Molecular Characterization of Carbonic Anhydrase Genes in Lotus japonicus and Their Potential Roles in Symbiotic Nitrogen Fixation
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of Carbonic Anhydrase Genes in Arabidopsis and Lotus
2.2. Expression Profiles of LjCAs across Different Tissues and Different Developmental Stages of Root Nodule
2.3. Spatiotemporal Expression Patterns of Nodule-Enhanced LjCAs in Root Nodule
2.4. Construction of LjCAs Mutants and Symbiotic Phenotypic Analysis
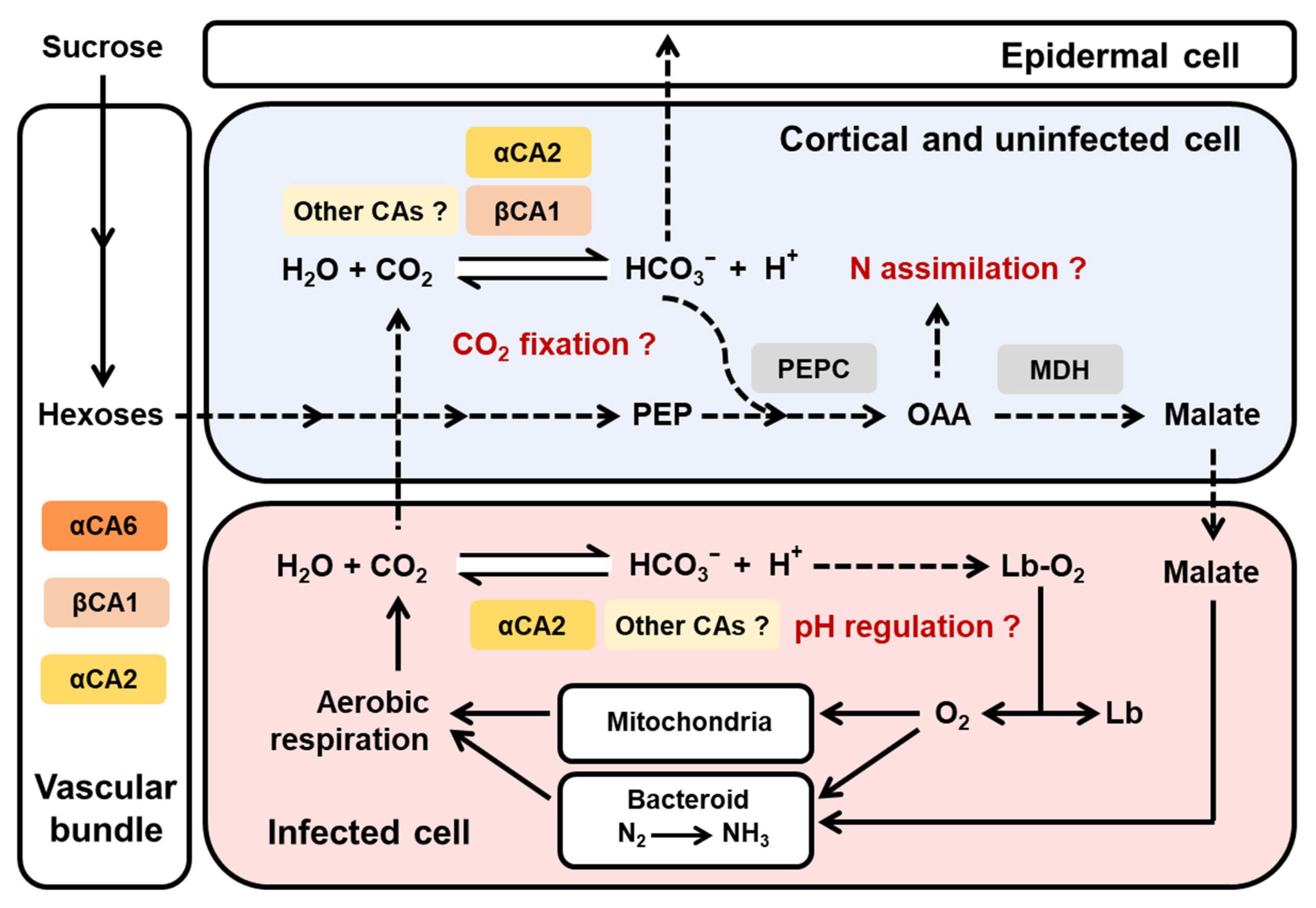
3. Discussion
4. Materials and Methods
4.1. Gene Identification and Phylogenetic Analysis
4.2. Plant Growth and Transformation
4.3. GUS Staining and tYFP-NLS Observation
4.4. Construction of LjCAs Knockout Mutants
4.5. RNA Extraction and qRT-PCR
4.6. Western Blot Analysis
4.7. Nitrogenase Activity Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ARA | Acetylene reduction activity |
| BCA | Bicinchoninic acid |
| CA | Carbonic anhydrase |
| CCM | CO2 concentrating mechanism |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| EDTA | Ethylenediaminetetraacetic acid |
| GRAVY | Grand average of hydropathicity |
| GUS | β-Glucuronidase |
| Lb | Leghemoglobin |
| MDH | Malate dehydrogenase |
| MW | Molecular weight |
| NLS | Nuclear localization signal |
| OAA | Oxaloacetate |
| PEP | Phosphoenolpyruvate |
| PEPC | Phosphoenolpyruvate carboxylase |
| pI | Isoelectric point |
| PVDF | Polyvinylidene difluoride |
| RBCs | Red blood cells |
| SDS | Sodium dodecyl sulfate |
| SNF | Symbiotic nitrogen fixation |
| tYFP | Triple yellow fluorescent protein |
References
- Hewett-Emmett, D.; Tashian, R.E. Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol. Phylogenet. Evol. 1996, 5, 50–77. [Google Scholar] [CrossRef] [PubMed]
- Chegwidden, W.R.; Carter, N.D. Introduction to the carbonic anhydrases. In The Carbonic Anhydrases: New Horizons; Chegwidden, W.R., Carter, N.D., Edwards, Y.H., Eds.; Birkhäuser Verlag: Basel, Switzerland, 2000; pp. 13–28. ISBN 978-3-0348-8446-4. [Google Scholar]
- DiMario, R.J.; Clayton, H.; Mukherjee, A.; Ludwig, M.; Moroney, J.V. Plant carbonic anhydrases—Structures, locations, evolution and physiological roles. Mol. Plant 2017, 10, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Fabre, N.; Reiter, I.M.; Becuwe-Linka, N.; Genty, B.; Rumeau, D. Characterization and expression analysis of genes encoding α and β carbonic anhydrases in Arabidopsis. Plant Cell Environ. 2007, 30, 617–629. [Google Scholar] [CrossRef]
