Common Muscle Metabolic Signatures Highlight Arginine and Lysine Metabolism as Potential Therapeutic Targets to Combat Unhealthy Aging
Abstract
:1. Introduction
2. Results
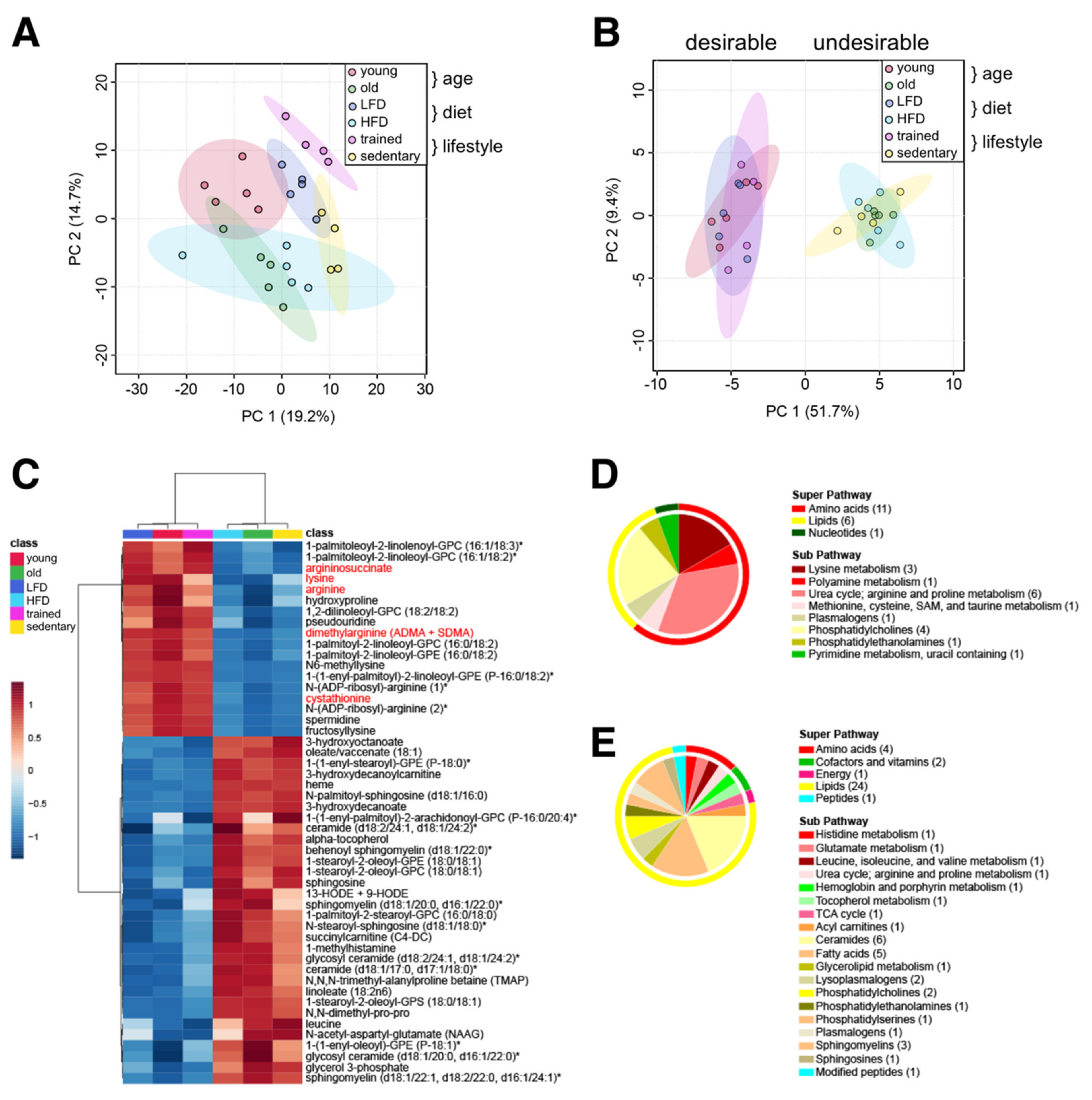
2.1. Changes in the Skeletal Muscle Metabolome Associated with Age, Diet and Lifestyle
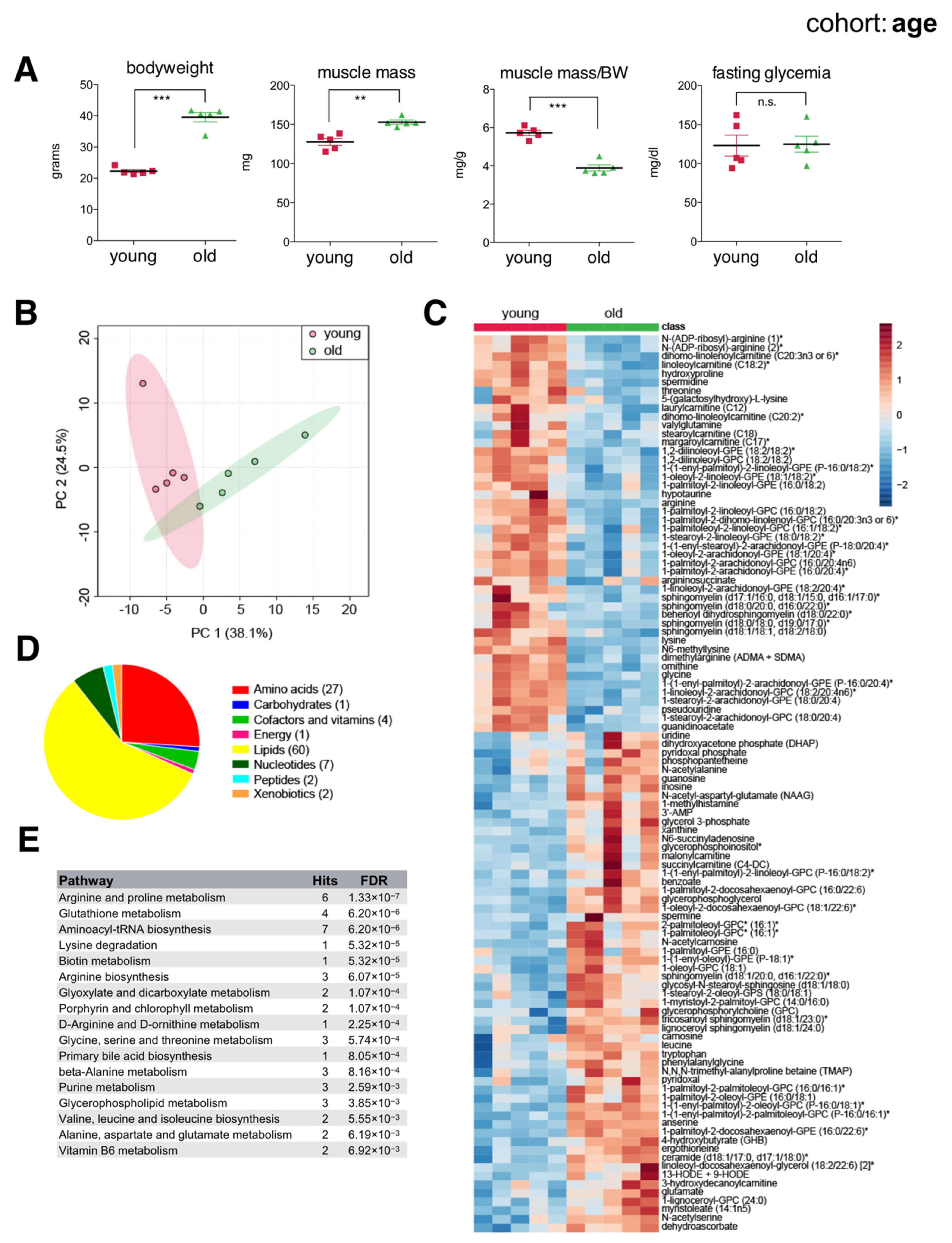
2.2. Age-Associated Changes in the Skeletal Muscle Metabolome
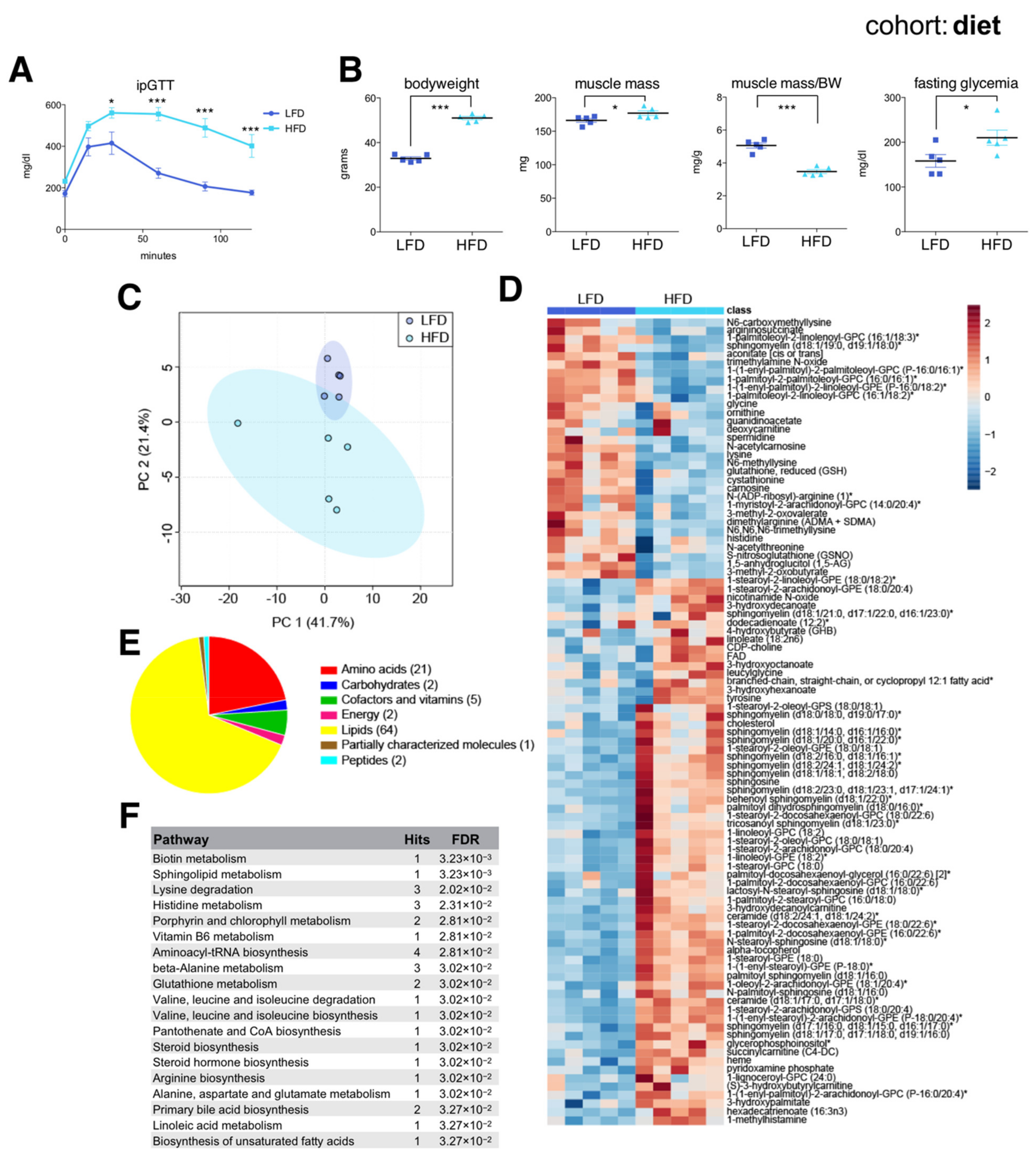
2.3. Diet-Associated Changes in the Skeletal Muscle Metabolome
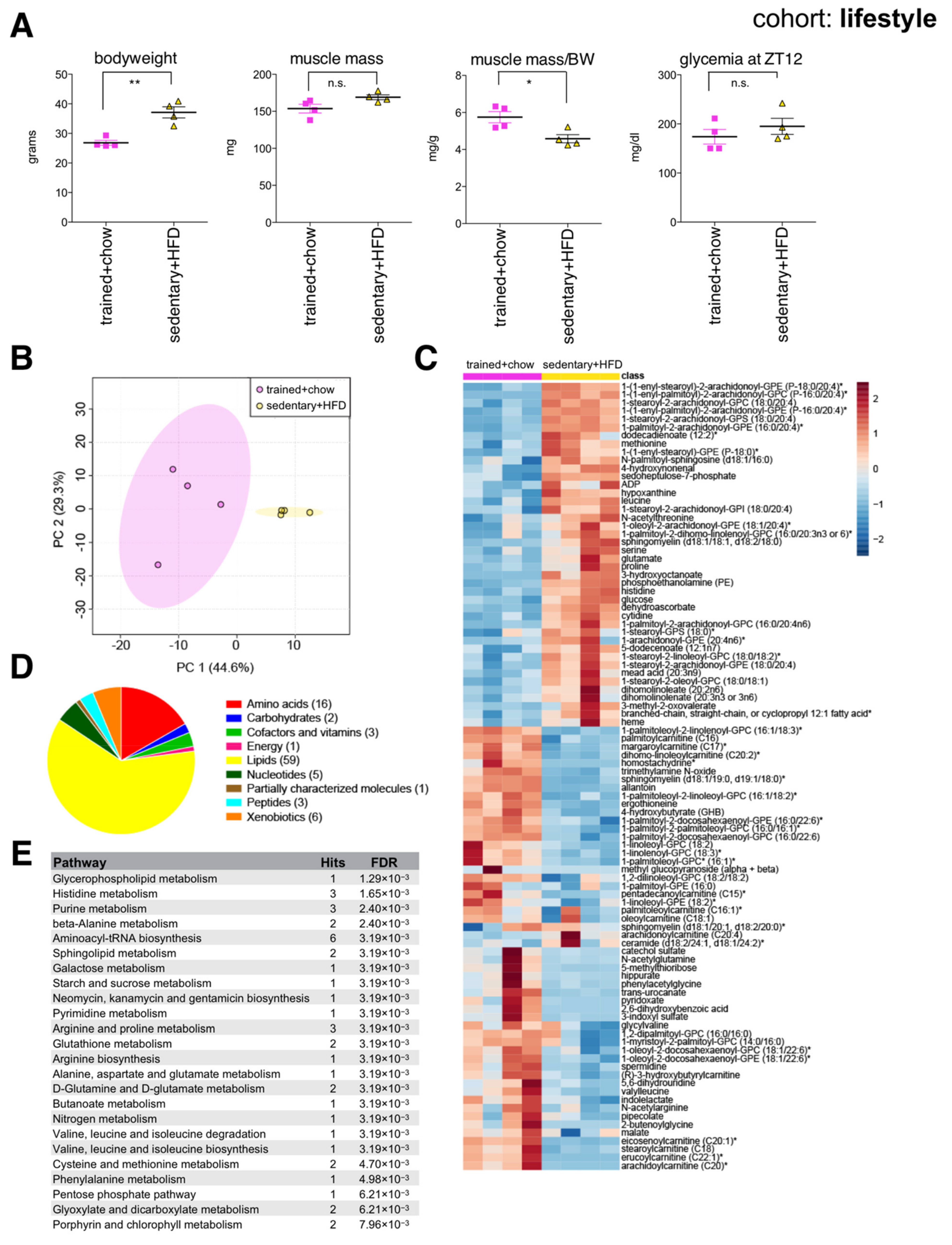
2.4. Lifestyle-Associated Changes in the Skeletal Muscle Metabolome
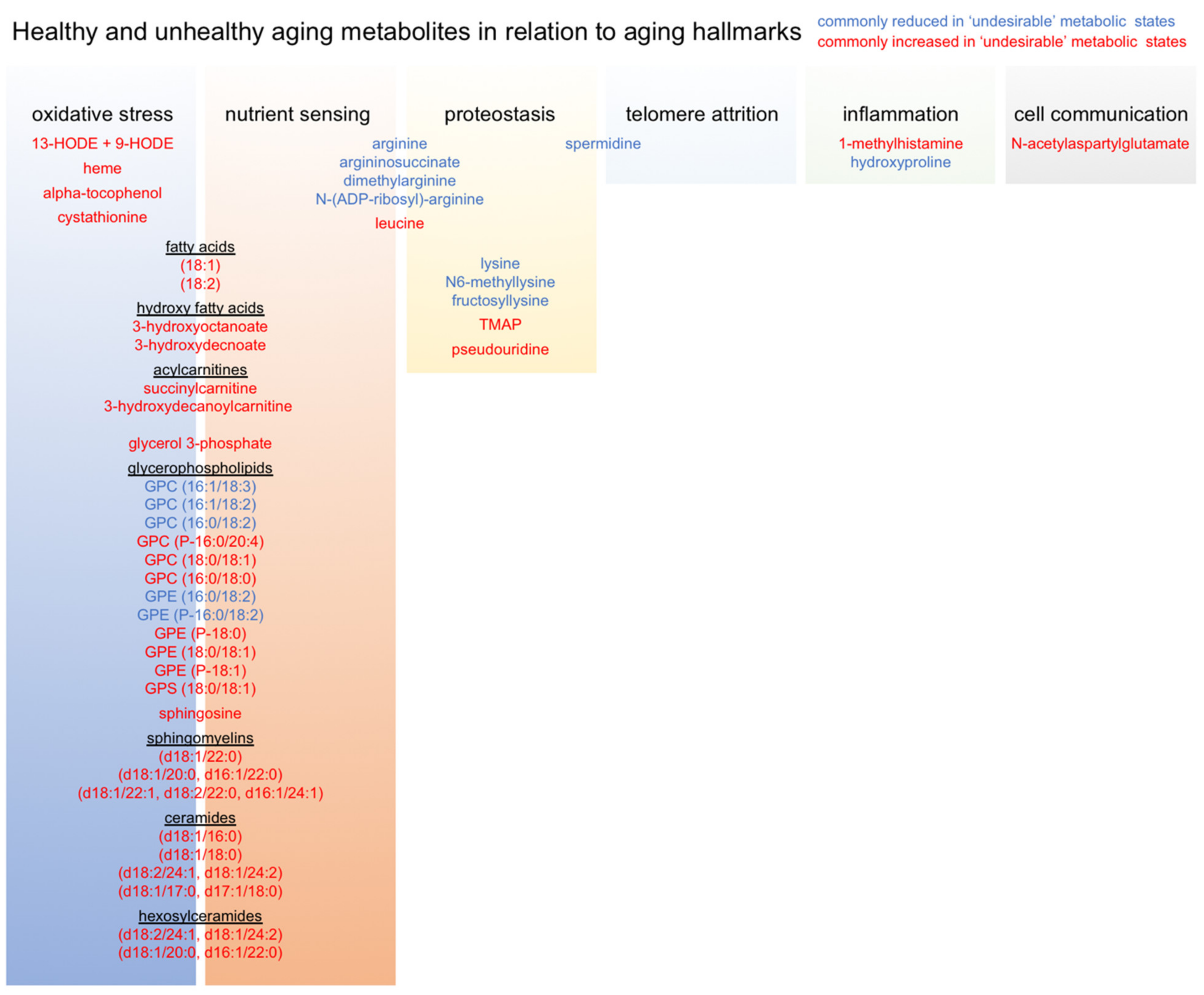
2.5. Common Skeletal Muscle Metabolite Signatures Distinguish ‘Desirable’ from ‘Undesirable’ Aging Trajectories
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Endurance Training
4.3. Intraperitoneal Glucose Tolerance Test
4.4. Muscle Tissue Collection and Preparation for Metabolomics
4.5. Nontargeted Metabolomics
4.6. Statistical Analysis and Quantitative Enrichment Analysis
4.7. Statistical Meta-Analysis
4.8. Generation of the AAMEMS-Library and Application in Quantitative Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [Green Version]
- United Nations. World Population Prospects 2019: Highlights; Statistical, Papers; United Nations (Ser. A), Population and Vital Statistics Report; UN: New York, NY, USA, 2019; ISBN 978-92-1-004235. [Google Scholar]
- World Health Organization. World Report on Ageing and Health; World Health Organization: Geneva, Switzerland, 2015; ISBN 978-92-4-156504-2. [Google Scholar]
- López-Otín, C.; Galluzzi, L.; Freije, J.M.P.; Madeo, F.; Kroemer, G. Metabolic Control of Longevity. Cell 2016, 166, 802–821. [Google Scholar] [CrossRef] [Green Version]
- Solanas, G.; Peixoto, F.O.; Perdiguero, E.; Jardí, M.; Ruiz-Bonilla, V.; Datta, D.; Symeonidi, A.; Castellanos, A.; Welz, P.-S.; Caballero, J.M.; et al. Aged Stem Cells Reprogram Their Daily Rhythmic Functions to Adapt to Stress. Cell 2017, 170, 678–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvestrini, V.; Sell, C.; Lorenzini, A. Obesity May Accelerate the Aging Process. Front. Endocrinol. 2019, 10, 266. [Google Scholar] [CrossRef]
- Balasubramanian, P.; Howell, P.R.; Anderson, R.M. Aging and Caloric Restriction Research: A Biological Perspective with Translational Potential. EBioMedicine 2017, 21, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohal, R.S.; Forster, M.J. Caloric Restriction and the Aging Process: A Critique. Free Radic. Biol. Med. 2014, 73, 366–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briggs, A.M.; Cross, M.J.; Hoy, D.G.; Sànchez-Riera, L.; Blyth, F.M.; Woolf, A.D.; March, L. Musculoskeletal Health Conditions Represent a Global Threat to Healthy Aging: A Report for the 2015 World Health Organization World Report on Ageing and Health. Gerontologist 2016, 56 (Suppl. 