The Impact of Graphite Oxide Nanocomposites on the Antibacterial Activity of Serum
Abstract
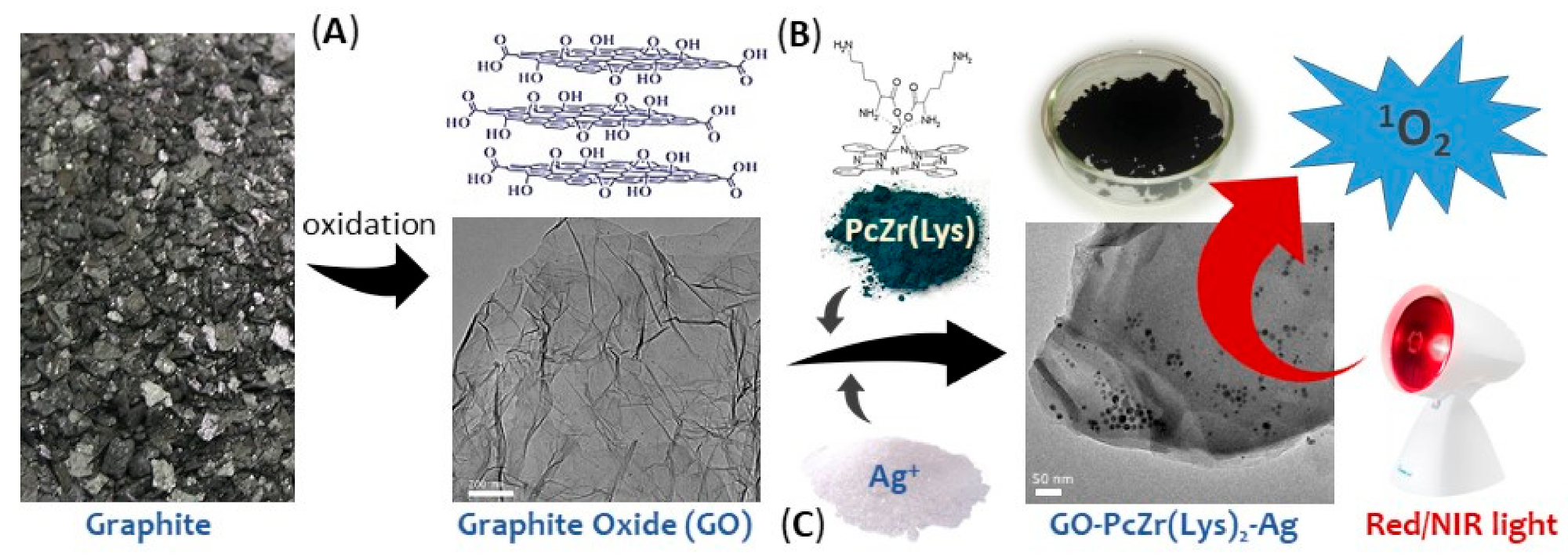
:1. Introduction
2. Results
2.1. Survival of E. coli 6.2E in NHS, iNHS (Inactivated Serum) and Nanocomposites (GO, GO-PcZr(Lys)2-Ag, GO-Ag, GO-PcZr(Lys)2) with and without IR Irradiation
2.2. The Influence of Nanocomposites (GO, GO-PcZr(Lys)2-Ag, GO-Ag, GO-PcZr(Lys)2) on the Bacterial Serum Survival
2.3. Serum Activity against Bacteria Treated with Nanocomposites and Nanocomposite Sensitivity of Bacteria Exposed to Serum
2.4. Galleria mellonella—In Vivo Cytotoxicity Tests
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Nanocomposites
4.3. IR Exposure
4.4. Normal Human Serum (NHS)
4.5. Inactivated Normal Human Serum (iNHS)
4.6. Serum Bactericidal Assay
4.7. Statistical Analysis
4.8. Galleria mellonella Treatment Assays
4.8.1. G. mellonella Larvae Acquisition
4.8.2. In Vivo Cytotoxicity Tests
5. Conclusions
- GO-PcZr(Lys)2, without IR irradiation enhance the antimicrobial efficacy of the human serum;
- IR irradiation enhances bactericidal activity of human serum in the case of the GO-PcZr(Lys)2-Ag sample;
- bacteria exposed to nanocomposites become more sensitive to the action of human serum;
- bacteria exposed to human serum become more sensitive to the GO-Ag sample;
- the influence of GO nanocomposites on the antibacterial activity of human serum and the impact on the bacterial sensitivity to human serum after their contact with GO nanocomposites strongly depend on the physicochemical properties of GO nanocomposites;
- the designed graphite nanocomposites showed no cytotoxic effect toward Galleria mellonella larvae;
- in comparison to that of E. coli J53, antimicrobial efficacy of human serum depends on individual properties of bacteria.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Replicate | T0 | T15 | T30 | T60 |
|---|---|---|---|---|
| 6.2E+NHS | 582,000,000 | 728,000,000 | 61,000,000 | 178,000,000 |
| 6.2E+NHS_2 | 795,000,000 | 603,000,000 | 608,000,000 | 153,000,000 |
| 6.2E+NHS_3 | 58,000,000 | 535,000,000 | 718,000,000 | 8,500,000 |
| 6.2E+iNHS | 463,000,000 | 623,000,000 | 453,000,000 | 545,000,000 |
| 6.2E+iNHS_2 | 573,000,000 | 493,000,000 | 383,000,000 | 825,000,000 |
| 6.2E+iNHS_3 | 508,000,000 | 493,000,000 | 355,000,000 | 84,000,000 |
| 6.2E+GO | 62,000 | 0 | 0 | 0 |
| 6.2E+GO_2 | 51,250 | 0 | 0 | 0 |
| 6.2E+GO_3 | 76,000 | 0 | 0 | 0 |
| 6.2E+IV | 3,680,000 | 2,330,000 | 2,300,000 | 1,500,000 |
| 6.2E+IV_2 | 4,580,000 | 3,530,000 | 3,180,000 | 2,600,000 |
| 6.2E+IV_3 | 4,680,000 | 3,800,000 | 2,600,000 | 2,370,000 |
| 6.2E+V | 3,330,000 | 2,030,000 | 2,380,000 | 1,330,000 |
| 6.2E+V_2 | 2,850,000 | 2,200,000 | 1,600,000 | 1,650,000 |
| 6.2E+V_3 | 2,000,000 | 1,880,000 | 1,580,000 | 1,950,000 |
| 6.2E+VI | 4,330,000 | 8,380,000 | 3,330,000 | 4,500,000 |
| 6.2E+VI_2 | 3,670,000 | 5,500,000 | 4,830,000 | 6,750,000 |
| 6.2E+VI_3 | 7,380,000 | 5,170,000 | 6,400,000 | 5,330,000 |
| 6.2E+NHS+GO | 3,830,000 | 4,830,000 | 5,170,000 | 4,330,000 |
| 6.2E+NHS+GO_2 | 9,630,000 | 4,170,000 | 7,630,000 | 6,330,000 |
| 6.2E+NHS+GO_3 | 8,130,000 | 10,200,000 | 4,670,000 | |
| 6.2E+NHS+IV | 655,000,000 | 645,000,000 | 438,000,000 | 145,000,000 |
| 6.2E+NHS+IV_2 | 713,000,000 | 560,000,000 | 463,000,000 | 173,000,000 |
| 6.2E+NHS+IV_3 | 680,000,000 | 538,000,000 | 408,000,000 | 75,000,000 |
| 6.2E+NHS+V | 603,000,000 | 298,000,000 | 205,000,000 | 160,000,000 |
| 6.2E+NHS+V_2 | 735,000,000 | 520,000,000 | 323,000,000 | 193,000,000 |
| 6.2E+NHS+V_3 | 580,000,000 | 540,000,000 | 623,000,000 | 185,000,000 |
| 6.2E+NHS+V_4 | 718,000,000 | 423,000,000 | 445,000,000 | 320,000,000 |
| 6.2E+NHS+VI | 6,630,000 | 3,500,000 | 2,920,000 | 1,290,000 |
| 6.2E+NHS+VI_2 | 6,000,000 | 8,500,000 | 2,920,000 | 1,140,000 |
| 6.2E+NHS+VI_3 | 8,000,000 | 3,990,000 | 525,000 | 100,000 |
| 6.2E+GO+IR | 2,333,333 | 0 | 0 | 0 |
| 6.2E+GO+IR_2 | 25,700 | 0 | 0 | 0 |
| 6.2E+GO+IR_3 | 19,500 | 0 | 0 | 0 |
| 6.2E+IV+IR | 2,670,000 | 2,680,000 | 2,080,000 | 650,000 |
| 6.2E+IV+IR_2 | 2,300,000 | 2,900,000 | 1,850,000 | 200,000 |
| 6.2E+IV+IR_3 | 2,230,000 | 2,630,000 | 1,800,000 | 400,000 |
| 6.2E+V+IR | 2,000,000 | 2,630,000 | 2,400,000 | 1,480,000 |
| 6.2E+V+IR_2 | 2,470,000 | 1,450,000 | 1,530,000 | 920,000 |
| 6.2E+V+IR_3 | 2,500,000 | 2,180,000 | 1,530,000 | 900,000 |
| 6.2E+VI+IR | 5,250,000 | 7,500,000 | 7,700,000 | 3,330,000 |
| 6.2E+VI+IR_2 | 6,750,000 | 7,080,000 | 4,670,000 | 8,700,000 |
| 6.2E+VI+IR_3 | 5,880,000 | 4,880,000 | 3,170,000 | 8,630,000 |
| (NHS+GO)+6.2E | 51,250 | 61,250 | 67,500 | 78,750 |
| (NHS+GO)+6.2E_2 | 45,000 | 107,500 | 65,000 | 100,000 |
| (NHS+GO)+6.2E_3 | 118,000 | 86,000 | 358,000 | 86,250 |
| (NHS+IV)+6.2E | 6,100,000 | 4,330,000 | 7,450,000 | 5,630,000 |
| (NHS+IV)+6.2E_2 | 7,800,000 | 8,030,000 | 8,430,000 | 7,880,000 |
| (NHS+IV)+6.2E_3 | 5,950,000 | 6,080,000 | 5,680,000 | 6,000,000 |
| (NHS+V)+6.2E | 5,280,000 | 4,850,000 | 1,780,000 | 50,000 |
| (NHS+V)+6.2E_2 | 6,720,000 | 4,900,000 | 4,350,000 | 0 |
| (NHS+V)+6.2E_3 | 5,870,000 | 6,030,000 | 1,730,000 | 4,000,000 |
| (NHS+VI)+6.2E | 88,000 | 75,000 | 25,750 | 13,300 |
| (NHS+VI)+6.2E_2 | 6,333,333 | 3,333,333 | 17,800 | 31,000 |
| (NHS+VI)+6.2E_3 | 72,500 | 61,250 | 29,000 | 12,625 |
| (iNHS+GO)+6.2E | 9,100,000 | 5,000,000 | 10,200,000 | 4,500,000 |
| (iNHS+GO)+6.2E_2 | 9,800,000 | 14,600,000 | 44,300,000 | 9,630,000 |
| (iNHS+IV)+6.2E | 475,000,000 | 665,000,000 | 510,000,000 | 410,000,000 |
| (iNHS+IV)+6.2E_2 | 662,000,000 | 475,000,000 | 578,000,000 | 427,000,000 |
| (iNHS+IV)+6.2E_3 | 388,000,000 | 585,000,000 | 618,000,000 | 297,000,000 |
| (iNHS+IV)+6.2E_4 | 253,000,000 | 410,000,000 | 425,000,000 | 393,000,000 |
| (iNHS+IV)+6.2E_5 | 395,000,000 | 443,000,000 | 470,000,000 | 330,000,000 |
| (iNHS+IV)+6.2E_6 | 583,000,000 | 318,000,000 | 315,000,000 | 370,000,000 |
| (iNHS+V)+6.2E | 535,000,000 | 510,000,000 | 630,000,000 | 278,000,000 |
| (iNHS+V)+6.2E_2 | 398,000,000 | 373,000,000 | 418,000,000 | 140,000,000 |
