Characterization of the Free and Membrane-Associated Fractions of the Thylakoid Lumen Proteome in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
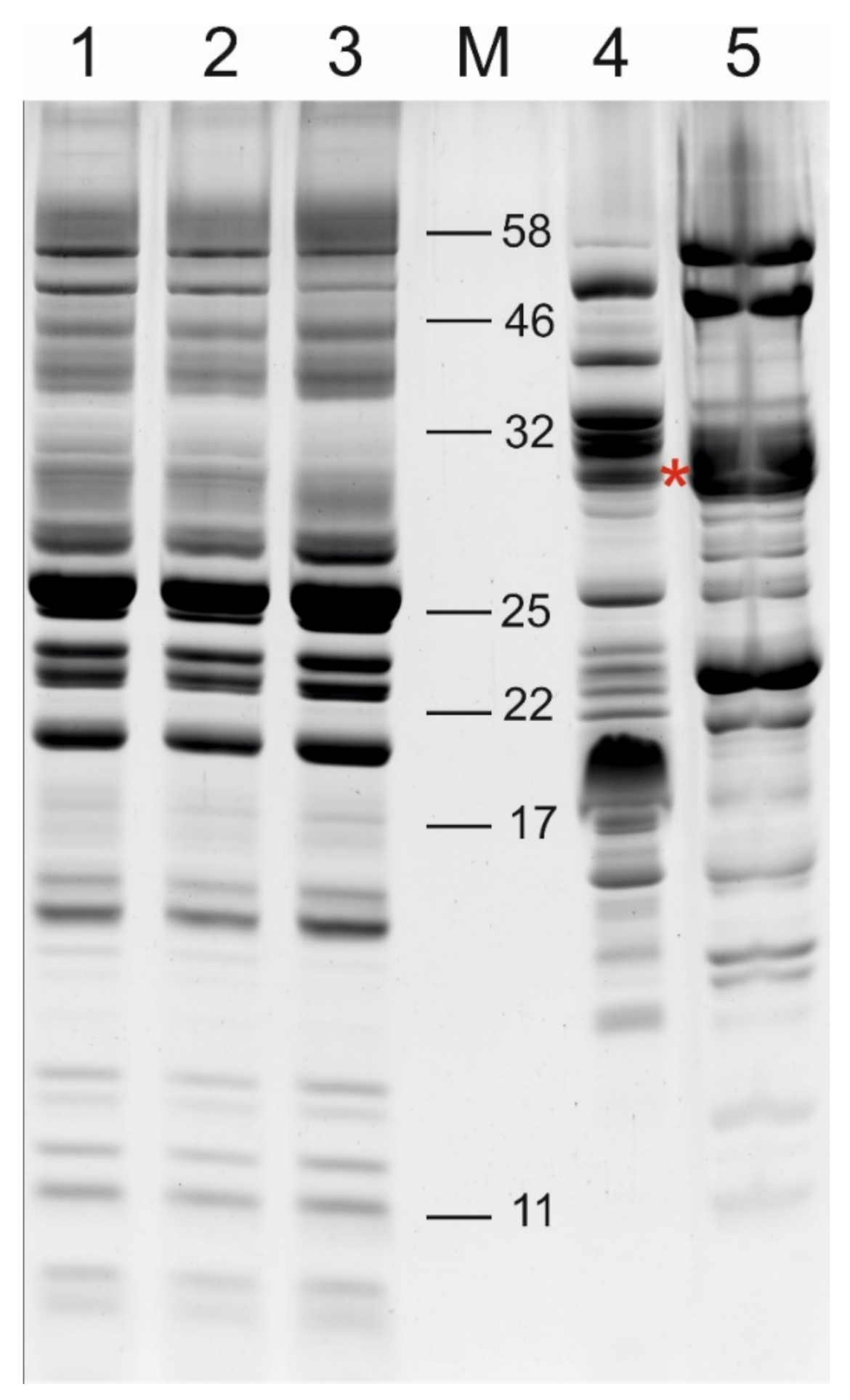
2.1. Comparison of the Free and Membrane-Associated Lumen Fraction
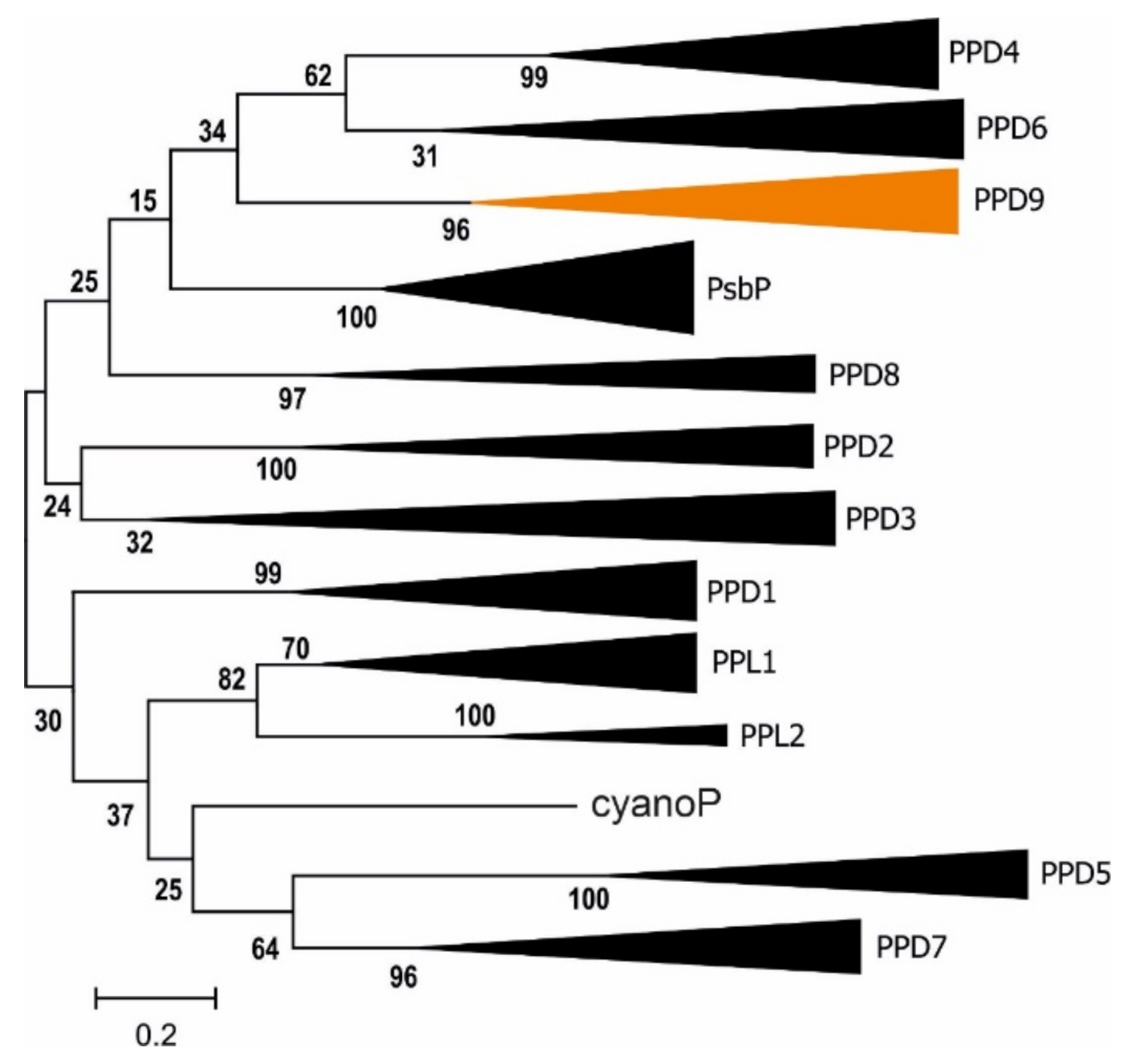
2.2. Novel Lumen Proteins
2.3. N-Termini
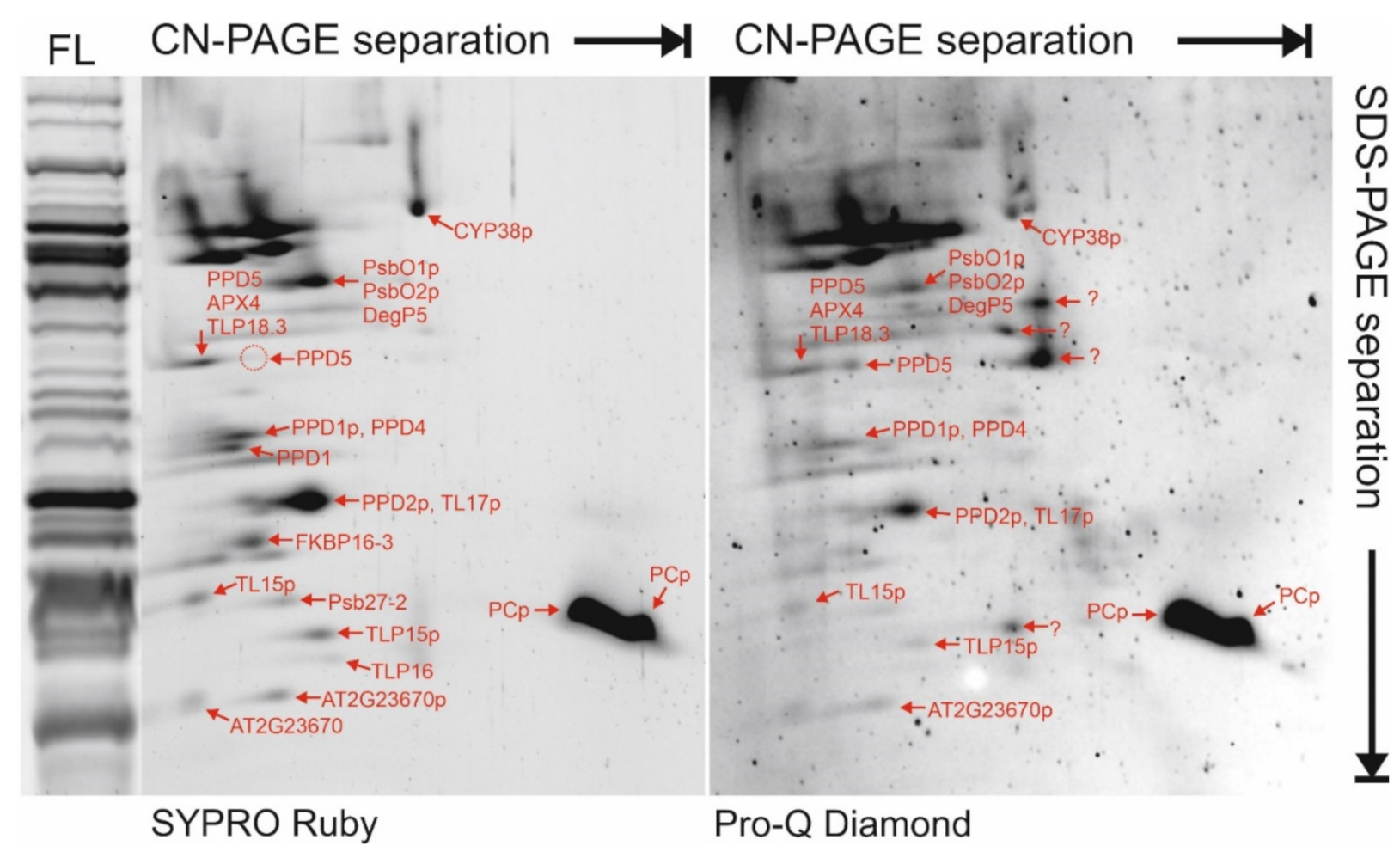
2.4. Protein Oligomer Complexes in the Lumen
2.5. Phosphorylated Proteins/Peptides in the Thylakoid Lumen
| Accession | Name | Phosphorylation Site(s) * | Phosphorylation Reported ** |
|---|---|---|---|
| AT3G01480.1 | CYP38 | Y109: (pY)ALPIDNKAIR S159:(pS)IIVAGFAESK T291: (pS)DGFVVQTGDPEGPAEGFIDPSTEKTR T298: SDGFVVQ(pT)GDPEGPAEGFIDPSTEKTR T318: (pT)VPLEImVTGEK S380: E(pS)ELTPSNSNILDGR | T318 experimentally detected (PhosPhAt, study not detailed) S159 [36] |
| AT5G66570.1 | PsbO1 | T108: G(pT)GTANQCPTIDGGSETFSFKPGK T108/T110: G(pT)G(pT)ANQCPTIDGGSETFSFKPGK S221: QLDA(pS)GKPDSFTGK T251: GGS(pT)GYDNAVALPAGGRGDEEELVK T278 ***: N(pT)AASVGEITLK T296 ***: SKPE(pT)GEVIGVFESLQPSDTDLGAK T305 ***: SKPETGEVIGVFE(pS)LQPSDTDLGAK | S226, S281 on the same peptides as S221, T278, respectively [35] S221 [36] |
| AT3G50820.1 | PsbO2 | T277 ***: ENVKN(pT)AASVGEITLK T295 ***: SKPE(pT)GEVIGVFESLQPSDTDLGAK | |
| AT4G15510.1 | PPD1 | T183: QYL(pT)EF(oM)STR | S28 [35] |
| AT2G28605.1 | PPD2 | S126: IK(pS)LDQFGSPQFVADK | S126 predicted (PhosPhAt) |
| AT1G77090.1 | PPD4 | no phosphopeptide detected | |
| AT5G11450.1 | PPD5 | no phosphopeptide detected | T283 [38] Y250 predicted (PhosPhAt) |
| AT1G20340.1 | PC major | Y103: NNAG(pY)PHNVVFDEDEIPSGVDVAK | Y103 predicted (PhosPhAt) |
| AT4G18370.1 | DegP5 | no phosphopeptide detected | |
| AT4G09010.1 | APX4 (TLP29) | no phosphopeptide detected | S155 [39] |
| AT2G44920.2 | TL15 | S99: GQDL(pS)GKDFSGQTLIR T164: VNL(pT)NANLEGATVTGNTSFK T190: GSNITGADF(pT)DVPLRDDQR | S99 predicted (PhosPhAt) |
| AT5G53490.3 | TL17 | T114: AFVGN(pT)IGQANGVYDKPLDLR T161: FDGAD(oM)(pT)EVV(oM)SK Y169: A(pY)AVEASFK T181: GVNF(pT)NAVIDR | T114 predicted (PhosPhAt) |
| AT2G43560.1 | FKBP16-3 | no phosphopeptide detected | |
| AT1G05385.1 | Psb27-2 (LPA19) | no phosphopeptide detected | |
| AT5G52970.1 | TLP15 | Y129: VLAQN(pY)PVTPGLAIK | |
| AT4G02530.1 | TLP16 | no phosphopeptide detected | |
| AT1G54780.1 | TLP18.3 | no phosphopeptide detected | |
| AT2G23670.1 | AT2G23670 | T100: EGFE(pT)AEKGVDAAEK | T100 predicted (PhosPhAt) |
3. Discussion
3.1. The Soluble and Membrane-Associated Fractions of the Thylakoid Lumen Are Distinct
3.2. A Major Role of the Lumen Proteome Is PSII Assembly and Maintenance
3.3. Protein Families Populate the Thylakoid Lumen
3.4. Post-Translational Modifications in the Thylakoid Lumen
3.5. Concluding Remarks
4. Materials and Methods
4.1. Plant Growth and Isolation of Thylakoid Lumen Fractions
4.2. Western Blotting
4.3. Clear Native PAGE Separation and Protein Visualization
4.4. Proteomics Methods
4.5. Bioinformatics Methods