2), S243–S255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brack, A.S.; Muñoz-Cánoves, P. The Ins and Outs of Muscle Stem Cell Aging. Skelet. Muscle 2016, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Hwang, A.B.; Brack, A.S. Muscle Stem Cells and Aging. Curr. Top. Dev. Biol. 2018, 126, 299–322. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Paschalis, V.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Redox Basis of Exercise Physiology. Redox Biol. 2020, 35, 101499. [Google Scholar] [CrossRef]
- Glass, D.; Roubenoff, R. Recent Advances in the Biology and Therapy of Muscle Wasting. Ann. N. Y. Acad. Sci. 2010, 1211, 25–36. [Google Scholar] [CrossRef]
- Welch, A.A.; Hayhoe, R.P.G.; Cameron, D. The Relationships between Sarcopenic Skeletal Muscle Loss during Ageing and Macronutrient Metabolism, Obesity and Onset of Diabetes. Proc. Nutr. Soc. 2020, 79, 158–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayer, A.A.; Robinson, S.M.; Patel, H.P.; Shavlakadze, T.; Cooper, C.; Grounds, M.D. New Horizons in the Pathogenesis, Diagnosis and Management of Sarcopenia. Age Ageing 2013, 42, 145–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertel, J.; Friedrich, N.; Wittfeld, K.; Pietzner, M.; Budde, K.; Van der Auwera, S.; Lohmann, T.; Teumer, A.; Völzke, H.; Nauck, M.; et al. Measuring Biological Age via Metabonomics: The Metabolic Age Score. J. Proteome Res. 2016, 15, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Kerbl-Knapp, J.; Akhmetshina, A.; Korbelius, M.; Kuentzel, K.B.; Vujić, N.; Hörl, G.; Paar, M.; Kratky, D.; Steyrer, E.; et al. Tissue-Specific Landscape of Metabolic Dysregulation during Ageing. Biomolecules 2021, 11, 235. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bartok, B.; Oler, E.; Liang, K.Y.H.; Budinski, Z.; Berjanskii, M.; Guo, A.; Cao, X.; Wilson, M. MarkerDB: An Online Database of Molecular Biomarkers. Nucleic Acids Res. 2021, 49, D1259–D1267. [Google Scholar] [CrossRef]
- Fazelzadeh, P.; Hangelbroek, R.W.J.; Tieland, M.; de Groot, L.C.P.G.M.; Verdijk, L.B.; van Loon, L.J.C.; Smilde, A.K.; Alves, R.D.A.M.; Vervoort, J.; Müller, M.; et al. The Muscle Metabolome Differs between Healthy and Frail Older Adults. J. Proteome Res. 2016, 15, 499–509. [Google Scholar] [CrossRef] [Green Version]
