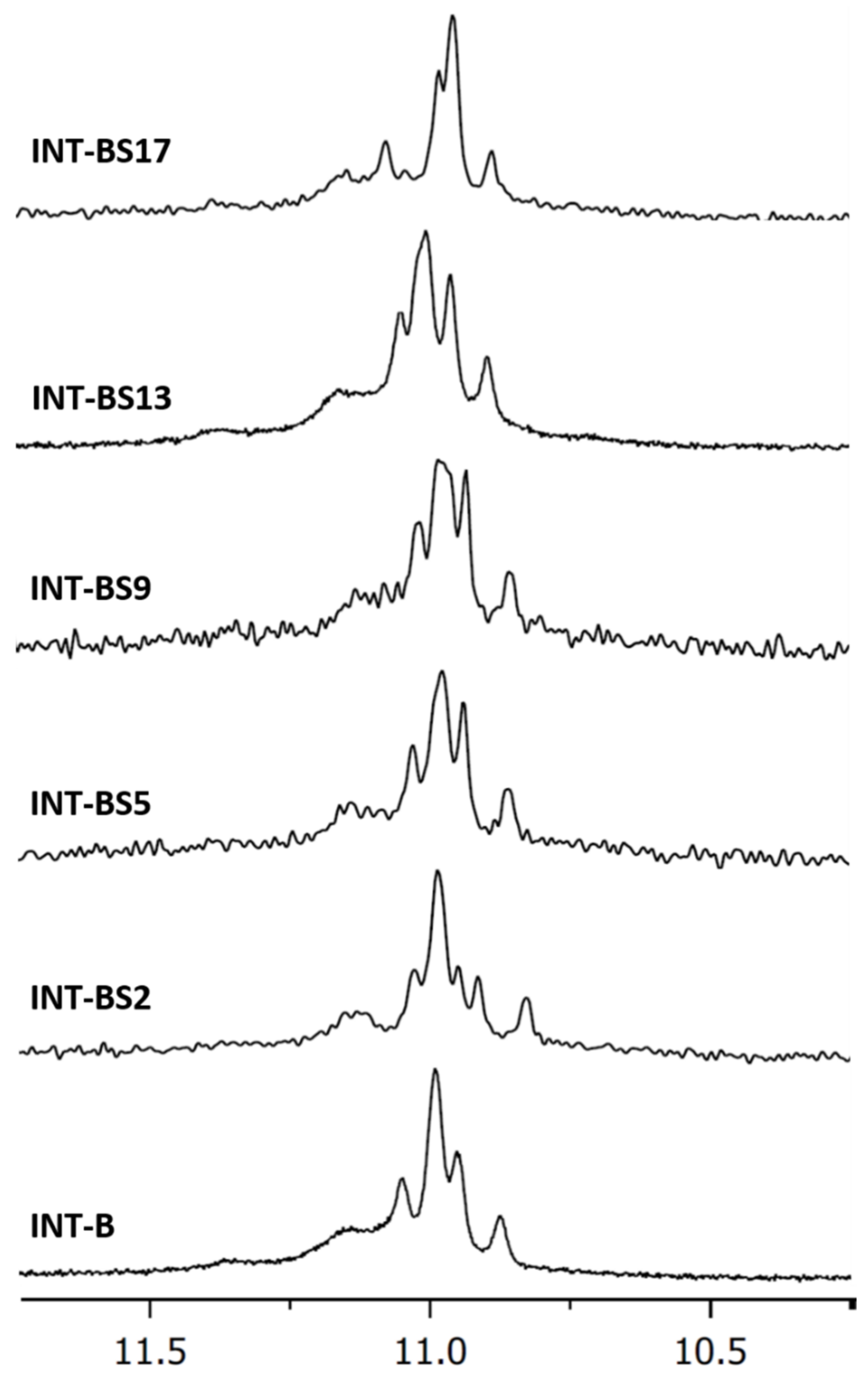
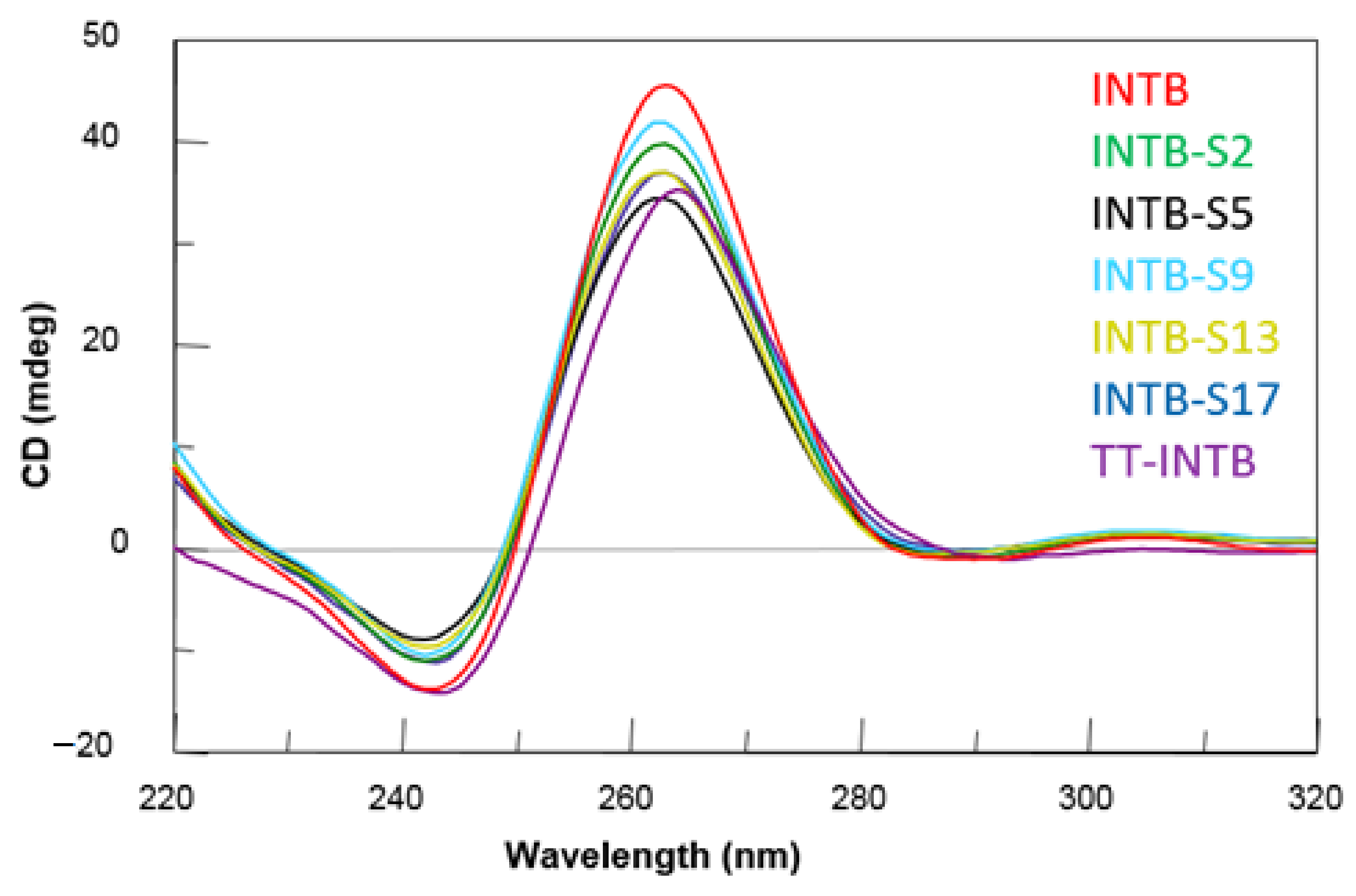
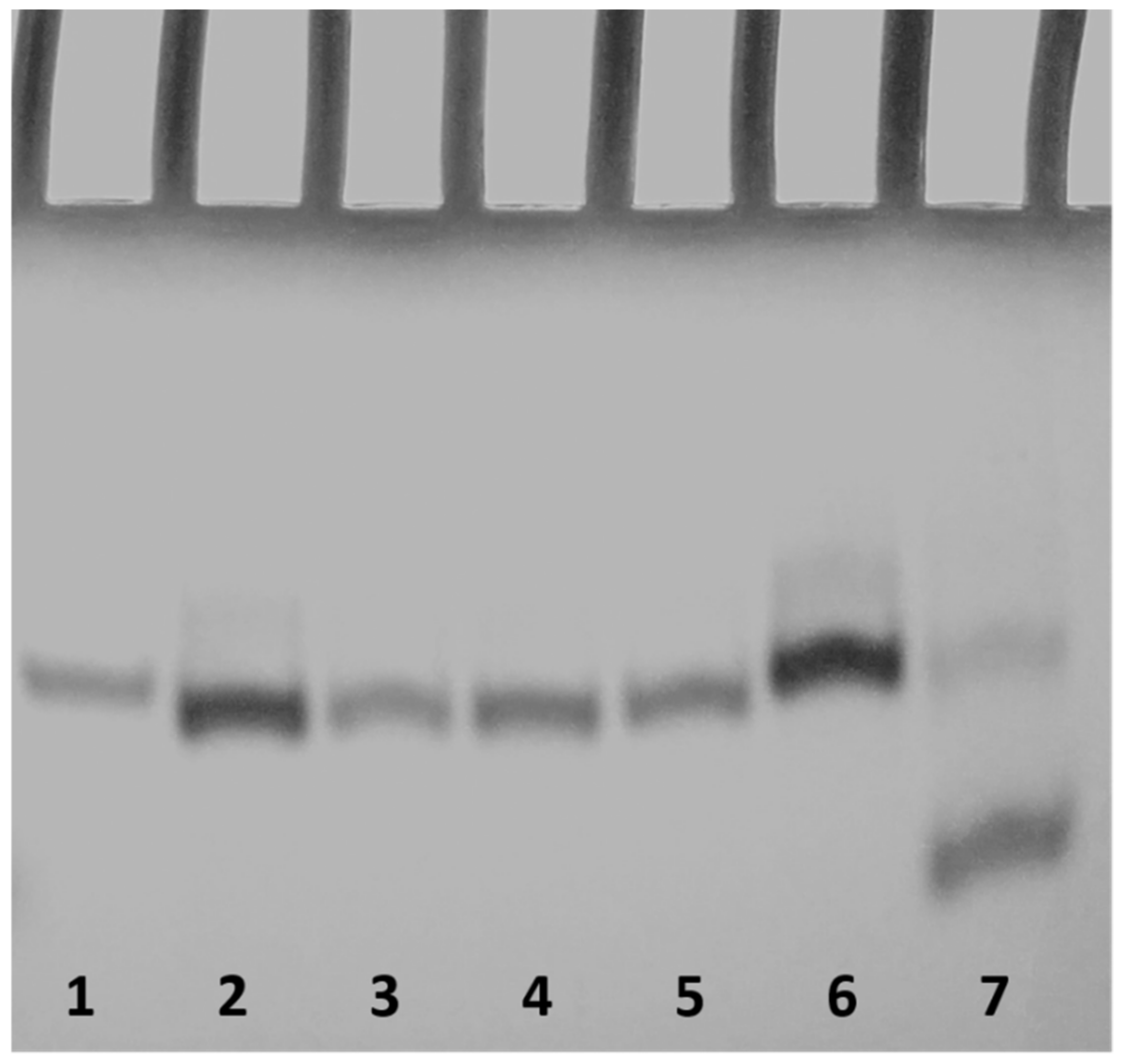

| Oligonucleotide | Sequence | Tm (°C) ± 1 |
|---|---|---|
| INT-B (T30175) | 5′-GTGGTGGGTGGGTGGGT-3′ | 86 |
| INT-BS2 | 5′-GSGGTGGGTGGGTGGGT-3′ | 86 |
| INT-BS5 | 5′-GTGGSGGGTGGGTGGGT-3′ | 84 |
| INT-BS9 | 5′-GTGGTGGGSGGGTGGGT-3′ | 85 |
| INT-BS13 | 5′-GTGGTGGGTGGGSGGGT-3′ | 84 |
| INT-BS17 | 5′-GTGGTGGGTGGGTGGGS-3′ | 83 |
| TT-INT-B | 5′-TTGTGGTGGGTGGGTGGGT-3′ | 