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Åkerlund, H.E.; Jansson, C.; Andersson, B. Reconstitution of photosynthetic water splitting in inside-out thylakoid vesicles and identification of a participating polypeptide. BBA Bioenerg. 1982, 681, 1–10. [Google Scholar] [CrossRef]
- Miyao, M.; Murata, N. Role of the 33-kDa polypeptide in preserving Mn in the photosynthetic oxygen-evolution system and its replacement by chloride ions. FEBS Lett. 1984, 170, 350–354. [Google Scholar] [CrossRef] [Green Version]
- Ettinger, W.F.; Theg, S.M. Physiologically active chloroplasts contain pools of unassembled extrinsic proteins of the photosynthetic oxygen-evolving enzyme complex in the thylakoid lumen. J. Cell Biol. 1991, 115, 321–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redinbo, M.R.; Yeates, T.O.; Merchant, S. Plastocyanin: Structural and functional analysis. J. Bioenerg. Biomembr. 1994, 26, 49–66. [Google Scholar] [CrossRef]
- Ifuku, K.; Endo, T.; Shikanai, T.; Aro, E.M. Structure of the chloroplast NADH dehydrogenase-like complex: Nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 2011, 52, 1560–1568. [Google Scholar] [CrossRef]
- Peltier, G.; Aro, E.-M.; Shikanai, T. NDH-1 and NDH-2 Plastoquinone Reductases in Oxygenic Photosynthesis. Annu. Rev. Plant Biol. 2016, 67, 55–80. [Google Scholar] [CrossRef]
- Tyystjärvi, E.; Aro, E.M. The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc. Natl. Acad. Sci. USA 1996, 93, 2213–2218. [Google Scholar] [CrossRef] [Green Version]
- Kato, Y.; Sun, X.; Zhang, L.; Sakamoto, W. Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 2012, 159, 1428–1439. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Peng, L.; Guo, J.; Chi, W.; Ma, J.; Lu, C.; Zhang, L. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. Plant Cell 2007, 19, 1347–1361. [Google Scholar] [CrossRef] [Green Version]
- Kapri-Pardes, E.; Naveh, L.; Adam, Z. The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 2007, 19, 1039–1047. [Google Scholar] [CrossRef] [Green Version]
- Anbudurai, P.R.; Mor, T.S.; Ohad, I.; Shestakov, S.V.; Pakrasi, H.B. The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. USA 1994, 91, 8082–8086. [Google Scholar] [CrossRef] [Green Version]
- Yin, S.M.; Sun, X.W.; Zhang, L.X. An Arabidopsis ctpA homologue is involved in the repair of photosystem II under high light. Chin. Sci. Bull. 2008, 53, 1021–1026. [Google Scholar] [CrossRef] [Green Version]
- Oelmüller, R.; Herrmann, R.G.; Pakrasi, H.B. Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J. Biol. Chem. 1996, 271, 21848–21852. [Google Scholar] [CrossRef] [Green Version]
- Järvi, S.; Gollan, P.J.; Aro, E.M. Understanding the roles of the thylakoid lumen in photosynthesis regulation. Front. Plant Sci. 2013, 4, 434. [Google Scholar] [CrossRef] [Green Version]
- Kieselbach, T.; Hagman, Å.; Andersson, B.; Schröder, W.P. The Thylakoid Lumen of Chloroplasts. J. Biol. Chem. 1998, 273, 6710–6716. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Petersson, U.A.; Haas, B.J.; Funk, C.; Schröder, W.P.; Kieselbach, T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 8354–8365. [Google Scholar] [CrossRef] [Green Version]