- Uchitomi, R.; Hatazawa, Y.; Senoo, N.; Yoshioka, K.; Fujita, M.; Shimizu, T.; Miura, S.; Ono, Y.; Kamei, Y. Metabolomic Analysis of Skeletal Muscle in Aged Mice. Sci. Rep. 2019, 9, 10425. [Google Scholar] [CrossRef] [Green Version]
- White, R.B.; Biérinx, A.-S.; Gnocchi, V.F.; Zammit, P.S. Dynamics of Muscle Fibre Growth during Postnatal Mouse Development. BMC Dev. Biol. 2010, 10, 21. [Google Scholar] [CrossRef] [Green Version]
- Gattazzo, F.; Laurent, B.; Relaix, F.; Rouard, H.; Didier, N. Distinct Phases of Postnatal Skeletal Muscle Growth Govern the Progressive Establishment of Muscle Stem Cell Quiescence. Stem Cell Rep. 2020, 15, 597–611. [Google Scholar] [CrossRef]
- Börsch, A.; Ham, D.J.; Mittal, N.; Tintignac, L.A.; Migliavacca, E.; Feige, J.N.; Rüegg, M.A.; Zavolan, M. Molecular and Phenotypic Analysis of Rodent Models Reveals Conserved and Species-Specific Modulators of Human Sarcopenia. Commun. Biol. 2021, 4, 194. [Google Scholar] [CrossRef]
- Dyar, K.A.; Hubert, M.J.; Mir, A.A.; Ciciliot, S.; Lutter, D.; Greulich, F.; Quagliarini, F.; Kleinert, M.; Fischer, K.; Eichmann, T.O.; et al. Transcriptional Programming of Lipid and Amino Acid Metabolism by the Skeletal Muscle Circadian Clock. PLoS Biol. 2018, 16, e2005886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila, J.J.; Kim, S.K.; Massett, M.P. Differences in Exercise Capacity and Responses to Training in 24 Inbred Mouse Strains. Front. Physiol. 2017, 8, 974. [Google Scholar] [CrossRef] [PubMed]
- Haskell, W.L.; Lee, I.-M.; Pate, R.R.; Powell, K.E.; Blair, S.N.; Franklin, B.A.; Macera, C.A.; Heath, G.W.; Thompson, P.D.; Bauman, A. Physical Activity and Public Health: Updated Recommendation for Adults from the American College of Sports Medicine and the American Heart Association. Med. Sci. Sports Exerc. 2007, 39, 1423–1434. [Google Scholar] [CrossRef] [Green Version]
- Garber, C.E.; Blissmer, B.; Deschenes, M.R.; Franklin, B.A.; Lamonte, M.J.; Lee, I.-M.; Nieman, D.C.; Swain, D.P.; American College of Sports Medicine Position Stand. Quantity and Quality of Exercise for Developing and Maintaining Cardiorespiratory, Musculoskeletal, and Neuromotor Fitness in Apparently Healthy Adults: Guidance for Prescribing Exercise. Med. Sci. Sports Exerc. 2011, 43, 1334–1359. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Cantó, C.; Jeninga, E.H.; Andreux, P.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The Metabolic Footprint of Aging in Mice. Sci. Rep. 2011, 1, 134. [Google Scholar] [CrossRef]