| (iNHS+V)+6.2E_3 | 347,000,000 | 505,000,000 | 390,000,000 | 85,000,000 |
| (iNHS+VI)+6.2E | 9,380,000 | 5,000,000 | 7,700,000 | 7,700,000 |
| (iNHS+VI)+6.2E_2 | 8,900,000 | 10,800,000 | 9,400,000 | 28,300,000 |
| (NHS+GO)+6.2E+IR | 95,000 | 132,500 | 284,000 | |
| (NHS+GO)+6.2E+IR_2 | 3,666,667 | 60,000 | 58,750 | 6,666,667 |
| (NHS+IV)+6.2E+IR | 4,350,000 | 2,480,000 | 3,730,000 | 1,530,000 |
| (NHS+IV)+6.2E+IR_2 | 4,670,000 | 6,730,000 | 4,400,000 | 2,430,000 |
| (NHS+IV)+6.2E+IR_3 | 4,100,000 | 1,950,000 | 2,600,000 | 1,680,000 |
| (NHS+V)+6.2E+IR | 3,980,000 | 1,830,000 | 2,100,000 | 1,470,000 |
| (NHS+V)+6.2E+IR_2 | 3,470,000 | 2,680,000 | 4,200,000 | 1,170,000 |
| (NHS+V)+6.2E+IR_3 | 2,530,000 | 1,580,000 | 2,670,000 | 1,930,000 |
| (NHS+VI)+6.2E+IR | 7,166,667 | 85,000 | 2,333,333 | 14,100 |
| (NHS+VI)+6.2E+IR_2 | 101,000 | 102,000 | 92,000 | 20,500 |
| (NHS+VI)+6.2E+IR_3 | 75,000 | 24,000 | 18,250 | 7,833,333 |
| (iNHS+GO)+6.2E+IR | 24,200,000 | 8,250,000 | 7,800,000 | 23,800,000 |
| (iNHS+GO)+6.2E+IR_2 | 8,130,000 | 14,600,000 | 9,380,000 | 7,000,000 |
| (iNHS+IV)+6.2E+IR | 370,000,000 | 368,000,000 | 373,000,000 | 687,000,000 |
| (iNHS+IV)+6.2E+IR_2 | 370,000,000 | 500,000,000 | 423,000,000 | 530,000,000 |
| (iNHS+IV)+6.2E+IR_3 | 398,000,000 | 508,000,000 | 473,000,000 | 517,000,000 |
| (iNHS+V)+6.2E+IR | 180,000,000 | 363,000,000 | 233,000,000 | 530,000,000 |
| (iNHS+V)+6.2E+IR_2 | 178,000,000 | 323,000,000 | 347,000,000 | 597,000,000 |
| (iNHS+V)+6.2E+IR_3 | 153,000,000 | 275,000,000 | 320,000,000 | 680,000,000 |
| (iNHS+VI)+6.2E+IR | 6,170,000 | 8,630,000 | 10,800,000 | 10,900,000 |
| (iNHS+VI)+6.2E+IR_2 | 8500,000 | 5,670,000 | 7,000,000 | 22,800,000 |
| (iNHS+VI)+6.2E+IR_3 | 7500,000 | 9,100,000 | 7,900,000 | 6,630,000 |
| (6.2E+GO)+NHS | 0 | 0 | 0 | 0 |
| (6.2E+GO)+NHS_2 | 0 | 0 | 0 | 0 |
| (6.2E+IV)+NHS | 270,000,000 | 180,000,000 | 15,000,000 | 25,500,000 |
| (6.2E+IV)+NHS_2 | 353,000,000 | 318,000,000 | 90,000,000 | 21,800,000 |
| (6.2E+IV)+NHS_3 | 177,000,000 | 153,000,000 | 100,000,000 | 33,500,000 |
| (6.2E+V)+NHS | 265,000,000 | 117,000,000 | 15,000,000 | 4,000,000 |
| (6.2E+V)+NHS_2 | 232,000,000 | 110,000,000 | 25,000,000 | 2,000,000 |
| (6.2E+V)+NHS_3 | 298,000,000 | 107,000,000 | 0 | 3,000,000 |
| (6.2E+VI)+NHS | 7,130,000 | 1,840,000 | 425,000 | 200,000 |
| (6.2E+VI)+NHS_2 | 7,000,000 | 1,290,000 | 0 | 0 |
| (6.2E+GO)+iNHS | 0 | 0 | 0 | 0 |
| (6.2E+GO)+iNHS_2 | 0 | 0 | 0 | 0 |
| (6.2E+GO)+iNHS_3 | 0 | 0 | 0 | 0 |
| (6.2E+IV)+iNHS | 325,000,000 | 168,000,000 | 18,000,000 | 3,500,000 |
| (6.2E+IV)+iNHS_2 | 348,000,000 | 298,000,000 | 488,000,000 | 623,000,000 |
| (6.2E+IV)+iNHS_3 | 144,000,000 | 227,000,000 | 18,000,000 | 275,000,000 |
| (6.2E+V)+iNHS | 265,000,000 | 117,000,000 | 1,500,000 | 40,000 |
| (6.2E+V)+iNHS_2 | 232,000,000 | 11,000,000 | 2,500,000 | 20,000 |
| (6.2E+V)+iNHS_3 | 298,000,000 | 107,000,000 | 0 | 30,000 |
| (6.2E+VI)+iNHS | 63,750,000 | 43,333,330,000 | 6,250,000 | 80,000 |
| (6.2E+VI)+iNHS_2 | 61,250,000 | 53,333,330,000 | 5,750,000 | 63,750,000 |
| (NHS+6.2E)+GO | 180,000 | 18,333,330,000 | 1,950,000 | 190,000 |
| (NHS+6.2E)+GO_2 | 19,250,000 | 42,583,330,000 | 14,333,330,000 | 1,580,000 |
| (NHS+6.2E)+GO_3 | 1,830,000 | 183,750,000 | 1740,000 | 1,670,000 |
| (NHS+6.2E)+IV | 365,000,000 | 338,000,000 | 245,000,000 | 133,000,000 |
| (NHS+6.2E)+IV_2 | 375,000,000 | 327,000,000 | 253,000,000 | 23,000,000 |
| (NHS+6.2E)+IV_3 | 233,000,000 | 20,500,0000 | 235,000,000 | 248,000,000 |
| (NHS+6.2E)+V | 43,000,000 | 42,500,000 | 4,500,000 | 650,000 |
| (NHS+6.2E)+V_2 | 58,000,000 | 32,500,000 | 300,000 | 90,000 |
| (NHS+6.2E)+V_3 | 533,000,000 | 2,500,000 | 2,500,000 | 250,000 |
| (NHS+6.2E)+VI | 1,240,000 | 1,390,000 | 9000 | 325,000 |
| (NHS+6.2E)+VI_2 | 3,520,000 | 1,740,000 | 140,000 | 3,140,000 |
| (NHS+6.2E)+VI_3 | 860,000 | 5,250,000 | 32,666,670,000 | 1,910,000 |
| (iNHS+6.2E)+GO | 840,000 | 730,000 | 5,750,000 | 48,333,330,000 |
| (iNHS+6.2E)+GO_2 | 91,250,000 | 12,700,000 | 83,750,000 | 70,000 |
| (iNHS+6.2E)+GO_3 | 950,000 | 9,250,000 | 260,833,300,000 | 81,250,000 |
| (iNHS+6.2E)+IV | 743,000,000 | 478,000,000 | 417,000,000 | 477,000,000 |
| (iNHS+6.2E)+IV_2 | 413,000,000 | 383,000,000 | 653,000,000 | 69,000,000 |
| (iNHS+6.2E)+IV_3 | 402,000,000 | 345,000,000 | 61,000,000 | 547,000,000 |
| (iNHS+6.2E)+V | 467,000,000 | 605,000,000 | 58,000,000 | 658,000,000 |
| (iNHS+6.2E)+V_2 | 692,000,000 | 44,000,000 | 133,000,000 | 565,000,000 |
| (iNHS+6.2E)+V_3 | 402,000,000 | 455,000,000 | 245,000,000 | 423,000,000 |
| (iNHS+6.2E)+VI | 166,250,000 | 920,000 | 950,000 | 770,000 |
| (iNHS+6.2E)+VI_2 | 68,333,330,000 | 262,500,000 | 213,571,400,000 | 7,250,000 |
| (iNHS+6.2E)+VI_3 | 960,000 | 84,166,670,000 | 990,000 | 225,714,300,000 |
| J53+NHS | 7,170,000 | 6,400,000 | 0 | 0 |
| J53+NHS_2 | 7,950,000 | 5,500,000 | 0 | 0 |
| J53+NHS_3 | 7,120,000 | 7,450,000 | 0 | 0 |
| J53+iNHS | 6,230,000 | 6,130,000 | 7,830,000 | 170,000,000 |
| J53+iNHS_2 | 7,900,000 | 5,950,000 | 6,380,000 | 1,450,000,000 |
| J53+iNHS_3 | 7,320,000 | 4,280,000 | 6,580,000 | 120,000,000 |
| J53+GO | 143,000 | 1,566,667 | 78,750 | 15,000 |
| J53+GO_2 | 140,000 | 0 | 0 | 0 |
| J53+GO_3 | 102,500 | 6,333,333 | 0 | 0 |
| J53+IV | 5,050,000 | 1,540,000,000 | 1,830,000,000 | 1,290,000,000 |
| J53+IV_2 | 6,830,000 | 1,140,000,000 | 1,860,000,000 | 1,290,000,000 |
| J53+IV_3 | 7,250,000 | 1,080,000,000 | 1,680,000,000 | 1,630,000,000 |
| J53+V | 8,750,000 | 2,390,000,000 | 4,430,000 | 4,750,000 |
| J53+V_2 | 5,750,000 | 1,050,000,000 | 6,130,000 | 5,850,000 |
| J53+V_3 | 7,580,000 | 1,580,000,000 | 8,850,000 | 6,000,000 |
| J53+VI | 1,583,333 | 130,000 | 106,250 | 193,000 |
| J53+VI_2 | 6,666,667 | 45,000 | 4,166,667 | 57,500 |
| J53+VI_3 | 129,000 | 85,000 | 98,750 | 93,000 |
| J53+NHS+GO | 85,000 | 0 | 0 | 0 |
| J53+NHS+GO_2 | 0 | 0 | 0 | 0 |
| J53+NHS+GO_3 | 109,000 | 61,250 | 0 | 0 |
| J53+NHS+IV | 5,580,000 | 1,090,000,000 | 1,900,000 | 7500 |
| J53+NHS+IV_2 | 7,720,000 | 1,280,000,000 | 6,230,000 | 155,000 |
| J53+NHS+IV_3 | 8,350,000 | 1,310,000,000 | 4,400,000 | 305,000 |