- Peltier, J.; Emanuelsson, O.; Kalume, D.E.; Ytterberg, J.; Friso, G.; Rudella, A.; Liberles, D.A.; Söderberg, L.; Roepstorff, P.; Heijne, G.V.; et al. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 2002, 14, 211–236. [Google Scholar] [CrossRef] [Green Version]
- Kieselbach, T.; Schröder, W.P. The proteome of the chloroplast lumen of higher plants. Photosynth. Res. 2003, 78, 249–264. [Google Scholar] [CrossRef]
- Friso, G.; Giacomelli, L.; Ytterberg, A.J.; Peltier, J.B.; Rudella, A.; Sun, Q.; Wijk, K.J. In-Depth Analysis of the Thylakoid Membrane Proteome of Arabidopsis thaliana. Plant Cell 2004, 16, 478–499. [Google Scholar] [CrossRef] [Green Version]
- Goulas, E.; Schubert, M.; Kieselbach, T.; Kleczkowski, L.A.; Gardeström, P.; Schröder, W.P.; Hurry, V. The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J. 2006, 47, 720–734. [Google Scholar] [CrossRef]
- Zybailov, B.; Rutschow, H.; Friso, G.; Rudella, A.; Emanuelsson, O.; Sun, Q.; van Wijk, K.J. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 2008, 3, e1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granlund, I.; Hall, M.; Kieselbach, T.; Schröder, W.P. Light induced changes in protein expression and uniform regulation of transcription in the thylakoid lumen of Arabidopsis thaliana. PLoS ONE 2009, 4, e5649. [Google Scholar] [CrossRef]
- Hall, M.; Mata-Cabana, A.; Åkerlund, H.E.; Florencio, F.J.; Schröder, W.P.; Lindahl, M.; Kieselbach, T. Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 2010, 10, 987–1001. [Google Scholar] [CrossRef] [PubMed]
- Cline, K. Mechanistic aspects of folded protein transport by the twin arginine translocase (Tat). J. Biol. Chem. 2015, 290, 16530–16538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef] [Green Version]
- Miyao, M.; Murata, N. Partial disintegration and reconstitution of the photosynthetic oxygen evolution system. Binding of 24 kilodalton and 18 kilodalton polypeptides. BBA Bioenerg. 1983, 725, 87–93. [Google Scholar] [CrossRef]
- Bricker, T.M. Oxygen Evolution in the Absence of the 33-Kilodalton Manganese-Stabilizing Protein. Biochemistry 1992, 31, 4623–4628. [Google Scholar] [CrossRef]
- Hall, M.; Mishra, Y.; Schröder, W.P. Preparation of stroma, thylakoid membrane, and lumen fractions from Arabidopsis thaliana chloroplasts for proteomic analysis. Methods Mol. Biol. 2011, 775, 207–222. [Google Scholar] [CrossRef]
- Rahikainen, M.; Trotta, A.; Alegre, S.; Pascual, J.; Vuorinen, K.; Overmyer, K.; Moffatt, B.; Ravanel, S.; Glawischnig, E.; Kangasjärvi, S. PP2A-B′γ modulates foliar trans-methylation capacity and the formation of 4-methoxy-indol-3-yl-methyl glucosinolate in Arabidopsis leaves. Plant J. 2017, 89, 112–127. [Google Scholar] [CrossRef] [Green Version]
- Ifuku, K.; Ishihara, S.; Shimamoto, R.; Ido, K.; Sato, F. Structure, function, and evolution of the PsbP protein family in higher plants. Photosynth. Res. 2008, 98, 427–437. [Google Scholar] [CrossRef]