- Vangaveti, V.; Baune, B.T.; Kennedy, R.L. Hydroxyoctadecadienoic Acids: Novel Regulators of Macrophage Differentiation and Atherogenesis. Ther. Adv. Endocrinol. Metab. 2010, 1, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Dyar, K.A.; Lutter, D.; Artati, A.; Ceglia, N.J.; Liu, Y.; Armenta, D.; Jastroch, M.; Schneider, S.; de Mateo, S.; Cervantes, M.; et al. Atlas of Circadian Metabolism Reveals System-Wide Coordination and Communication between Clocks. Cell 2018, 174, 1571–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schader, J.F.; Haid, M.; Cecil, A.; Schoenfeld, J.; Halle, M.; Pfeufer, A.; Prehn, C.; Adamski, J.; Nieman, D.C.; Scherr, J. Metabolite Shifts Induced by Marathon Race Competition Differ between Athletes Based on Level of Fitness and Performance: A Substudy of the Enzy-MagIC Study. Metabolites 2020, 10, 87. [Google Scholar] [CrossRef] [Green Version]
- Strimbu, K.; Tavel, J.A. What Are Biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Ham, D.J.; Börsch, A.; Lin, S.; Thürkauf, M.; Weihrauch, M.; Reinhard, J.R.; Delezie, J.; Battilana, F.; Wang, X.; Kaiser, M.S.; et al. The Neuromuscular Junction Is a Focal Point of MTORC1 Signaling in Sarcopenia. Nat. Commun. 2020, 11, 4510. [Google Scholar] [CrossRef] [PubMed]
- Melzer, T.M.; Manosso, L.M.; Yau, S.-Y.; Gil-Mohapel, J.; Brocardo, P.S. In Pursuit of Healthy Aging: Effects of Nutrition on Brain Function. Int. J. Mol. Sci. 2021, 22, 5026. [Google Scholar] [CrossRef] [PubMed]
- Thyfault, J.P.; Du, M.; Kraus, W.E.; Levine, J.A.; Booth, F.W. Physiology of Sedentary Behavior and Its Relationship to Health Outcomes. Med. Sci. Sports Exerc. 2015, 47, 1301–1305. [Google Scholar] [CrossRef] [Green Version]
- Carapeto, P.V.; Aguayo-Mazzucato, C. Effects of Exercise on Cellular and Tissue Aging. Aging 2021, 13, 14522–14543. [Google Scholar] [CrossRef] [PubMed]
- Garvey, S.M.; Dugle, J.E.; Kennedy, A.D.; McDunn, J.E.; Kline, W.; Guo, L.; Guttridge, D.C.; Pereira, S.L.; Edens, N.K. Metabolomic Profiling Reveals Severe Skeletal Muscle Group-Specific Perturbations of Metabolism in Aged FBN Rats. Biogerontology 2014, 15, 217–232. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, D.J.; Rodriguez-Blanco, G.; Dunn, W.B.; Phillips, B.E.; Williams, J.P.; Greenhaff, P.L.; Smith, K.; Gallagher, I.J.; Atherton, P.J. Untargeted Metabolomics for Uncovering Biological Markers of Human Skeletal Muscle Ageing. Aging 2020, 12, 12517–12533. [Google Scholar] [CrossRef]
- Van der Kolk, B.W.; Saari, S.; Lovric, A.; Arif, M.; Alvarez, M.; Ko, A.; Miao, Z.; Sahebekhtiari, N.; Muniandy, M.; Heinonen, S.; et al. Molecular Pathways behind Acquired Obesity: Adipose Tissue and Skeletal Muscle Multiomics in Monozygotic Twin Pairs Discordant for BMI. Cell Rep. Med. 2021, 2, 100226. [Google Scholar] [CrossRef]