| J53+NHS+IV_4 | 4,730,000 | 3,830,000 | 0 | 0 |
| J53+NHS+IV_5 | 6,200,000 | 7,030,000 | 0 | 0 |
| J53+NHS+IV_6 | 7,620,000 | 6,530,000 | 0 | 0 |
| J53+NHS+V | 6,130,000 | 0 | 0 | 0 |
| J53+NHS+V_2 | 7,770,000 | 0 | 0 | 0 |
| J53+NHS+V_3 | 7,340,000 | 0 | 0 | 0 |
| J53+NHS+V_4 | 7,330,000 | 0 | 0 | 0 |
| J53+NHS+VI | 138,000 | 0 | 0 | 0 |
| J53+NHS+VI_2 | 8,166,667 | 0 | 0 | 0 |
| J53+NHS+VI_3 | 75,000 | 0 | 0 | 0 |
| J53+GO+IR | 1,991,667 | 139,000 | 67,500 | 1,666,667 |
| J53+GO+IR_2 | 1,383,333 | 0 | 0 | 0 |
| J53+GO+IR_3 | 116,250 | 35,000 | 45,000 | 0 |
| J53+IV+IR | 7,400,000 | 1,330,000,000 | 1,350,000,000 | 2,010,000,000 |
| J53+IV+IR_2 | 6,370,000 | 1,210,000,000 | 1,280,000,000 | 1,850,000,000 |
| J53+IV+IR_3 | 7,870,000 | 1,760,000,000 | 1,450,000,000 | 1,610,000,000 |
| J53+V+IR | 6,870,000 | 6,300,000 | 7,750,000 | 6,000,000 |
| J53+V+IR_2 | 4,950,000 | 6,480,000 | 7,330,000 | 4,960,000 |
| J53+V+IR_3 | 6,300,000 | 4,730,000 | 7,300,000 | 5,940,000 |
| J53+VI+IR | 1,733,333 | 145,000 | 139,000 | 83,000 |
| J53+VI+IR_2 | 74,000 | 60,000 | 67,500 | 35,000 |
| J53+VI+IR_3 | 60,000 | 76,250 | 60,000 | 4,333,333 |
| (NHS+GO)+J53 | 129,000 | 0 | 0 | 0 |
| (NHS+GO)+J53_2 | 0 | 0 | 0 | 0 |
| (NHS+GO)+J53_3 | 154,000 | 0 | 0 | 0 |
| (NHS+IV)+J53 | 7,970,000 | 6,930,000 | 75,000 | 0 |
| (NHS+IV)+J53_2 | 8,350,000 | 7,480,000 | 25,000 | 0 |
| (NHS+IV)+J53_3 | 8,630,000 | 7,980,000 | 50,000 | 0 |
| (NHS+V)+J53 | 6,200,000 | 0 | 0 | 0 |
| (NHS+V)+J53_2 | 6,100,000 | 0 | 0 | 0 |
| (NHS+V)+J53_3 | 7,080,000 | 0 | 0 | 0 |
| (NHS+V)+J53_4 | 7,530,000 | 0 | 0 | 0 |
| (NHS+V)+J53_5 | 7,430,000 | 3,480,000 | 550,000 | |
| (NHS+V)+J53_6 | 6,800,000 | 3,250,000 | 4,130,000 | 500,000 |
| (NHS+VI)+J53 | 2,058,333 | 0 | 0 | 0 |
| (NHS+VI)+J53_2 | 114,000 | 0 | 0 | 0 |
| (NHS+VI)+J53_3 | 87,500 | 0 | 0 | 0 |
| (iNHS+GO)+J53 | 237,500 | 2,466,667 | 2,333,333 | |
| (iNHS+GO)+J53_2 | 105,000 | 1,358,333 | 150,000 | 3,833,333 |
| (iNHS+GO)+J53_3 | 83,750 | 760,00 | 146,000 | 145,000 |
| (iNHS+IV)+J53 | 6,870,000 | 6,500,000 | 5,400,000 | 8,880,000 |
| (iNHS+IV)+J53_2 | 7,300,000 | 7,550,000 | 4,500,000 | 3,350,000 |
| (iNHS+IV)+J53_3 | 6,250,000 | 6,880,000 | 4,330,000 | 6,100,000 |
| (iNHS+V)+J53 | 5,700,000 | 1,490,000,000 | 1,870,000,000 | 3,250,000 |
| (iNHS+V)+J53_2 | 7,830,000 | 1,280,000,000 | 1,950,000,000 | 2,600,000 |
| (iNHS+V)+J53_3 | 7,080,000 | 1,290,000,000 | 1,290,000,000 | 3,600,000 |
| (iNHS+V)+J53_4 | 6,350,000 | 1,550,000,000 | 1,260,000,000 | 2,700,000 |
| (iNHS+V)+J53_5 | 4,480,000 | 1,460,000,000 | 1,570,000,000 | 2,150,000 |
| (iNHS+V)+J53_6 | 4,650,000 | 1,200,000,000 | 1,270,000,000 | 2,700,000 |
| (iNHS+VI)+J53 | 1,666,667 | 132,000 | 123,750 | 155,000 |
| (iNHS+VI)+J53_2 | 8,666,667 | 5,833,333 | 99,000 | 130,000 |
| (iNHS+VI)+J53_3 | 89,000 | 5,833,333 | 89,000 | 130,000 |
| (NHS+GO)+J53+IR | 0 | 0 | 0 | 0 |
| (NHS+GO)+J53+IR_2 | 0 | 0 | 0 | 0 |
| (NHS+GO)+J53+IR_3 | 9,083,333 | 0 | 0 | 0 |
| (NHS+IV)+J53+IR | 5,420,000 | 5,000,000 | 3,500,000 | 3,010,000 |
| (NHS+IV)+J53+IR_2 | 5,200,000 | 1,000,000 | 500,000 | 653,000 |
| (NHS+IV)+J53+IR_3 | 6,000,000 | 0 | 0 | 0 |
| (NHS+IV)+J53+IR_4 | 5,180,000 | 0 | 0 | 0 |
| (NHS+IV)+J53+IR_5 | 6,100,000 | 0 | 0 | 0 |
| (NHS+V)+J53+IR | 333,000 | 0 | 0 | 0 |
| (NHS+V)+J53+IR_2 | 133,000 | 0 | 0 | 0 |
| (NHS+V)+J53+IR_3 | 533,000 | 0 | 0 | 0 |
| (NHS+VI)+J53+IR | 157,500 | 0 | 0 | 0 |
| (NHS+VI)+J53+IR_2 | 90,000 | 1,108,333 | 62,500 | 51,250 |
| (NHS+VI)+J53+IR_3 | 82,500 | 0 | 0 | 0 |
| (iNHS+GO)+J53+IR | 218,000 | 256,250 | 175,000 | 5,166,667 |
| (iNHS+GO)+J53+IR_2 | 99,000 | 137,000 | 187,000 | 350,000 |
| (iNHS+GO)+J53+IR_3 | 102,000 | 132,000 | 1,658,333 | 1,766,667 |
| (iNHS+IV)+J53+IR | 6,600,000 | 2,700,000,000 | 2,650,000,000 | 6,930,000 |
| (iNHS+IV)+J53+IR_2 | 6,570,000 | 2,350,000,000 | 2,300,000,000 | 4,700,000 |
| (iNHS+IV)+J53+IR_3 | 6,830,000 | 1,300,000,000 | 1,450,000,000 | 6,730,000 |
| (iNHS+IV)+J53+IR_4 | 1,590,000,000 | 5,950,000,000 | 3,750,000,000 | 7,200,000,000 |
| (iNHS+IV)+J53+IR_5 | 1,680,000,000 | 4,450,000,000 | 3,850,000,000 | 6,000,000,000 |
| (iNHS+IV)+J53+IR_6 | 2,030,000,000 | 2,100,000,000 | 3,500,000,000 | 5,000,000,000 |
| (iNHS+V)+J53+IR | 4,980,000,000 | 1,430,000,000 | 2,100,000,000 | 2,400,000,000 |
| (iNHS+V)+J53+IR_2 | 3,650,000,000 | 1,730,000,000 | 1,780,000,000 | 1,000,000,000 |
| (iNHS+V)+J53+IR_3 | 3,980,000,000 | 1,700,000,000 | 2,150,000,000 | 1,000,000,000 |
| (iNHS+VI)+J53+IR | 173,000 | 8,916,667 | 1,683,333 | 164,000 |
| (iNHS+VI)+J53+IR_2 | 4,666,667 | 101,250 | 110,000 | 155,000 |
| (iNHS+VI)+J53+IR_3 | 71,000 | 105,000 | 1,216,667 | 157,500 |
| (J53+GO)+NHS | 10,000 | 0 | 0 | 0 |
| (J53+GO)+NHS_2 | 0 | 0 | 0 | 0 |
| (J53+GO)+NHS_3 | 0 | 0 | 0 | 0 |
| (J53+IV)+NHS | 7,500,000 | 4,200,000 | 147,000 | 0 |
| (J53+IV)+NHS_2 | 7,780,000 | 4,730,000 | 280,000 | 0 |
| (J53+IV)+NHS_3 | 7,470,000 | 5,800,000 | 237,000 | 0 |
| (J53+V)+NHS | 2,170,000 | 0 | 0 | 0 |
| (J53+V)+NHS_2 | 2,300,000 | 0 | 0 | 0 |
| (J53+V)+NHS_3 | 2,920,000 | 0 | 0 | 0 |
| (J53+VI)+NHS | 136,000 | 0 | 0 | 0 |
| (J53+VI)+NHS_2 | 65,000 | 0 | 0 | 0 |
| (J53+VI)+NHS_3 | 4,333,333 | 0 | 0 | 0 |
| (J53+GO)+iNHS | 0 | 0 | 10,000 | 20,000 |
| (J53+GO)+iNHS_2 | 17,500 | 150,00 | 20,000 | 3,333,333 |
| (J53+GO)+iNHS_3 | 0 | 0 | 0 | 0 |
| (J53+IV)+iNHS | 6,700,000 | 6,380,000 | 8,270,000 | 6,350,000 |
| (J53+IV)+iNHS_2 | 6,420,000 | 6,680,000 | 8,970,000 | 5,300,000 |
| (J53+IV)+iNHS_3 | 6,200,000 | 6,250,000 | 7,630,000 | 6,580,000 |
| (J53+V)+iNHS | 3,020,000 | 1,420,000,000 | 1,440,000,000 | 2,650,000,000 |
| (J53+V)+iNHS_2 | 3,220,000 | 1,450,000,000 | 1,550,000,000 | 270,000,000 |
| (J53+V)+iNHS_3 | 2,930,000 | 1,550,000,000 | 1,350,000,000 | 260,000,000 |
| (J53+VI)+iNHS | 1,708,333 | 1,466,667 | 1,566,667 | 147,500 |
| (J53+VI)+iNHS_2 | 80,000 | 102,500 | 127,000 | 256,000 |
| (J53+VI)+iNHS_3 | 81,250 | 70,000 | 71,000 | |
| (NHS+J53)+GO | 0 | 0 | 0 | 0 |
| (NHS+J53)+GO_2 | 0 | 0 | 0 | 0 |
| (NHS+J53)+GO_3 | 0 | 0 | 0 | 0 |
| (NHS+J53)+IV | 0 | 0 | 0 | 0 |