- Floryszak-Wieczorek, J.; Arasimowicz-Jelonek, M. The multifunctional face of plant carbonic anhydrase. Plant Physiol. Biochem. 2017, 112, 362–368. [Google Scholar] [CrossRef]
- Fromm, S.; Senkler, J.; Zabaleta, E.; Peterhansel, C.; Braun, H.P. The carbonic anhydrase domain of plant mitochondrial complex I. Physiol. Plant. 2016, 157, 289–296. [Google Scholar] [CrossRef]
- Ignatova, L.; Rudenko, N.; Zhurikova, E.; Borisova-Mubarakshina, M.; Ivanov, B. Carbonic anhydrases in photosynthesizing cells of C3 higher plants. Metabolites 2019, 9, 73. [Google Scholar] [CrossRef]
- Studer, A.J.; Gandin, A.; Kolbe, A.R.; Wang, L.; Cousins, A.B.; Brutnell, T.P. A limited role for carbonic anhydrase in C4 photosynthesis as revealed by a ca1ca2 double mutant in maize. Plant Physiol. 2014, 165, 608–617. [Google Scholar] [CrossRef]
- Hu, H.; Boisson-Dernier, A.; Israelsson-Nordstrom, M.; Bohmer, M.; Xue, S.; Ries, A.; Godoski, J.; Kuhn, J.M.; Schroeder, J.I. Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 2010, 12, 87–93. [Google Scholar] [CrossRef]
- Engineer, C.B.; Ghassemian, M.; Anderson, J.C.; Peck, S.C.; Hu, H.; Schroeder, J.I. Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 2014, 513, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wu, H.; Wu, J.; Fan, X.; Li, X.; Lin, Y. Absence of OsβCA1 causes CO2 deficit and affects leaf photosynthesis and stomatal response to CO2 in rice. Plant J. 2017, 90, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, Z.; Biener, G.; Xiong, E.; Malik, S.; Eaton, N.; Zhao, C.Z.; Raicu, V.; Kong, H.; Zhao, D. Carbonic anhydrases function in anther cell differentiation downstream of the receptor-like kinase EMS1. Plant Cell 2017, 29, 1335–1356. [Google Scholar] [CrossRef]
- Hu, Z.; Ma, Q.; Foyer, C.H.; Lei, C.; Choi, H.W.; Zheng, C.; Li, J.; Zuo, J.; Mao, Z.; Mei, Y.; et al. High CO2- and pathogen-driven expression of the carbonic anhydrase βCA3 confers basal immunity in tomato. New Phytol. 2021, 229, 2827–2843. [Google Scholar] [CrossRef]
- Atkins, C.A. Occurrence and some properties of carbonic anhydrases from legume root nodules. Phytochemistry 1974, 13, 93–98. [Google Scholar] [CrossRef]