- Chorner, Z.; Barbeau, P.-A.; Castellani, L.; Wright, D.C.; Chabowski, A.; Holloway, G.P. Dietary α-Linolenic Acid Supplementation Alters Skeletal Muscle Plasma Membrane Lipid Composition, Sarcolemmal FAT/CD36 Abundance, and Palmitate Transport Rates. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R1234–R1242. [Google Scholar] [CrossRef] [Green Version]
- Herbst, E.A.F.; Paglialunga, S.; Gerling, C.; Whitfield, J.; Mukai, K.; Chabowski, A.; Heigenhauser, G.J.F.; Spriet, L.L.; Holloway, G.P. Omega-3 Supplementation Alters Mitochondrial Membrane Composition and Respiration Kinetics in Human Skeletal Muscle. J. Physiol. 2014, 592, 1341–1352. [Google Scholar] [CrossRef]
- Andersson, A.; Sjödin, A.; Olsson, R.; Vessby, B. Effects of Physical Exercise on Phospholipid Fatty Acid Composition in Skeletal Muscle. Am. J. Physiol. 1998, 274, E432–E438. [Google Scholar] [CrossRef]
- Pollard, A.K.; Ortori, C.A.; Stöger, R.; Barrett, D.A.; Chakrabarti, L. Mouse Mitochondrial Lipid Composition Is Defined by Age in Brain and Muscle. Aging 2017, 9, 986–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeromson, S.; Gallagher, I.J.; Galloway, S.D.R.; Hamilton, D.L. Omega-3 Fatty Acids and Skeletal Muscle Health. Mar. Drugs 2015, 13, 6977–7004. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Zhang, H.; Wei, J.; Tian, X.Y.; Luan, H.; Li, S.; Zhao, H.; Cao, G.; Chung, A.C.K.; Yang, C.; et al. Metabolomics Studies on Db/Db Diabetic Mice in Skeletal Muscle Reveal Effective Clearance of Overloaded Intermediates by Exercise. Anal. Chim. Acta 2018, 1037, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Starnes, J.W.; Parry, T.L.; O’Neal, S.K.; Bain, J.R.; Muehlbauer, M.J.; Honcoop, A.; Ilaiwy, A.; Christopher, P.M.; Patterson, C.; Willis, M.S. Exercise-Induced Alterations in Skeletal Muscle, Heart, Liver, and Serum Metabolome Identified by Non-Targeted Metabolomics Analysis. Metabolites 2017, 7, 40. [Google Scholar] [CrossRef] [Green Version]
- Courtney-Martin, G.; Ball, R.O.; Pencharz, P.B.; Elango, R. Protein Requirements during Aging. Nutrients 2016, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Averous, J.; Lambert-Langlais, S.; Mesclon, F.; Carraro, V.; Parry, L.; Jousse, C.; Bruhat, A.; Maurin, A.-C.; Pierre, P.; Proud, C.G.; et al. GCN2 Contributes to MTORC1 Inhibition by Leucine Deprivation through an ATF4 Independent Mechanism. Sci. Rep. 2016, 6, 27698. [Google Scholar] [CrossRef]
- Valvezan, A.J.; Manning, B.D. Molecular Logic of MTORC1 Signalling as a Metabolic Rheostat. Nat. Metab. 2019, 1, 321–333. [Google Scholar] [CrossRef]