- Sato, N. Phylogenomic and structural modeling analyses of the PsbP superfamily reveal multiple small segment additions in the evolution of photosystem II-associated PsbP protein in green plants. Mol. Phylogenet. Evol. 2010, 56, 176–186. [Google Scholar] [CrossRef]
- Yabuta, S.; Ifuku, K.; Takabayashi, A.; Ishihara, S.; Ido, K.; Ishikawa, N.; Endo, T.; Sato, F. Three PsbQ-like proteins are required for the function of the chloroplast NAD(P)H dehydrogenase complex in arabidopsis. Plant Cell Physiol. 2010, 51, 866–876. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Li, L.; Luan, S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in arabidopsis. Plant Physiol. 2004, 134, 1248–1267. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, P.F.; Glaser, E. Processing peptidases in mitochondria and chloroplasts. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 360–370. [Google Scholar] [CrossRef] [Green Version]
- Roitinger, E.; Hofer, M.; Köcher, T.; Pichler, P.; Novatchkova, M.; Yang, J.; Schlögelhofer, P.; Mechtler, K. Quantitative phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and ataxia telangiectasia-mutated and Rad3-related (ATR) dependent DNA damage response in Arabidopsis thaliana. Mol. Cell. Proteom. 2015, 14, 556–571. [Google Scholar] [CrossRef] [Green Version]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef]
- Heazlewood, J.I.; Durek, P.; Hummel, J.; Selbig, J.; Weckwerth, W.; Walther, D.; Schulze, W.X. PhosPhAt: A database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008, 36, D1015–D1021. [Google Scholar] [CrossRef]
- Hong, Y.; Wang, Z.; Liu, X.; Yao, J.; Kong, X.; Shi, H.; Zhu, J.-K. Two Chloroplast Proteins Negatively Regulate Plant Drought Resistance Through Separate Pathways. Plant Physiol. 2020, 182, 1007–1021. [Google Scholar] [CrossRef]
- Aryal, U.K.; Krochko, J.E.; Ross, A.R.S. Identification of Phosphoproteins in Arabidopsis thaliana Leaves Using Polyethylene Glycol Fractionation, Immobilized Metal-ion Affinity Chromatography, Two-Dimensional Gel Electrophoresis and Mass Spectrometry. J. Proteome Res. 2011, 11, 425–437. [Google Scholar] [CrossRef]
- Hashimoto, A.; Yamamoto, Y.; Theg, S.M. Unassembled subunits of the photosynthetic oxygen-evolving complex present in the thylakoid lumen are long-lived and assembly-competent. FEBS Lett. 1996, 391, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Haldrup, A.; Naver, H.; Scheller, H.V. The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J. 1999, 17, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Levesque-Tremblay, G.; Havaux, M.; Ouellet, F. The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J. 2009, 60, 691–702. [Google Scholar] [CrossRef]