- Coba de la Pena, T.; Frugier, F.; McKhann, H.I.; Bauer, P.; Brown, S.; Kondorosi, A.; Crespi, M. A carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobium during development. Plant J. 1997, 11, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, S.; Hirsch, A.M.; Wycoff, K.L.; Hunt, S.; Layzell, D.B.; Kondorosi, A.; Crespi, M. Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol. 2000, 124, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Flemetakis, E.; Dimou, M.; Cotzur, D.; Aivalakis, G.; Efrose, R.C.; Kenoutis, C.; Udvardi, M.; Katinakis, P. A Lotus japonicus β-type carbonic anhydrase gene expression pattern suggests distinct physiological roles during nodule development. Biochim. Biophys. Acta 2003, 1628, 186–194. [Google Scholar] [CrossRef]
- Kavroulakis, N.; Flemetakis, E.; Aivalakis, G.; Katinakis, P. Carbon metabolism in developing soybean root nodules: The role of carbonic anhydrase. Mol. Plant Microbe Interact. 2000, 13, 14–22. [Google Scholar] [CrossRef]
- Kavroulakis, N.; Flemetakis, E.; Aivalakis, G.; Dahiya, P.; Brewin, N.J.; Fasseas, K.; Hatzopoulos, P.; Katinakis, P. Tissue distribution and subcellular localization of carbonic anhydrase in mature soybean root nodules indicates a role in CO2 diffusion. Plant Physiol. Biochem. 2003, 41, 479–484. [Google Scholar] [CrossRef]
- Tsikou, D.; Stedel, C.; Kouri, E.D.; Udvardi, M.K.; Wang, T.L.; Katinakis, P.; Labrou, N.E.; Flemetakis, E. Characterization of two novel nodule-enhanced α-type carbonic anhydrases from Lotus japonicus. Biochim. Biophys. Acta 2011, 1814, 496–504. [Google Scholar] [CrossRef]
- Kalloniati, C.; Tsikou, D.; Lampiri, V.; Fotelli, M.N.; Rennenberg, H.; Chatzipavlidis, I.; Fasseas, C.; Katinakis, P.; Flemetakis, E. Characterization of a Mesorhizobium loti α-type carbonic anhydrase and its role in symbiotic nitrogen fixation. J. Bacteriol. 2009, 191, 2593–2600. [Google Scholar] [CrossRef]
- Vullo, D.; Flemetakis, E.; Scozzafava, A.; Capasso, C.; Supuran, C.T. Anion inhibition studies of two α-carbonic anhydrases from Lotus japonicus, LjCAA1 and LjCAA2. J. Inorg. Biochem. 2014, 136, 67–72. [Google Scholar] [CrossRef]
- Fischinger, S.A.; Hristozkova, M.; Mainassara, Z.A.; Schulze, J. Elevated CO2 concentration around alfalfa nodules increases N2 fixation. J. Exp. Bot. 2010, 61, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Fischinger, S.A.; Schulze, J. The importance of nodule CO2 fixation for the efficiency of symbiotic nitrogen fixation in pea at vegetative growth and during pod formation. J. Exp. Bot. 2010, 61, 2281–2291. [Google Scholar] [CrossRef]