- Sandri, M.; Barberi, L.; Bijlsma, A.Y.; Blaauw, B.; Dyar, K.A.; Milan, G.; Mammucari, C.; Meskers, C.G.M.; Pallafacchina, G.; Paoli, A.; et al. Signalling Pathways Regulating Muscle Mass in Ageing Skeletal Muscle: The Role of the IGF1-Akt-MTOR-FoxO Pathway. Biogerontology 2013, 14, 303–323. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T. MTOR as Regulator of Lifespan, Aging, and Cellular Senescence: A Mini-Review. Gerontology 2018, 64, 127–134. [Google Scholar] [CrossRef]
- Jin, C.-L.; Ye, J.-L.; Yang, J.; Gao, C.-Q.; Yan, H.-C.; Li, H.-C.; Wang, X.-Q. MTORC1 Mediates Lysine-Induced Satellite Cell Activation to Promote Skeletal Muscle Growth. Cells 2019, 8, 1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Tsun, Z.-Y.; Wolfson, R.L.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W.; et al. Metabolism. Lysosomal Amino Acid Transporter SLC38A9 Signals Arginine Sufficiency to MTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR Proteins Are Arginine Sensors for the MTORC1 Pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, B.; Maetzel, D.; Maddocks, O.D.; Otten, G.; Ratcliff, M.; Smith, G.R.; Dunlop, E.A.; Passos, J.F.; Davies, O.R.; Jaenisch, R.; et al. Control of TSC2-Rheb Signaling Axis by Arginine Regulates MTORC1 Activity. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Arnau, F.M.; Fonfría-Vivas, R.; Buigues, C.; Castillo, Y.; Molina, P.; Hoogland, A.J.; van Doesburg, F.; Pruimboom, L.; Fernández-Garrido, J.; Cauli, O. Effects of Leucine Administration in Sarcopenia: A Randomized and Placebo-Controlled Clinical Trial. Nutrients 2020, 12, 932. [Google Scholar] [CrossRef] [Green Version]
- Komar, B.; Schwingshackl, L.; Hoffmann, G. Effects of Leucine-Rich Protein Supplements on Anthropometric Parameter and Muscle Strength in the Elderly: A Systematic Review and Meta-Analysis. J. Nutr. Health Aging 2015, 19, 437–446. [Google Scholar] [CrossRef]
- Martínez-Arnau, F.M.; Fonfría-Vivas, R.; Cauli, O. Beneficial Effects of Leucine Supplementation on Criteria for Sarcopenia: A Systematic Review. Nutrients 2019, 11, 2504. [Google Scholar] [CrossRef] [Green Version]
- Børsheim, E.; Bui, Q.-U.T.; Tissier, S.; Kobayashi, H.; Ferrando, A.A.; Wolfe, R.R. Effect of Amino Acid Supplementation on Muscle Mass, Strength and Physical Function in Elderly. Clin. Nutr. Edinb. Scotl. 