- Malnoë, A.; Schultink, A.; Shahrasbi, S.; Rumeau, D.; Havaux, M.; Niyogi, K.K. The Plastid Lipocalin LCNP Is Required for Sustained Photoprotective Energy Dissipation in Arabidopsis. Plant Cell 2018, 30, 196–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahns, P.; Holzwarth, A.R. The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 182–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aro, E.M.; Virgin, I.; Andersson, B. Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. BBA Bioenerg. 1993, 1143, 113–134. [Google Scholar] [CrossRef]
- Meurer, J.; Plücken, H.; Kowallik, K.V.; Westhoff, P. A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 1998, 17, 5286–5297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komenda, J.; Nickelsen, J.; Tichý, M.; Prášil, O.; Eichacker, L.A.; Nixon, P.J. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 2008, 283, 22390–22399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, A.; He, Z.; Hye, S.C.; Lima, A.; Buchanan, B.B.; Luan, S. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 15947–15952. [Google Scholar] [CrossRef] [Green Version]
- Sirpiö, S.; Khrouchtchova, A.; Allahverdiyeva, Y.; Hansson, M.; Fristedt, R.; Vener, A.V.; Scheller, H.V.; Jensen, P.E.; Haldrup, A.; Aro, E.M. AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 2008, 55, 639–651. [Google Scholar] [CrossRef]
- Ishihara, S.; Takabayashi, A.; Ido, K.; Endo, T.; Ifuku, K.; Sato, F. Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 2007, 145, 668–679. [Google Scholar] [CrossRef] [Green Version]
- Butenko, Y.; Lin, A.; Naveh, L.; Kupervaser, M.; Levin, Y.; Reich, Z.; Adam, Z. Differential Roles of the Thylakoid Lumenal Deg Protease Homologs in Chloroplast Proteostasis. Plant Physiol. 2018, 178, 1065–1080. [Google Scholar] [CrossRef] [Green Version]
- Satoh, K.; Yamamoto, Y. The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosynth. Res. 2007, 94, 203–215. [Google Scholar] [CrossRef]
- Granlund, I.; Storm, P.; Schubert, M.; García-Cerádn, J.G.; Funk, C.; Schröder, W.P. The TL29 protein is lumen located, associated with psii and not an ascorbate peroxidase. Plant Cell Physiol. 2009, 50, 1898–1910. [Google Scholar] [CrossRef] [Green Version]
- Lazzarotto, F.; Menguer, P.K.; Del-Bem, L.-E.; Zámocký, M.; Margis-Pinheiro, M. Ascorbate Peroxidase Neofunctionalization at the Origin of APX-R and APX-L: Evidence from Basal Archaeplastida. Antioxidants 2021, 10, 597. [Google Scholar] [CrossRef]
- Bricker, T.M.; Roose, J.L.; Zhang, P.; Frankel, L.K. The PsbP family of proteins. Photosynth. Res. 2013, 116, 235–250. [Google Scholar] [CrossRef]
- Ifuku, K. The PsbP and PsbQ family proteins in the photosynthetic machinery of chloroplasts. Plant Physiol. Biochem. 2014, 81, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yang, H.; Lu, Q.; Wen, X.; Chen, F.; Peng, L.; Zhang, L.; Lu, C. PSBP-DOMAIN PROTEIN1, a Nuclear-Encoded Thylakoid Lumenal Protein, Is Essential for Photosystem I Assembly in Arabidopsis. Plant Cell 2012, 24, 4992–5006. [Google Scholar] [CrossRef] [Green Version]
- Gollan, P.J.; Bhave, M.; Aro, E.M. The FKBP families of higher plants: Exploring the structures and functions of protein interaction specialists. FEBS Lett. 2012, 586, 3539–3547. [Google Scholar] [CrossRef] [Green Version]