| (NHS+J53)+IV_2 | 0 | 0 | 0 | 0 |
| (NHS+J53)+IV_3 | 0 | 0 | 0 | 0 |
| (NHS+J53)+V | 2,680,000 | 0 | 0 | 0 |
| (NHS+J53)+V_2 | 3,250,000 | 0 | 0 | 0 |
| (NHS+J53)+V_3 | 5,080,000 | 0 | 0 | 0 |
| (NHS+J53)+VI | 0 | 0 | 0 | 0 |
| (NHS+J53)+VI_2 | 0 | 0 | 0 | 0 |
| (NHS+J53)+VI_3 | 0 | 0 | 0 | 0 |
| (iNHS+J53)+GO | 1,400,000 | 1,483,333 | 1,400,000 | 1,600,000 |
| (iNHS+J53)+GO_2 | 194,000 | 284,000 | 5,333,333 | 8,166,667 |
| (iNHS+J53)+IV | 8,100,000 | 1,970,000,000 | 3,900,000,000 | 3,700,000,000 |
| (iNHS+J53)+IV_2 | 3,630,000 | 1,380,000,000 | 2,750,000,000 | 1,300,000,000 |
| (iNHS+J53)+IV_3 | 1,430,000,000 | 1,620,000,000 | 3,760,000,000 | 2,400,000,000 |
| (iNHS+J53)+V | 1,910,000,000 | 1,970,000,000 | 1,970,000,000 | 1,700,000,000 |
| (iNHS+J53)+V_2 | 1,600,000,000 | 3,060,000,000 | 1,750,000,000 | 1,450,000,000 |
| (iNHS+J53)+V_3 | 1,840,000,000 | 2,290,000,000 | 1,500,000,000 | 1,200,000,000 |
| (iNHS+J53)+VI | 1,458,333 | 1,791,667 | 186,250 | |
| (iNHS+J53)+VI_2 | 160,000 | 1,533,333 | 400,000 | 7,333,333 |
| (iNHS+J53)+VI_3 | 98,750 | 150,000 | 235,000 | 550,000 |
Appendix B




Appendix C









| Experiments Comparison | p-Value |
|---|---|
| J53+IV vs. J53+NHS+IV | 0.0369 |
| J53+IV vs. (NHS+IV)+J53 | 0.0367 |
| J53+IV vs. (NHS+IV)+J53+IR | 0.0141 |
| J53+IV vs. (J53+IV)+NHS | 0.0367 |
| J53+NHS+IV vs. (J53+IV)+iNHS | 0.025 |
| J53+IV+IR vs. (NHS+IV)+J53+IR | 0.033 |
| (NHS+IV)+J53 vs. (J53+IV)+iNHS | 0.0255 |
| (NHS+IV)+J53+IR vs. (J53+IV)+iNHS | 0.011 |
| (J53+IV)+NHS vs. (J53+IV)+iNHS | 0.0255 |
| J53+V vs. (J53+V)+iNHS | <0.0001 |
| J53+NHS+V vs. J53+V+IR | 0.0103 |
| J53+NHS+V vs. (iNHS+V)+J53 | 0.0008 |
| J53+NHS+V vs. (J53+V)+iNHS | <0.0001 |
| J53+V+IR vs. (NHS+V)+J53 | 0.003 |
| J53+V+IR vs. (iNHS+V)+J53 | 0.0028 |
| J53+V+IR vs. (NHS+V)+J53+IR | 0.0103 |
| J53+V+IR vs. (iNHS+V)+J53+IR | 0.0429 |
| J53+V+IR vs. (J53+V)+NHS | 0.0103 |
| J53+V+IR vs. (J53+V)+iNHS | <0.0001 |
| J53+V+IR vs. (NHS+J53)+V | 0.0103 |
| (NHS+V)+J53 vs. (iNHS+V)+J53 | 0.0003 |
| (NHS+V)+J53 vs. (J53+V)+iNHS | <0.0001 |
| (NHS+V)+J53 vs. (iNHS+J53)+V | 0.0491 |
| (iNHS+V)+J53 vs. (NHS+V)+J53+IR | 0.0008 |
| (iNHS+V)+J53 vs. (J53+V)+NHS | 0.0008 |
| (iNHS+V)+J53 vs. (J53+V)+iNHS | <0.0001 |
| (iNHS+V)+J53 vs. (NHS+J53)+V | 0.0008 |
| (NHS+V)+J53+IR vs. (J53+V)+iNHS | <0.0001 |
| (iNHS+V)+J53+IR vs. (J53+V)+iNHS | <0.0001 |
| (J53+V)+NHS vs. (J53+V)+iNHS | <0.0001 |
| (J53+V)+iNHS vs. (NHS+J53)+V | <0.0001 |
| (J53+V)+iNHS vs. (iNHS+J53)+V | <0.0001 |
Appendix D
| Experiments | Fixed Effects (Type III) | p Value | F (DFn, DFd) |
|---|---|---|---|
| bacteria+ NHS/iNHS | Time | 0.2859 | F (1.649, 6.594) = 1.491 |
| NHS activity | 0.0257 | F (1, 4) = 12.00 | |
| Time × NHS activity | <0.0001 | F (3, 12) = 17.98 | |
| bacteria+ nanocomposite | Time | 0.0011 | F (2.001, 16.01) = 10.75 |
| Nanocomposite | 0.0012 | F (3, 8) = 14.86 | |
| Time × Nanocomposite | 0.0014 | F (9, 24) = 4.556 | |
| bacteria+ NHS+ nanocomposite | Time | <0.0001 | F (1.865, 16.16) = 24.99 |
| Nanocomposite | 0.076 | F (3, 9) = 3.210 | |
| Time × Nanocomposite | 0.0724 | F (9, 26) = 2.062 | |
| bacteria+ nanocomposite+ IR | Time | 0.0028 | F (1.319, 10.55) = 13.03 |
| Nanocomposite | 0.0003 | F (3, 8) = 22.32 | |
| Time × Nanocomposite | 0.0005 | F (9, 24) = 5.372 | |
| (NHS+ nanocomposite)+ bacteria | Time | 0.3299 | F (1.231, 13.13) = 1.097 |
| Nanocomposite | <0.0001 | F (3, 32) = 12.50 | |
| Time × Nanocomposite | 0.0501 | F (9, 32) = 2.188 | |
| (iNHS+ nanocomposite)+ bacteria | Time | 0.083 | F (2.000, 18.00) = 2.866 |
| Nanocomposite | 0.5903 | F (3, 9) = 0.6723 | |
| Time × Nanocomposite | 0.0059 | F (9, 27) = 3.458 | |
| (NHS+ nanocomposite)+ bacteria+ IR | Time | 0.6662 | F (2.311, 15.41) = 0.4609 |
| Nanocomposite | 0.0028 | F (3, 7) = 13.40 | |
| Time × Nanocomposite | 0.0007 | F (9, 20) = 5.543 | |
| (iNHS+ nanocomposite)+ bacteria+ IR | Time | 0.0074 | F (1.498, 10.49) = 9.169 |
| Nanocomposite | 0.0042 | F (3, 7) = 11.61 | |
| Time × Nanocomposite | 0.013 | F (9, 21) = 3.224 | |
| (bacteria+ nanocomposite)+ NHS | Time | <0.0001 | F (1.520, 7.600) = 218.3 |
| Nanocomposite | 0.0215 | F (2, 5) = 9.123 | |
| Time × Nanocomposite | 0.0014 | F (6, 15) = 6.644 | |
| (bacteria+ nanocomposite)+ iNHS | Time | 0.2459 | F (1.460, 7.301) = 1.677 |
| Nanocomposite | 0.071 | F (2, 5) = 4.701 | |
| Time × Nanocomposite | 0.0316 | F (6, 15) = 3.197 | |
| (NHS+ bacteria)+ nanocomposite | Time | 0.0033 | F (1.533, 16.36) = 9.414 |
| Nanocomposite | <0.0001 | F (3, 32) = 21.17 | |
| Time × Nanocomposite | 0.0161 | F (9, 32) = 2.772 | |
| (iNHS+ bacteria)+ nanocomposite | Time | 0.1946 | F (2.089, 16.71) = 1.803 |
| Nanocomposite | 0.0943 | F (3, 8) = 3.013 | |
| Time × Nanocomposite | 0.1167 | F (9, 24) = 1.819 | |
| All experiments with GO | Time | 0.3581 | F (1.791, 25.67) = 1.048 |
| Assay type | 0.0008 | F (8, 15) = 6.686 | |
| Time × Assay type | 0.0158 | F (24, 43) = 2.115 | |
| All experiments with nanocomposite IV: GO-PcZr(Lys)2-Ag | Time | <0.0001 | F (2.547, 63.66) = 9.845 |
| Assay type | 0.0147 | F (10, 25) = 2.910 | |
| Time × Assay type | <0.0001 | F (30, 75) = 3.616 | |
| All experiments with nanocomposite V: GO-Ag | Time | <0.0001 | F (2.398, 52.75) = 20.64 |
| Assay type | <0.0001 | F (10, 23) = 27.47 | |
| Time × Assay type | <0.0001 | F (30, 66) = 15.30 | |
| All experiments with nanocomposite VI: GO-PcZr(Lys)2 | Time | 0.6084 | F (2.108, 40.06) = 0.5192 |
| Assay type | 0.005 | F (10, 19) = 3.939 | |
| Time × Assay type | 0.0102 | F (30, 57) = 2.042 | |
| bacteria+ NHS/iNHS | Time | 0.0168 | F (1.367, 5.468) = 10.51 |
| NHS activity | 0.0072 | F (1, 4) = 25.52 | |
| Time × NHS activity | <0.0001 | F (3, 12) = 44.22 | |
| bacteria+ nanocomposite | Time | 0.0319 | F (1.965, 15.72) = 4.349 |
| Nanocomposite | 0.0003 | F (3, 8) = 22.21 | |
| Time × Nanocomposite | <0.0001 | F (9, 24) = 11.60 | |
| bacteria+ NHS+ nanocomposite | Time | <0.0001 | F (1.457, 16.02) = 83.74 |
| Nanocomposite | 0.0015 | F (3, 11) = 10.46 | |
| Time × Nanocomposite | <0.0001 | F (9, 33) = 13.92 | |