2008, 27, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maykish, A.; Sikalidis, A.K. Utilization of Hydroxyl-Methyl Butyrate, Leucine, Glutamine and Arginine Supplementation in Nutritional Management of Sarcopenia-Implications and Clinical Considerations for Type 2 Diabetes Mellitus Risk Modulation. J. Pers. Med. 2020, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Flakoll, P.; Sharp, R.; Baier, S.; Levenhagen, D.; Carr, C.; Nissen, S. Effect of Beta-Hydroxy-Beta-Methylbutyrate, Arginine, and Lysine Supplementation on Strength, Functionality, Body Composition, and Protein Metabolism in Elderly Women. Nutr. Burbank Los Angel. Cty. Calif. 2004, 20, 445–451. [Google Scholar] [CrossRef]
- Sato, T.; Ito, Y.; Nagasawa, T. Regulation of Skeletal Muscle Protein Degradation and Synthesis by Oral Administration of Lysine in Rats. J. Nutr. Sci. Vitaminol. Tokyo 2013, 59, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Ito, Y.; Nagasawa, T. L-Lysine Suppresses Myofibrillar Protein Degradation and Autophagy in Skeletal Muscles of Senescence-Accelerated Mouse Prone 8. Biogerontology 2017, 18, 85–95. [Google Scholar] [CrossRef]
- Chen, W.W.; Freinkman, E.; Wang, T.; Birsoy, K.; Sabatini, D.M. Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell 2016, 166, 1324–1337. [Google Scholar] [CrossRef]
- Ibebunjo, C.; Chick, J.M.; Kendall, T.; Eash, J.K.; Li, C.; Zhang, Y.; Vickers, C.; Wu, Z.; Clarke, B.A.; Shi, J.; et al. Genomic and Proteomic Profiling Reveals Reduced Mitochondrial Function and Disruption of the Neuromuscular Junction Driving Rat Sarcopenia. Mol. Cell. Biol. 2013, 33, 194–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatori, M.; Panda, S. Response of Peripheral Rhythms to the Timing of Food Intake. Methods Enzymol. 2015, 552, 145–161. [Google Scholar] [CrossRef]
- Manoogian, E.N.C.; Panda, S. Circadian Rhythms, Time-Restricted Feeding, and Healthy Aging. Ageing Res. Rev. 2017, 39, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Paddon-Jones, D.; Børsheim, E.; Wolfe, R.R. Potential Ergogenic Effects of Arginine and Creatine Supplementation. J. Nutr. 2004, 134, 2888S–2894S. [Google Scholar] [CrossRef] [Green Version]
- Morris, S.M. Arginine Metabolism: Boundaries of Our Knowledge. J. Nutr. 2007, 137, 1602S–1609S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madeo, F.; Eisenberg, T.; Büttner, S.; Ruckenstuhl, C.; Kroemer, G. Spermidine: A Novel Autophagy Inducer and Longevity Elixir. Autophagy 2010, 6, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Wirth, A.; Wolf, B.; Huang, C.-K.; Glage, S.; Hofer, S.J.; Bankstahl, M.; Bär, C.; Thum, T.; Kahl, K.G.; Sigrist, S.J.; et al. Novel Aspects of Age-Protection by Spermidine Supplementation Are Associated with Preserved Telomere Length. GeroScience 2021, 43, 673–690. [Google Scholar] [CrossRef]