- Vasudevan, D.; Gopalan, G.; Kumar, A.; Garcia, V.J.; Luan, S.; Swaminathan, K. Plant immunophilins: A review of their structure-function relationship. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 2145–2158. [Google Scholar] [CrossRef] [Green Version]
- Lima, A.; Lima, S.; Wong, J.H.; Phillips, R.S.; Buchanan, B.B.; Luan, S. A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2006, 103, 12631–12636. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.; Yamamoto, H.; Shikanai, T. Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 945–953. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Li, Q.P.; Wang, J.; Yan, Y.; Zhang, G.L.; Zhang, H.; Wu, J.; Chen, F.; Wang, X.; Kang, Z.; et al. YR36/WKS1-Mediated Phosphorylation of PsbO, an Extrinsic Member of Photosystem II, Inhibits Photosynthesis and Confers Stripe Rust Resistance in Wheat. Mol. Plant 2019, 12, 1639–1650. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Ni, S.; Kennedy, M.A. NMR Analysis of Amide Hydrogen Exchange Rates in a Pentapeptide-Repeat Protein from A. thaliana. Biophys. J. 2017, 112, 2075–2088. [Google Scholar] [CrossRef] [Green Version]
- Spetea, C.; Lundin, B. Evidence for nucleotide-dependent processes in the thylakoid lumen of plant chloroplasts—An update. FEBS Lett. 2012, 586, 2946–2954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.Y.; Liu, M.S.; Lin, T.P.; Cheng, Y.S. Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen. Plant Physiol. 2011, 157, 1015–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bienvenut, W.V.; Brünje, A.; Boyer, J.; Mühlenbeck, J.S.; Bernal, G.; Lassowskat, I.; Dian, C.; Linster, E.; Dinh, T.V.; Koskela, M.M.; et al. Dual lysine and N-terminal acetyltransferases reveal the complexity underpinning protein acetylation. Mol. Syst. Biol. 2020, 16, e9464. [Google Scholar] [CrossRef] [PubMed]
- Markwell, M.A.K.; Haas, S.M.; Bieber, L.L.; Tolbert, N.E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978, 87, 206–210. [Google Scholar] [CrossRef]
- Blum, H.; Beier, H.; Gross, H.J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 1987, 8, 93–99. [Google Scholar] [CrossRef]
- Trotta, A.; Bajwa, A.A.; Mancini, I.; Paakkarinen, V.; Pribil, M.; Aro, E.M. The role of phosphorylation dynamics of curvature thylakoid 1B in plant thylakoid membranes. Plant Physiol. 2019, 181, 1615–1631. [Google Scholar] [CrossRef] [Green Version]
- Taus, T.; Köcher, T.; Pichler, P.; Paschke, C.; Schmidt, A.; Henrich, C.; Mechtler, K. Universal and Confident Phosphorylation Site Localization Using phosphoRS. J. Proteome Res. 2011, 10, 5354–5362. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]


| Accession | Name * | Description | Pred. MW (Mature) | Ave. PSMs FL ** | Std. Dev. PSMs FL | Ave. PSMs MAL ** | Std. Dev. PSMs MAL | N Terminus *** | Pub. N Terminus **** | Proteomic Localis. Study |
|---|---|---|---|---|---|---|---|---|---|---|
| AT5G23120.1 | HCF136 (YCF48) | Photosystem II stability/assembly factor | 35.9 | 671.25 | 46.00 | 150.00 | 48.51 | 79 | 79 | [16,17] |