84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Virgilio, A.; Benigno, D.; Pecoraro, A.; Russo, A.; Russo, G.; Esposito, V.; Galeone, A. Exploring New Potential Anticancer Activities of the G-Quadruplexes Formed by [(GTG2T(G3T)3] and Its Derivatives with an Abasic Site Replacing Single Thymidine. Int. J. Mol. Sci. 2021, 22, 7040. https://doi.org/10.3390/ijms22137040
Virgilio A, Benigno D, Pecoraro A, Russo A, Russo G, Esposito V, Galeone A. Exploring New Potential Anticancer Activities of the G-Quadruplexes Formed by [(GTG2T(G3T)3] and Its Derivatives with an Abasic Site Replacing Single Thymidine. International Journal of Molecular Sciences. 2021; 22(13):7040. https://doi.org/10.3390/ijms22137040
Chicago/Turabian StyleVirgilio, Antonella, Daniela Benigno, Annalisa Pecoraro, Annapina Russo, Giulia Russo, Veronica Esposito, and Aldo Galeone. 2021. "Exploring New Potential Anticancer Activities of the G-Quadruplexes Formed by [(GTG2T(G3T)3] and Its Derivatives with an Abasic Site Replacing Single Thymidine" International Journal of Molecular Sciences 22, no. 13: 7040. https://doi.org/10.3390/ijms22137040