- Van de Poll, M.C.G.; Siroen, M.P.C.; van Leeuwen, P.A.M.; Soeters, P.B.; Melis, G.C.; Boelens, P.G.; Deutz, N.E.P.; Dejong, C.H.C. Interorgan Amino Acid Exchange in Humans: Consequences for Arginine and Citrulline Metabolism. Am. J. Clin. Nutr. 2007, 85, 167–172. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef] [PubMed]

| Experiment | Group | Strain | Diet | Age | Time of Sac | Nutritional State |
|---|---|---|---|---|---|---|
| Age | Young | C57BL/6N | chow (Altromin 1314) | 2 months | ZT6 | 4 h fasted |
| Old | C57BL/6N | chow (Altromin 1314) | 21 months | ZT6 | 4 h fasted | |
| Diet | Low fat diet (LFD) | C57BL/6J | 20 weeks 10% kcal fat (D12450Ji Research diets) | 7 months | ZT6 | 4 h fasted |
| High fat diet (HFD) | C57BL/6J | 20 weeks 60% kcal fat (D12492i Research diets) | 7 months | ZT6 | 4 h fasted | |
| Lifestyle | Trained + chow | C57BL/6J | chow (4RF21 Mucedola, Settimo Milanese, Italy) | 7 months | ZT11 | End of the physiological fasting phase |
| Sedentary + HFD | C57BL/6J | 12 weeks 60% kcal fat (PF4051/D, Mucedola, Settimo Milanese, Italy) | 7 months | ZT11 | End of the physiological fasting phase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokarz, J.; Möller, G.; Artati, A.; Huber, S.; Zeigerer, A.; Blaauw, B.; Adamski, J.; Dyar, K.A. Common Muscle Metabolic Signatures Highlight Arginine and Lysine Metabolism as Potential Therapeutic Targets to Combat Unhealthy Aging. Int. J. Mol. Sci. 2021, 22, 7958. https://doi.org/10.3390/ijms22157958
Tokarz J, Möller G, Artati A, Huber S, Zeigerer A, Blaauw B, Adamski J, Dyar KA. Common Muscle Metabolic Signatures Highlight Arginine and Lysine Metabolism as Potential Therapeutic Targets to Combat Unhealthy Aging. International Journal of Molecular Sciences. 2021; 22(15):7958. https://doi.org/10.3390/ijms22157958
Chicago/Turabian StyleTokarz, Janina, Gabriele Möller, Anna Artati, Simone Huber, Anja Zeigerer, Bert Blaauw, Jerzy Adamski, and Kenneth Allen Dyar. 2021. "Common Muscle Metabolic Signatures Highlight Arginine and Lysine Metabolism as Potential Therapeutic Targets to Combat Unhealthy Aging" International Journal of Molecular Sciences 22, no. 15: 7958. https://doi.org/10.3390/ijms22157958
APA StyleTokarz, J., Möller, G., Artati, A., Huber, S., Zeigerer, A., Blaauw, B., Adamski, J., & Dyar, K. A. (2021). Common Muscle Metabolic Signatures Highlight Arginine and Lysine Metabolism as Potential Therapeutic Targets to Combat Unhealthy Aging. International Journal of Molecular Sciences, 22(15), 7958. https://doi.org/10.3390/ijms22157958