- Fotelli, M.N.; Tsikou, D.; Kolliopoulou, A.; Aivalakis, G.; Katinakis, P.; Udvardi, M.K.; Rennenberg, H.; Flemetakis, E. Nodulation enhances dark CO2 fixation and recycling in the model legume Lotus japonicus. J. Exp. Bot. 2011, 62, 2959–2971. [Google Scholar] [CrossRef]
- King, B.J.; Layzell, D.B.; Canvin, D.T. The role of dark carbon dioxide fixation in root nodules of soybean. Plant Physiol. 1986, 81, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Mai, H.T.; Fujii, M.; Hata, S.; Izui, K.; Tajima, S. Phosphoenolpyruvate carboxylase plays a crucial role in limiting nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol. 2006, 47, 613–621. [Google Scholar] [CrossRef]
- Miller, S.S.; Driscoll, B.T.; Gregerson, R.G.; Gantt, J.S.; Vance, C.P. Alfalfa malate dehydrogenase (MDH): Molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J. 1998, 15, 173–184. [Google Scholar] [CrossRef]
- Schwember, A.R.; Schulze, J.; Del Pozo, A.; Cabeza, R.A. Regulation of symbiotic nitrogen fixation in legume root nodules. Plants 2019, 8, 333. [Google Scholar] [CrossRef]
- White, J.; Prell, J.; James, E.K.; Poole, P. Nutrient sharing between symbionts. Plant Physiol. 2007, 144, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, K.M. Transport and exchange of respiratory gases in the blood, carbonic anhydrase in gas transport and exchange. In Encyclopedia of Fish Physiology: From Genome to Environment; Farrell, A.P., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 899–908. [Google Scholar] [CrossRef]
- Knight, K. Carbonic anhydrase short-circuit could release root haemoglobin oxygen. J. Exp. Biol. 2011, 214, 2319–2328. [Google Scholar] [CrossRef][Green Version]
- Okonjo, K.O. Bohr effect of human hemoglobin: Separation of tertiary and quaternary contributions based on the Wyman equation. Biophys. Chem. 2017, 228, 87–97. [Google Scholar] [CrossRef]
- Appleby, C.A. Leghemoglobin and rhizobium respiration. Annu. Rev. Physiol. 1984, 35, 443–478. [Google Scholar] [CrossRef]
- Ott, T.; van Dongen, J.T.; Gunther, C.; Krusell, L.; Desbrosses, G.; Vigeolas, H.; Bock, V.; Czechowski, T.; Geigenberger, P.; Udvardi, M.K. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 2005, 15, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rubio, M.C.; Xin, X.; Zhang, B.; Fan, Q.; Wang, Q.; Ning, G.; Becana, M.; Duanmu, D. CRISPR/Cas9 knockout of leghemoglobin genes in Lotus japonicus uncovers their synergistic roles in symbiotic nitrogen fixation. New Phytol. 2019, 224, 818–832. [Google Scholar] [CrossRef]