| bacteria+ nanocomposite+ IR | Time | 0.6116 | F (1.678, 13.43) = 0.4535 |
| Nanocomposite | <0.0001 | F (3, 8) = 74.37 | |
| Time × Nanocomposite | <0.0001 | F (9, 24) = 18.47 | |
| (NHS+ nanocomposite)+ bacteria | Time | <0.0001 | F (1.460, 14.11) = 263.7 |
| Nanocomposite | 0.0449 | F (3, 10) = 3.872 | |
| Time × Nanocomposite | <0.0001 | F (9, 29) = 14.72 | |
| (iNHS+ nanocomposite)+ bacteria | Time | 0.0921 | F (1.881, 20.06) = 2.729 |
| Nanocomposite | 0.009 | F (3, 11) = 6.420 | |
| Time × Nanocomposite | <0.0001 | F (9, 32) = 11.52 | |
| (NHS+ nanocomposite)+ bacteria+ IR | Time | 0.0001 | F (1.020, 8.162) = 48.04 |
| Nanocomposite | 0.7289 | F (3, 8) = 0.4427 | |
| Time × Nanocomposite | 0.8765 | F (9, 24) = 0.4758 | |
| (iNHS+ nanocomposite)+ bacteria+ IR | Time | 0.1098 | F (1.608, 17.69) = 2.614 |
| Nanocomposite | 0.0059 | F (3, 11) = 7.242 | |
| Time × Nanocomposite | 0.0518 | F (9, 33) = 2.161 | |
| (bacteria+ nanocomposite)+ NHS | Time | <0.0001 | F (1.008, 6.049) = 1725 |
| Nanocomposite | <0.0001 | F (3, 6) = 69.34 | |
| Time × Nanocomposite | <0.0001 | F (9, 18) = 67.55 | |
| (bacteria+ nanocomposite)+ iNHS | Time | <0.0001 | F (1.396, 7.909) = 61.74 |
| Nanocomposite | <0.0001 | F (3, 6) = 127.6 | |
| Time × Nanocomposite | <0.0001 | F (9, 17) = 51.46 | |
| (NHS+ bacteria)+ nanocomposite | NA | ||
| (iNHS+ bacteria)+ nanocomposite | Time | 0.0093 | F (1.957, 13.05) = 6.904 |
| Nanocomposite | 0.1302 | F (3, 7) = 2.650 | |
| Time × Nanocomposite | 0.0292 | F (9, 20) = 2.736 | |
| All experiments with GO | Time | 0.0767 | F (1.560, 16.64) = 3.198 |
| Assay type | 0.0046 | F (9, 11) = 5.667 | |
| Time × Assay type | 0.0095 | F (27, 32) = 2.393 | |
| All experiments with nanocomposite IV: GO-PcZr(Lys)2-Ag | Time | 0.0093 | F (1.936, 54.20) = 5.188 |
| Assay type | <0.0001 | F (9, 28) = 15.78 | |
| Time × Assay type | <0.0001 | F (27, 84) = 6.024 | |
| All experiments with nanocomposite V: GO-Ag | Time | 0.0542 | F (2.134, 61.18) = 2.994 |
| Assay type | <0.0001 | F (10, 29) = 244.4 | |
| Time × Assay type | <0.0001 | F (30, 86) = 88.45 | |
| All experiments with nanocomposite VI: GO-PcZr(Lys)2 | Time | 0.0112 | F (1.310, 25.32) = 6.566 |
| Assay type | <0.0001 | F (9, 20) = 8.579 | |
| Time × Assay type | <0.0001 | F (27, 58) = 4.693 | |
Appendix E
| Replicate | Weak Growth/Decline | Rapid Decline | Slight Decline | Growth | Moderate Decline |
|---|---|---|---|---|---|
| 6.2E+NHS | 0.393 | 0.168 | 0.252 | 0.095 | 0.092 |
| 6.2E+NHS_2 | 0.009 | 0.018 | 0.943 | 0.004 | 0.026 |
| 6.2E+NHS_3 | 0.422 | 0.215 | 0.224 | 0.068 | 0.071 |
| 6.2E+iNHS | 0.154 | 0.033 | 0.075 | 0.708 | 0.031 |
| 6.2E+iNHS_2 | 0.179 | 0.111 | 0.189 | 0.400 | 0.121 |
| 6.2E+iNHS_3 | 0.183 | 0.102 | 0.187 | 0.408 | 0.120 |
| 6.2E+GO | 0.042 | 0.701 | 0.147 | 0.013 | 0.097 |
| 6.2E+GO_2 | 0.042 | 0.701 | 0.147 | 0.013 | 0.097 |
| 6.2E+GO_3 | 0.042 | 0.701 | 0.147 | 0.013 | 0.097 |
| 6.2E+IV | 0.240 | 0.078 | 0.338 | 0.033 | 0.311 |
| 6.2E+IV_2 | 0.201 | 0.081 | 0.519 | 0.073 | 0.126 |
| 6.2E+IV_3 | 0.362 | 0.114 | 0.282 | 0.047 | 0.196 |
| 6.2E+V | 0.181 | 0.077 | 0.465 | 0.035 | 0.242 |
| 6.2E+V_2 | 0.325 | 0.113 | 0.313 | 0.064 | 0.185 |
| 6.2E+V_3 | 0.163 | 0.055 | 0.174 | 0.554 | 0.055 |
| 6.2E+VI | 0.224 | 0.090 | 0.183 | 0.402 | 0.101 |
| 6.2E+VI_2 | 0.275 | 0.080 | 0.133 | 0.436 | 0.076 |
| 6.2E+VI_3 | 0.253 | 0.079 | 0.350 | 0.198 | 0.120 |
| 6.2E+NHS+GO | 0.225 | 0.031 | 0.064 | 0.652 | 0.029 |
| 6.2E+NHS+GO_2 | 0.314 | 0.100 | 0.271 | 0.101 | 0.215 |
| 6.2E+NHS+GO_3 | 0.575 | 0.052 | 0.199 | 0.081 | 0.092 |
| 6.2E+NHS+IV | 0.040 | 0.051 | 0.783 | 0.011 | 0.115 |
| 6.2E+NHS+IV_2 | 0.024 | 0.037 | 0.863 | 0.008 | 0.069 |
| 6.2E+NHS+IV_3 | 0.048 | 0.169 | 0.378 | 0.015 | 0.390 |
| 6.2E+NHS+V | 0.113 | 0.214 | 0.320 | 0.012 | 0.341 |
| 6.2E+NHS+V_2 | 0.082 | 0.143 | 0.544 | 0.015 | 0.217 |
| 6.2E+NHS+V_3 | 0.435 | 0.188 | 0.236 | 0.067 | 0.075 |
| 6.2E+NHS+V_4 | 0.336 | 0.067 | 0.239 | 0.033 | 0.325 |
| 6.2E+NHS+VI | 0.076 | 0.099 | 0.124 | 0.008 | 0.693 |
| 6.2E+NHS+VI_2 | 0.273 | 0.168 | 0.200 | 0.036 | 0.323 |
| 6.2E+NHS+VI_3 | 0.019 | 0.299 | 0.057 | 0.004 | 0.621 |
| 6.2E+GO+IR | 0.042 | 0.701 | 0.147 | 0.013 | 0.097 |
| 6.2E+GO+IR_2 | 0.042 | 0.701 | 0.147 | 0.013 | 0.097 |
| 6.2E+GO+IR_3 | 0.042 | 0.701 | 0.147 | 0.013 | 0.097 |
| 6.2E+IV+IR | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 |
| 6.2E+IV+IR_2 | 0.131 | 0.191 | 0.490 | 0.047 | 0.141 |
| 6.2E+IV+IR_3 | 0.104 | 0.125 | 0.665 | 0.031 | 0.076 |
| 6.2E+V+IR | 0.590 | 0.036 | 0.099 | 0.231 | 0.044 |
| 6.2E+V+IR_2 | 0.224 | 0.068 | 0.277 | 0.026 | 0.405 |
| 6.2E+V+IR_3 | 0.172 | 0.139 | 0.433 | 0.036 | 0.220 |
| 6.2E+VI+IR | 0.518 | 0.071 | 0.126 | 0.187 | 0.097 |
| 6.2E+VI+IR_2 | 0.173 | 0.069 | 0.155 | 0.511 | 0.091 |
| 6.2E+VI+IR_3 | 0.218 | 0.132 | 0.176 | 0.309 | 0.165 |
| (NHS+GO)+6.2E | 0.261 | 0.050 | 0.095 | 0.546 | 0.047 |
| (NHS+GO)+6.2E_2 | 0.262 | 0.119 | 0.168 | 0.336 | 0.115 |
| (NHS+GO)+6.2E_3 | 0.294 | 0.132 | 0.198 | 0.235 | 0.141 |
| (NHS+IV)+6.2E | 0.329 | 0.039 | 0.110 | 0.467 | 0.056 |
| (NHS+IV)+6.2E_2 | 0.008 | 0.001 | 0.003 | 0.987 | 0.001 |
| (NHS+IV)+6.2E_3 | 0.061 | 0.010 | 0.031 | 0.888 | 0.009 |
| (NHS+V)+6.2E | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| (NHS+V)+6.2E_2 | 0.095 | 0.080 | 0.625 | 0.025 | 0.175 |
| (NHS+V)+6.2E_3 | 0.364 | 0.183 | 0.226 | 0.090 | 0.137 |
| (NHS+VI)+6.2E | 0.026 | 0.388 | 0.518 | 0.007 | 0.060 |
| (NHS+VI)+6.2E_2 | 0.331 | 0.127 | 0.283 | 0.024 | 0.236 |
| (NHS+VI)+6.2E_3 | 0.049 | 0.389 | 0.404 | 0.012 | 0.146 |
| (iNHS+GO)+6.2E | 0.512 | 0.082 | 0.177 | 0.106 | 0.124 |
| (iNHS+GO)+6.2E_2 | 0.258 | 0.138 | 0.183 | 0.288 | 0.134 |
| (iNHS+IV)+6.2E | 0.291 | 0.041 | 0.109 | 0.517 | 0.042 |
| (iNHS+IV)+6.2E_2 | 0.275 | 0.086 | 0.387 | 0.143 | 0.109 |
| (iNHS+IV)+6.2E_3 | 0.398 | 0.076 | 0.161 | 0.277 | 0.087 |
| (iNHS+IV)+6.2E_4 | 0.275 | 0.087 | 0.141 | 0.414 | 0.083 |
| (iNHS+IV)+6.2E_5 | 0.385 | 0.024 | 0.079 | 0.486 | 0.027 |
| (iNHS+IV)+6.2E_6 | 0.541 | 0.064 | 0.107 | 0.039 | 0.249 |