| AT4G09010.1 | APX4 (TLP29) | Non-functional ascorbate peroxidase | 29.3 | 411.50 | 56.13 | 231.00 | 31.46 | NF | 83 | [16,17] |
| AT5G66570.1 | PsbO1 | Photosystem II subunit O-1 | 26.6 | 280.00 | 27.54 | 342.75 | 51.32 | NF | 86 | [16,17] |
| AT3G50820.1 | PsbO2 | Photosystem II subunit O-2 | 26.6 | 271.00 | 35.97 | 341.75 | 58.53 | NF | 85 | [16,17] |
| AT1G06680.1 | PsbP1 | Photosystem II subunit P-1 | 21.0 | 294.75 | 171.22 | 406.50 | 58.16 | (70) 78 | 78 | [16,17] |
| AT4G21280.1 | PsbQ1 | Photosystem II subunit Q-1 | 17.0 | 42.00 | 4.55 | 70.00 | 7.26 | NF | 76 | [16,17] |
| AT4G05180.1 | PsbQ2 | Photosystem II subunit Q-2 | 16.4 | 52.75 | 6.29 | 41.50 | 4.43 | 83 (N-ac), 88 | 83 | [17] |
| AT3G27925.1 | DegP1 | DegP protease 1 | 35.2 | 215.50 | 20.14 | 34.25 | 10.11 | 106 | 106 | [16,17] |
| AT4G18370.1 | DegP5 | DegP protease 5 | 27.1 | 27.00 | 2.16 | 3.75 | 1.50 | 74 | 74 | [16,17] |
| AT5G39830.1 | DegP8 | Trypsin family protein with PDZ domain | 37.5 | 111.50 | 19.96 | 10.25 | 4.03 | 91 | 91 | [16] |
| AT4G17740.1 | D1 protease | Peptidase S41 family protein | 41.9 | 60.50 | 10.54 | 13.00 | 4.97 | 127 | 127 | [16] |
| AT5G46390.2 | CTPA1 (D1 protease-like) | Peptidase S41 family protein | 46.0 | 49.25 | 5.32 | 9.00 | 3.46 | NF | 68 | [16] |
| AT3G57680.1 | CTPA3 | Peptidase S41 family protein | 43.9 | 0.25 ** | NA | NF ** | NA | NF | none | current study, below cut-off |
| AT1G20340.1 | PC major | Plastocyanin major form | 10.5 | 16.75 | 4.99 | 4.25 | 2.22 | 69 | 69 | [16,17] |
| AT1G76100.1 | PC minor | Plastocyanin minor form | 10.5 | 4.50 | 1.00 | 1.75 | 0.50 | 73 | 73 | [16,17] |
| AT1G08550.1 | VDE | Violaxanthin deepoxidase | 39.8 | 134.50 | 30.96 | 20.00 | 6.38 | NF | 114 | [16] |
| AT3G47860.1 | LCNP (CHL) | Chloroplastic lipocalin | 28.4 | 8.50 | 1.29 | 21.00 | 8.91 | K_101 | none | current study |
| AT5G64040.1 | PsaN | Photosystem I reaction centre subunit PSI-N | 10.6 | 12.75 | 4.27 | 39.50 | 15.84 | 87 | 85/87 (N-ac) | [17] |
| AT3G55330.1 | PPL1 | Photosystem II reaction centre PsbP family protein | 17.8 | 111.25 | 24.90 | 39.50 | 14.75 | NF | 75 | [16,17] |
| AT2G39470.1 | PPL2 | Photosystem II reaction centre PsbP family protein | 18.7 | 0.50 ** | NA | 0.25 ** | NA | NF | 87 | [17] |
| AT4G15510.1 | PPD1 | Photosystem II reaction centre PsbP family protein | 21.3 | 109.25 | 14.86 | 12.00 | 6.48 | NF | 105 | [16,17] |
| AT2G28605.1 | PPD2 | Photosystem II reaction centre PsbP family protein | 14.7 | 22.75 | 1.89 | 4.50 | 3.11 | R_78 | 99 | |
| AT1G76450.1 | PPD3 | Photosystem II reaction centre PsbP family protein | 18.8 | 41.75 | 34.38 | 11.50 | 2.38 | 81 | 81 | [16,17] |
| AT1G77090.1 | PPD4 | Photosystem II reaction centre PsbP family protein | 20.8 | 28.50 | 3.70 | 11.75 | 2.06 | NF | 72 | [16,17] |
| AT5G11450.1 | PPD5 | Photosystem II reaction centre PsbP family protein | 24.9 | 76.75 | 17.46 | 23.00 | 8.29 | 80 | 80 | [16] |
| AT3G56650.1 | PPD6 | Photosystem II reaction centre PsbP family protein | 21.6 | 113.25 | 35.17 | 21.50 | 8.39 | (61) 68 | 68 | [16,17] |