- Pierre, O.; Engler, G.; Hopkins, J.; Brau, F.; Boncompagni, E.; Herouart, D. Peribacteroid space acidification: A marker of mature bacteroid functioning in Medicago truncatula nodules. Plant Cell Environ. 2013, 36, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Polishchuk, O.V. Stress-related changes in the expression and activity of plant carbonic anhydrases. Planta 2021, 253, 58. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kawaguchi, M. Lotus japonicus Miyakojima MG-20: An early-flowering accession suitable for indoor handling. J. Plant Res. 2000, 113, 507–509. [Google Scholar] [CrossRef]
- Lombari, P.; Ercolano, E.; Alaoui, H.; Chiurazzi, M. Agrobacterium-mediated in vitro transformation. In Lotus japonicus Handbook; Márquez, A.J., Ed.; Springer: Dordrecht, The Netherlands, 2005; pp. 251–259. ISBN 978-1-4020-3735-1. [Google Scholar]
- Broughton, W.; Dilworth, M. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef]
- Saeki, K.; Kouchi, H. The Lotus symbiont, Mesorhizobium loti: Molecular genetic techniques and application. J. Plant Res. 2000, 113, 457–465. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Asamizu, E.; Kato, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Ishikawa, A.; Kawashima, K. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000, 7, 331–338. [Google Scholar] [CrossRef]
- Wang, L.X.; Wang, L.L.; Tan, Q.; Fang, Q.L.; Zhu, H.; Hong, Z.L.; Zhang, Z.M.; Duanmu, D.Q. Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus using CRISPR-Cas9. Front. Plant Sci. 2016, 7, 1333. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Wang, L.X.; Zhou, Y.; Duanmu, D.Q. Use of CRISPR/Cas9 for symbiotic nitrogen fixation research in legumes. Prog. Mol. Biol. Transl. Sci. 2017, 149, 187–213. [Google Scholar] [CrossRef] [PubMed]
- Duanmu, D.; Casero, D.; Dent, R.M.; Gallaher, S.; Yang, W.; Rockwell, N.C.; Martin, S.S.; Pellegrini, M.; Niyogi, K.K.; Merchant, S.S.; et al. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc. Natl. Acad. Sci. USA 2013, 110, 3621–3626. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.W.; Holsten, R.; Jackson, E.; Burns, R. The acetylene-ethylene assay for N2 fixation: Laboratory and field evaluation. Plant Physiol. 1968, 43, 1185–1207. [Google Scholar] [CrossRef] [PubMed]