| (iNHS+V)+6.2E | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| (iNHS+V)+6.2E_2 | 0.456 | 0.172 | 0.222 | 0.066 | 0.084 |
| (iNHS+V)+6.2E_3 | 0.379 | 0.166 | 0.254 | 0.105 | 0.096 |
| (iNHS+VI)+6.2E | 0.273 | 0.078 | 0.255 | 0.200 | 0.194 |
| (iNHS+VI)+6.2E_2 | 0.238 | 0.111 | 0.166 | 0.378 | 0.107 |
| (NHS+GO)+6.2E+IR | 0.264 | 0.115 | 0.165 | 0.345 | 0.111 |
| (NHS+GO)+6.2E+IR_2 | 0.273 | 0.094 | 0.148 | 0.395 | 0.090 |
| (NHS+IV)+6.2E+IR | 0.127 | 0.132 | 0.486 | 0.041 | 0.215 |
| (NHS+IV)+6.2E+IR_2 | 0.449 | 0.100 | 0.174 | 0.128 | 0.149 |
| (NHS+IV)+6.2E+IR_3 | 0.297 | 0.086 | 0.189 | 0.027 | 0.402 |
| (NHS+V)+6.2E+IR | 0.290 | 0.100 | 0.116 | 0.018 | 0.477 |
| (NHS+V)+6.2E+IR_2 | 0.480 | 0.128 | 0.262 | 0.063 | 0.066 |
| (NHS+V)+6.2E+IR_3 | 0.338 | 0.063 | 0.179 | 0.319 | 0.102 |
| (NHS+VI)+6.2E+IR | 0.226 | 0.341 | 0.254 | 0.025 | 0.153 |
| (NHS+VI)+6.2E+IR_2 | 0.056 | 0.188 | 0.704 | 0.021 | 0.031 |
| (NHS+VI)+6.2E+IR_3 | 0.036 | 0.549 | 0.234 | 0.007 | 0.174 |
| (iNHS+GO)+6.2E+IR | 0.248 | 0.243 | 0.215 | 0.124 | 0.170 |
| (iNHS+GO)+6.2E+IR_2 | 0.352 | 0.066 | 0.138 | 0.377 | 0.067 |
| (iNHS+IV)+6.2E+IR | 0.185 | 0.066 | 0.129 | 0.558 | 0.062 |
| (iNHS+IV)+6.2E+IR_2 | 0.230 | 0.044 | 0.087 | 0.598 | 0.041 |
| (iNHS+IV)+6.2E+IR_3 | 0.230 | 0.032 | 0.067 | 0.641 | 0.030 |
| (iNHS+V)+6.2E+IR | 0.260 | 0.123 | 0.171 | 0.328 | 0.119 |
| (iNHS+V)+6.2E+IR_2 | 0.252 | 0.135 | 0.179 | 0.303 | 0.131 |
| (iNHS+V)+6.2E+IR_3 | 0.243 | 0.148 | 0.185 | 0.280 | 0.144 |
| (iNHS+VI)+6.2E+IR | 0.274 | 0.090 | 0.143 | 0.408 | 0.085 |
| (iNHS+VI)+6.2E+IR_2 | 0.189 | 0.128 | 0.220 | 0.297 | 0.166 |
| (iNHS+VI)+6.2E+IR_3 | 0.215 | 0.024 | 0.069 | 0.669 | 0.023 |
| (6.2E+GO)+NHS | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| (6.2E+IV)+NHS | 0.034 | 0.185 | 0.265 | 0.010 | 0.506 |
| (6.2E+IV)+NHS_2 | 0.016 | 0.768 | 0.175 | 0.005 | 0.036 |
| (6.2E+IV)+NHS_3 | 0.066 | 0.168 | 0.281 | 0.015 | 0.470 |
| (6.2E+V)+NHS | 0.021 | 0.602 | 0.060 | 0.005 | 0.312 |
| (6.2E+V)+NHS_2 | 0.021 | 0.298 | 0.065 | 0.005 | 0.611 |
| (6.2E+V)+NHS_3 | 0.016 | 0.791 | 0.055 | 0.004 | 0.134 |
| (6.2E+VI)+NHS | 0.020 | 0.785 | 0.101 | 0.005 | 0.089 |
| (6.2E+VI)+NHS_2 | 0.021 | 0.822 | 0.089 | 0.006 | 0.063 |
| (6.2E+GO)+iNHS | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 |
| (6.2E+IV)+iNHS | 0.000 | 0.000 | 0.000 | 0.000 | 1.000 |
| (6.2E+IV)+iNHS_2 | 0.262 | 0.078 | 0.131 | 0.449 | 0.080 |
| (6.2E+IV)+iNHS_3 | 0.275 | 0.083 | 0.136 | 0.426 | 0.079 |
| (6.2E+V)+iNHS | 0.021 | 0.602 | 0.060 | 0.005 | 0.312 |
| (6.2E+V)+iNHS_2 | 0.021 | 0.298 | 0.065 | 0.005 | 0.611 |
| (6.2E+V)+iNHS_3 | 0.043 | 0.380 | 0.048 | 0.007 | 0.522 |
| (6.2E+VI)+iNHS | 0.185 | 0.051 | 0.132 | 0.557 | 0.076 |
| (6.2E+VI)+iNHS_2 | 0.100 | 0.020 | 0.052 | 0.807 | 0.020 |
| (NHS+6.2E)+GO | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 |
| (NHS+6.2E)+GO_2 | 0.281 | 0.115 | 0.219 | 0.253 | 0.132 |
| (NHS+6.2E)+GO_3 | 0.153 | 0.026 | 0.085 | 0.713 | 0.023 |
| (NHS+6.2E)+IV | 0.150 | 0.126 | 0.547 | 0.036 | 0.141 |
| (NHS+6.2E)+IV_2 | 0.299 | 0.111 | 0.332 | 0.109 | 0.149 |
| (NHS+6.2E)+IV_3 | 0.037 | 0.007 | 0.017 | 0.930 | 0.007 |
| (NHS+6.2E)+V | 0.045 | 0.621 | 0.202 | 0.013 | 0.119 |
| (NHS+6.2E)+V_2 | 0.042 | 0.678 | 0.165 | 0.012 | 0.103 |
| (NHS+6.2E)+V_3 | 0.041 | 0.685 | 0.160 | 0.013 | 0.101 |
| (NHS+6.2E)+VI | 0.085 | 0.067 | 0.765 | 0.020 | 0.064 |
| (NHS+6.2E)+VI_2 | 0.364 | 0.144 | 0.169 | 0.107 | 0.217 |
| (NHS+6.2E)+VI_3 | 0.083 | 0.148 | 0.433 | 0.012 | 0.323 |
| (iNHS+6.2E)+GO | 0.291 | 0.104 | 0.357 | 0.091 | 0.156 |
| (iNHS+6.2E)+GO_2 | 0.324 | 0.077 | 0.188 | 0.337 | 0.075 |
| (iNHS+6.2E)+GO_3 | 0.297 | 0.108 | 0.182 | 0.303 | 0.110 |
| (iNHS+6.2E)+IV | 0.416 | 0.085 | 0.191 | 0.063 | 0.244 |
| (iNHS+6.2E)+IV_2 | 0.266 | 0.078 | 0.125 | 0.458 | 0.074 |
| (iNHS+6.2E)+IV_3 | 0.254 | 0.062 | 0.110 | 0.511 | 0.063 |
| (iNHS+6.2E)+V | 0.254 | 0.044 | 0.086 | 0.575 | 0.041 |
| (iNHS+6.2E)+V_2 | 0.221 | 0.135 | 0.346 | 0.090 | 0.208 |
| (iNHS+6.2E)+V_3 | 0.257 | 0.056 | 0.107 | 0.492 | 0.088 |
| (iNHS+6.2E)+VI | 0.221 | 0.176 | 0.196 | 0.234 | 0.174 |
| (iNHS+6.2E)+VI_2 | 0.245 | 0.145 | 0.184 | 0.285 | 0.141 |
| (iNHS+6.2E)+VI_3 | 0.222 | 0.099 | 0.155 | 0.426 | 0.099 |

References
- Lukowiak, A.; Kedziora, A.; Strek, W. Antimicrobial Graphene Family Materials: Progress, Advances, Hopes and Fears. Adv. Colloid Interface Sci. 2016, 236, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Kim, Y.-K.; Shin, D.; Ryoo, S.-R.; Hong, B.H.; Min, D.-H. Biomedical Applications of Graphene and Graphene Oxide. Acc. Chem. Res. 2013, 46, 2211–2224. [Google Scholar] [CrossRef] [PubMed]
- Goenka, S.; Sant, V.; Sant, S. Graphene-Based Nanomaterials for Drug Delivery and Tissue Engineering. J. Control. Release 2014, 173, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.R.; Deshmukh, K.; Sadasivuni, K.K.; Pasha, S.K.K. Graphene Quantum Dot Based Materials for Sensing, Bio-Imaging and Energy Storage Applications: A Review. RSC Adv. 2020, 10, 23861–23898. [Google Scholar] [CrossRef]
- Tajik, S.; Dourandish, Z.; Zhang, K.; Beitollahi, H.; Le, Q.V.; Jang, H.W.; Shokouhimehr, M. Carbon and Graphene Quantum Dots: A Review on Syntheses, Characterization, Biological and Sensing Applications for Neurotransmitter Determination. RSC Adv. 2020, 10, 15406–15429. [Google Scholar] [CrossRef] [Green Version]
- Saladino, M.L.; Markowska, M.; Carmone, C.; Cancemi, P.; Alduina, R.; Presentato, A.; Scaffaro, R.; Biały, D.; Hasiak, M.; Hreniak, D.; et al. Graphene Oxide Carboxymethylcellulose Nanocomposite for Dressing Materials. Materials 2020, 13, 1980. [Google Scholar] [CrossRef] [PubMed]
- Vijay Kumar, S.; Huang, N.M.; Lim, H.N.; Marlinda, A.R.; Harrison, I.; Chia, C.H. One-Step Size-Controlled Synthesis of Functional Graphene Oxide/silver Nanocomposites at Room Temperature. Chem. Eng. J. 2013, 219, 217–224. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, X.; Wang, X. Green Synthesis of Graphene Oxide Sheets Decorated by Silver Nanoprisms and Their Anti-Bacterial Properties. J. Inorg. Biochem. 2011, 105, 1181–1186. [Google Scholar] [CrossRef] [PubMed]
- Kedziora, A.; Gerasymchuk, Y.; Sroka, E.; Bugla-Płoskońska, G.; Doroszkiewicz, W.; Rybak, Z.; Hreniak, D.C.; Wilgusz, R.; Strek, W.A. Use of the materials based on partially reduced graphene-oxide with silver nanoparticle as bacteriostatic and bactericidal agent. Polim. Med. 2013, 43, 129–134. [Google Scholar]
- de Faria, A.F.; Martinez, D.S.T.; Meira, S.M.M.; de Moraes, A.C.M.; Brandelli, A.; Filho, A.G.S.; Alves, O.L. Anti-Adhesion and Antibacterial Activity of Silver Nanoparticles Supported on Graphene Oxide Sheets. Colloids Surf. B Biointerfaces 2014, 113, 115–124. [Google Scholar] [CrossRef]
- Zhu, Z.; Su, M.; Ma, L.; Ma, L.; Liu, D.; Wang, Z. Preparation of Graphene Oxide-Silver Nanoparticle Nanohybrids with Highly Antibacterial Capability. Talanta 2013, 117, 449–455. [Google Scholar] [CrossRef]
- Swimberghe, R.C.D.; Coenye, T.; Moor, R.J.G.D.; Meire, M.A. Biofilm Model Systems for Root Canal Disinfection: A Literature Review. Int. Endod. J. 2019, 52, 604–628. [Google Scholar] [CrossRef] [Green Version]
- Sakko, M.; Tjäderhane, L.; Rautemaa-Richardson, R. Microbiology of Root Canal Infections. Prim. Dent. J. 2016, 5, 84–89. [Google Scholar] [CrossRef]
- Szeto, G.L.; Lavik, E.B. Materials Design at the Interface of Nanoparticles and Innate Immunity. J. Mater. Chem. B 2016, 4, 1610–1618. [Google Scholar] [CrossRef] [Green Version]
- Kononenko, V.; Narat, M.; Drobne, D. Nanoparticle Interaction with the Immune System/Interakcije Nanodelcev Z Imunskim Sistemom. Arch. Ind. Hyg. Toxicol. 2015, 66. [Google Scholar] [CrossRef] [Green Version]
- Capriotti, A.L.; Caracciolo, G.; Caruso, G.; Cavaliere, C.; Pozzi, D.; Samperi, R.; Laganà, A. Label-Free Quantitative Analysis for Studying the Interactions between Nanoparticles and Plasma Proteins. Anal. Bioanal. Chem. 2013, 405, 635–645. [Google Scholar] [CrossRef]
- Bugla-Płoskońska, G.; Kiersnowski, A.; Futoma-Kołoch, B.; Doroszkiewicz, W. Killing of Gram-Negative Bacteria with Normal Human Serum and Normal Bovine Serum: Use of Lysozyme and Complement Proteins in the Death of Salmonella Strains O48. Microb. Ecol. 2009, 58, 276–289. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Zhang, Y.; Ye, Y.; Gao, Y.; Li, J. In Vitro and in Vivo Activity of Ciprofloxacin/fosfomycin Combination Therapy against Ciprofloxacin-Resistant Shigella Flexneri Isolates. Infect. Drug Resist. 2019, 12, 1619–1628. [Google Scholar] [CrossRef] [Green Version]
- Asai, M.; Li, Y.; Khara, J.S.; Robertson, B.D.; Langford, P.R.; Newton, S.M. Galleria Mellonella: An Infection Model for Screening Compounds Against the Mycobacterium Tuberculosis Complex. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Desbois, A.P.; Coote, P.J. Wax Moth Larva (Galleria mellonella): An in Vivo Model for Assessing the Efficacy of Antistaphylococcal Agents. J. Antimicrob. Chemother. 2011, 66, 1785–1790. [Google Scholar] [CrossRef] [Green Version]
- Kay, S.; Edwards, J.; Brown, J.; Dixon, R. Galleria Mellonella Infection Model Identifies Both High and Low Lethality of Clostridium Perfringens Toxigenic Strains and Their Response to Antimicrobials. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Harding, C.R.; Schroeder, G.N.; Reynolds, S.; Kosta, A.; Collins, J.W.; Mousnier, A.; Frankel, G. Legionella Pneumophila Pathogenesis in the Galleria Mellonella Infection Model. Infect. Immun. 2012, 80, 2780–2790. [Google Scholar] [CrossRef] [Green Version]
- Alghoribi, M.F.; Gibreel, T.M.; Dodgson, A.R.; Beatson, S.A.; Upton, M. Galleria Mellonella Infection Model Demonstrates High Lethality of ST69 and ST127 Uropathogenic E. coli. PLoS ONE 2014, 9, e101547. [Google Scholar] [CrossRef] [Green Version]
- Kavanagh, K.; Sheehan, G. The Use of Galleria Mellonella Larvae to Identify Novel Antimicrobial Agents against Fungal Species of Medical Interest. J. Fungi Basel Switz. 2018, 4, 113. [Google Scholar] [CrossRef] [Green Version]
- Nyamu, S.N.; Ombaka, L.; Masika, E.; Ng’ang’a, M. Antimicrobial Photodynamic Activity of Phthalocyanine Derivatives. Adv. Chem. 2018, 2018, 2598062. [Google Scholar] [CrossRef]
- Gao, Y.; Mai, B.; Wang, A.; Li, M.; Wang, X.; Zhang, K.; Liu, Q.; Wei, S.; Wang, P. Antimicrobial Properties of a New Type of Photosensitizer Derived from Phthalocyanine against Planktonic and Biofilm Forms of Staphylococcus Aureus. Photodiagnosis Photodyn. Ther. 2018, 21, 316–326. [Google Scholar] [CrossRef]
- Mantareva, V.N.; Angelov, I.; Wöhrle, D.; Borisova, E.; Kussovski, V. Metallophthalocyanines for Antimicrobial Photodynamic Therapy: An Overview of Our Experience. J. Porphyr. Phthalocyanines 2013, 17, 399–416. [Google Scholar] [CrossRef]
- Gerasymchuk, Y.; Łukowiak, A.; Kędziora, A.; Wedzyńska, A.; Bugla-Płoskońska, G.; Piątek, D.; Bachanek, T.; Chernii, V.; Tomachynski, L.; Stręk, W. New Antibacterial Photoactive Nanocomposite Additives for Endodontic Cements and Fillings; Di Bartolo, B., Collins, J., Silvestri, L., Eds.; Springer: Amsterdam, The Netherlands, 2017; pp. 507–509. [Google Scholar]
- Gerasymchuk, Y.; Lukowiak, A.; Wedzynska, A.; Kedziora, A.; Bugla-Ploskonska, G.; Piatek, D.; Bachanek, T.; Chernii, V.; Tomachynski, L.; Strek, W. New Photosensitive Nanometric Graphite Oxide Composites as Antimicrobial Material with Prolonged Action. J. Inorg. Biochem. 2016, 159, 142–148. [Google Scholar] [CrossRef]
- Husain, M.; Wu, D.; Saber, A.T.; Decan, N.; Jacobsen, N.R.; Williams, A.; Yauk, C.L.; Wallin, H.; Vogel, U.; Halappanavar, S. Intratracheally Instilled Titanium Dioxide Nanoparticles Translocate to Heart and Liver and Activate Complement Cascade in the Heart of C57BL/6 Mice. Nanotoxicology 2015, 9, 1013–1022. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.; Lai, B.F.L.; Foley, J.H.; Krisinger, M.J.; Conway, E.M.; Kizhakkedathu, J.N. Modulation of Complement Activation and Amplification on Nanoparticle Surfaces by Glycopolymer Conformation and Chemistry. ACS Nano 2014, 8, 7687–7703. [Google Scholar] [CrossRef]