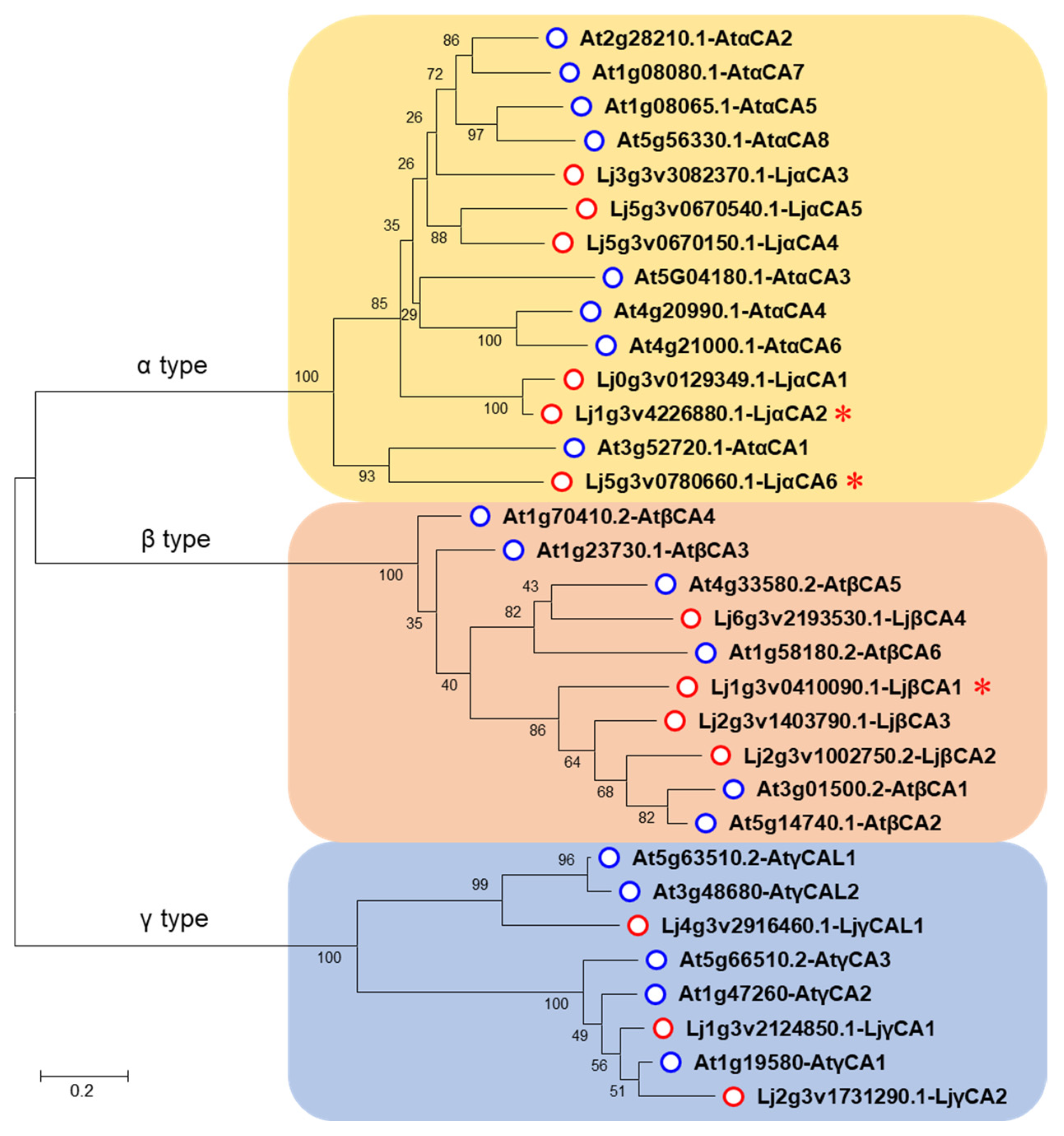




| Type | Gene | Chr. | Transcript ID | Former Name | NO. of AA | MW (kDa) | pI | GRAVY |
|---|---|---|---|---|---|---|---|---|
| α type | LjαCA1 | Lj0g3v0129349.1 | 269 | 30.47 | 8.79 | −0.454 | ||
| LjαCA2 | Chr 1 | Lj1g3v4226880.1 | LjCAA1 [20] | 269 | 30.29 | 9.05 | −0.453 | |
| LjαCA3 | Chr 3 | Lj3g3v3082370.1 | 218 | 24.65 | 5.92 | −0.456 | ||
| LjαCA4 | Chr 5 | Lj5g3v0670150.1 | 280 | 32.01 | 9.66 | −0.619 | ||
| LjαCA5 | Chr 5 | Lj5g3v0670540.1 | 266 | 30.61 | 6.95 | −0.639 | ||
| LjαCA6 | Chr 5 | Lj5g3v0780660.1 | LjCAA2 [20] | 274 | 30.74 | 6.63 | −0.374 | |
| β type | LjβCA1 | Chr 1 | Lj1g3v0410090.1 | LjCA1 [17] | 263 | 29.87 | 6.00 | −0.303 |
| LjβCA2 | Chr 2 | Lj2g3v1002750.2 | 324 | 34.90 | 6.54 | −0.058 | ||
| LjβCA3 | Chr 2 | Lj2g3v1403790.1 | 256 | 27.91 | 5.49 | −0.129 | ||
| LjβCA4 | Chr 6 | Lj6g3v2193530.1 | 263 | 29.05 | 6.44 | −0.186 | ||
| γ type | LjγCA1 | Chr 1 | Lj1g3v2124850.1 | 273 | 29.58 | 6.23 | −0.095 | |
| LjγCA2 | Chr 2 | Lj2g3v1731290.1 | 271 | 29.46 | 6.07 | −0.101 | ||
| LjγCAL1 | Chr 4 | Lj4g3v2916460.1 | 186 | 20.20 | 9.44 | 0.176 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Liang, J.; Zhou, Y.; Tian, T.; Zhang, B.; Duanmu, D. Molecular Characterization of Carbonic Anhydrase Genes in Lotus japonicus and Their Potential Roles in Symbiotic Nitrogen Fixation. Int. J. Mol. Sci. 2021, 22, 7766. https://doi.org/10.3390/ijms22157766
Wang L, Liang J, Zhou Y, Tian T, Zhang B, Duanmu D. Molecular Characterization of Carbonic Anhydrase Genes in Lotus japonicus and Their Potential Roles in Symbiotic Nitrogen Fixation. International Journal of Molecular Sciences. 2021; 22(15):7766. https://doi.org/10.3390/ijms22157766
Chicago/Turabian StyleWang, Longlong, Jianjun Liang, Yu Zhou, Tao Tian, Baoli Zhang, and Deqiang Duanmu. 2021. "Molecular Characterization of Carbonic Anhydrase Genes in Lotus japonicus and Their Potential Roles in Symbiotic Nitrogen Fixation" International Journal of Molecular Sciences 22, no. 15: 7766. https://doi.org/10.3390/ijms22157766
APA StyleWang, L., Liang, J., Zhou, Y., Tian, T., Zhang, B., & Duanmu, D. (2021). Molecular Characterization of Carbonic Anhydrase Genes in Lotus japonicus and Their Potential Roles in Symbiotic Nitrogen Fixation. International Journal of Molecular Sciences, 22(15), 7766. https://doi.org/10.3390/ijms22157766