- Kaiser, J.-P.; Roesslein, M.; Diener, L.; Wick, P. Human Health Risk of Ingested Nanoparticles That Are Added as Multifunctional Agents to Paints: An In Vitro Study. PLoS ONE 2013, 8, e83215. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lai, W.; Cui, M.; Liang, L.; Lin, Y.; Fang, Q.; Liu, Y.; Xie, L. An Evaluation of Blood Compatibility of Silver Nanoparticles. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sladowski, D.; Wasiutyński, A.; Wilczyński, G.; Grabska-Liberek, I.; Coecke, S.; Kinsner, A.; Kochanowska, I. Expression of the Membrane Complement Regulatory Proteins (CD55 and CD59) in Human Thymus. Folia Histochem. Cytobiol. 2006, 44, 263–267. [Google Scholar] [PubMed]
- Moghimi, S.M.; Simberg, D. Complement Activation Turnover on Surfaces of Nanoparticles. Nano Today 2017, 15, 8–10. [Google Scholar] [CrossRef] [Green Version]
- Fornaguera, C.; Calderó, G.; Mitjans, M.; Vinardell, M.P.; Solans, C.; Vauthier, C. Interactions of PLGA Nanoparticles with Blood Components: Protein Adsorption, Coagulation, Activation of the Complement System and Hemolysis Studies. Nanoscale 2015, 7, 6045–6058. [Google Scholar] [CrossRef]
- Cao, W.; He, L.; Cao, W.; Huang, X.; Jia, K.; Dai, J. Recent Progress of Graphene Oxide as a Potential Vaccine Carrier and Adjuvant. Acta Biomater. 2020, 112, 14–28. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H.; Yi, C.; Chen, G.; Li, Y.; Zhou, Y.; Chen, G.; Li, Y.; He, Y.; Yu, D. Host Immune Response Triggered by Graphene Quantum-Dot-Mediated Photodynamic Therapy for Oral Squamous Cell Carcinoma. Int. J. Nanomed. 2020, 15, 9627–9638. [Google Scholar] [CrossRef]
- Kędziora, A.; Wernecki, M.; Korzekwa, K.; Speruda, M.; Gerasymchuk, Y.; Łukowiak, A.; Bugla-Płoskońska, G. Consequences Of Long-Term Bacteria’s Exposure To Silver Nanoformulations With Different PhysicoChemical Properties. Int. J. Nanomed. 2020, 15, 199–213. [Google Scholar] [CrossRef] [Green Version]
- Allegra, E.; Titball, R.W.; Carter, J.; Champion, O.L. Galleria Mellonella Larvae Allow the Discrimination of Toxic and Non-Toxic Chemicals. Chemosphere 2018, 198, 469–472. [Google Scholar] [CrossRef]
- Megaw, J.; Thompson, T.P.; Lafferty, R.A.; Gilmore, B.F. Galleria Mellonella as a Novel in Vivo Model for Assessment of the Toxicity of 1-Alkyl-3-Methylimidazolium Chloride Ionic Liquids. Chemosphere 2015, 139, 197–201. [Google Scholar] [CrossRef]
- Maguire, R.; Duggan, O.; Kavanagh, K. Evaluation of Galleria Mellonella Larvae as an in Vivo Model for Assessing the Relative Toxicity of Food Preservative Agents. Cell Biol. Toxicol. 2016, 32, 209–216. [Google Scholar] [CrossRef]
- Randall, C.P.; Gupta, A.; Jackson, N.; Busse, D.; O’Neill, A.J. Silver Resistance in Gram-Negative Bacteria: A Dissection of Endogenous and Exogenous Mechanisms. J. Antimicrob. Chemother. 2015, 70, 1037–1046. [Google Scholar] [CrossRef] [Green Version]
- Lukowiak, A.; Gerasymchuk, Y.; Wedzynska, A.; Tahershamsi, L.; Tomala, R.; Strek, W.; Piatek, D.; Korona-Glowniak, I.; Speruda, M.; Kedziora, A.; et al. Light-Activated Zirconium(IV) Phthalocyanine Derivatives Linked to Graphite Oxide Flakes and Discussion on Their Antibacterial Activity. Appl. Sci. 2019, 9, 4447. [Google Scholar] [CrossRef] [Green Version]
- Tahershamsi, L.; Gerasymchuk, Y.; Wedzynska, A.; Ptak, M.; Tretyakova, I.; Lukowiak, A. Synthesis, Spectroscopic Characterization and Photoactivity of Zr(IV) Phthalocyanines Functionalized with Aminobenzoic Acids and Their GO-Based Composites. C 2020, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Human Serum P2918. Available online: https://www.sigmaaldrich.com/catalog/product/sigma/p2918 (accessed on 25 August 2019).
- Skorek, K.; Raczkowska, A.; Dudek, B.; Miętka, K.; Guz-Regner, K.; Pawlak, A.; Klausa, E.; Bugla-Płoskońska, G.; Brzostek, K. Regulatory Protein OmpR Influences the Serum Resistance of Yersinia Enterocolitica O:9 by Modifying the Structure of the Outer Membrane. PLoS ONE 2013, 8, e79525. [Google Scholar] [CrossRef]
- Moritz, S.; Bartz-Beielstein, T. imputeTS: Time Series Missing Value Imputation in R. R J. 2017, 9, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Sardá-Espinosa, A. Time-Series Clustering in R Using the Dtwclust Package. R J. 2019, 11, 22–43. [Google Scholar] [CrossRef]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]









| Larvae Survival in 24 h after Nanocomposite Administration Number of Live Larvae [n] (Percentage of Live Larvae [%]) | ||||
|---|---|---|---|---|
| Nanocomposites | Run 1 | Run 2 | Run 3 | Medium |
| GO-PcZr(Lys)2-Ag (IV) | 9 (90%) | 10 (100%) | 9 (90%) | 9.3 (93%) |
| GO-Ag (V) | 10 (100%) | 10 (100%) | 10 (100%) | 10 (100%) |
| GO-PcZr(Lys)2 (VI) | 9 (90%) | 10 (100%) | 10 (100%) | 9.6 (96%) |
| rGO | 8 (100%) | 10 (100%) | 9 (100%) | 9 (90%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morka, K.D.; Wernecki, M.; Kędziora, A.; Książczyk, M.; Dudek, B.; Gerasymchuk, Y.; Lukowiak, A.; Bystroń, J.; Bugla-Płoskońska, G. The Impact of Graphite Oxide Nanocomposites on the Antibacterial Activity of Serum. Int. J. Mol. Sci. 2021, 22, 7386. https://doi.org/10.3390/ijms22147386
Morka KD, Wernecki M, Kędziora A, Książczyk M, Dudek B, Gerasymchuk Y, Lukowiak A, Bystroń J, Bugla-Płoskońska G. The Impact of Graphite Oxide Nanocomposites on the Antibacterial Activity of Serum. International Journal of Molecular Sciences. 2021; 22(14):7386. https://doi.org/10.3390/ijms22147386
Chicago/Turabian StyleMorka, Katarzyna Dorota, Maciej Wernecki, Anna Kędziora, Marta Książczyk, Bartłomiej Dudek, Yuriy Gerasymchuk, Anna Lukowiak, Jarosław Bystroń, and Gabriela Bugla-Płoskońska. 2021. "The Impact of Graphite Oxide Nanocomposites on the Antibacterial Activity of Serum" International Journal of Molecular Sciences 22, no. 14: 7386. https://doi.org/10.3390/ijms22147386
APA StyleMorka, K. D., Wernecki, M., Kędziora, A., Książczyk, M., Dudek, B., Gerasymchuk, Y., Lukowiak, A., Bystroń, J., & Bugla-Płoskońska, G. (2021). The Impact of Graphite Oxide Nanocomposites on the Antibacterial Activity of Serum. International Journal of Molecular Sciences, 22(14), 7386. https://doi.org/10.3390/ijms22147386