| AT3G05410.2 | PPD7 | Photosystem II reaction centre PsbP family protein | 23.2 | 3.00 | 1.83 | 2.00 | 0.82 | K_87 | none | current study |
| AT5G27390.1 | PPD8 | Photosystem II reaction centre PsbP family protein | 17.5 | 27.50 | 5.26 | 8.25 | 2.22 | R_70 | none | [22] |
| AT3G63525.1 | PPD9 (TL19) | Photosystem II reaction centre PsbP family protein | 20.8 | 76.00 | 15.64 | 13.25 | 4.57 | 43 | none | [16], current study |
| AT1G14150.1 | PQL1 | photosystem II subunit Q-like | 17.4 | 2.25 | 1.50 | 1.75 | 0.96 | K_79 | 79 | [19] |
| AT3G01440.1 | PQL2 | photosystem II subunit Q-like | 13.8 | 2.25 | 1.50 | 1.75 | 0.96 | 75 | 126 | [19] |
| AT2G01918.1 | PQL3 | photosystem II subunit Q-like | 14.1 | 1.50 | 1.00 | 0.75 ** | NA | 67 | none | current study |
| AT3G01480.1 | CYP38 | Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein | 38.3 | 330.50 | 69.95 | 126.00 | 13.29 | 93 (N-ac) | 93 | [16,17] |
| AT3G15520.1 | CYP37 | Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein | 37.9 | 132.75 | 38.59 | 53.00 | 8.12 | 115 | 115 | [16] |
| AT5G35100.1 | CYP28 | Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein | 28.4 | 92.50 | 11.00 | 18.25 | 3.59 | 20, 21, 24, 25, 30, 60 | 25 | [16] |
| AT1G74070.1 | CYP26-2 | Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein | 26.2 | 37.00 | 9.56 | 31.25 | 4.27 | 75 | none | current study |
| AT5G13120.1 | CYP20-2 | Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein | 20.9 | 83.75 | 31.19 | 33.00 | 3.16 | 73, 74 (±N-ac), 77 | 77 | [16,17] |
| AT5G45680.1 | FKBP13 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 13.6 | 17.75 | 2.99 | 4.75 | 3.59 | 80 | 80 | [16] |
| AT4G26555.1 | FKBP16-1 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 15.5 | 16.00 | 5.35 | 2.25 | 0.96 | 68 | none | [20] |
| AT4G39710.1 | FKBP16-2 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 15.5 | 9.00 | 2.71 | 1.33 | 0.58 | 76 | 94 | [17] |
| AT2G43560.1 | FKBP16-3 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 15.7 | 25.00 | 2.94 | 13.00 | 4.97 | 77 | 77 | [16,17] |
| AT3G10060.1 | FKBP16-4 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 14.4 | 16.75 | 3.86 | 17.75 | 2.99 | R_92 | 95 | [17] |
| AT4G19830.1 | FKBP17-1 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 16.7 | 3.25 | 0.50 | 0.25 | 0.50 | 79 | none | current study |
| AT1G18170.1 | FKBP17-2 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 16.9 | 3.25 | 0.50 | 10.25 | 3.30 | 29 (±N-ac), 30, 61, 90 | 61 | [21] |
| AT1G20810.1 | FKBP18 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 17.8 | 42.00 | 5.77 | 14.00 | 1.83 | NF | 72 | [16] |
| AT5G13410.1 | FKBP19 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 18.8 | 44.25 | 9.43 | 16.50 | 2.38 | 89 | 89 | [16,17] |
| AT3G60370.1 | FKBP20-2 | FKBP-like peptidyl-prolyl cis-trans isomerase family protein | 19.9 | 28.75 | 9.74 | 17.50 | 3.87 | NF | 68 | [16] |
| AT2G44920.2 | TL15 PRP | Pentatricopeptide repeat superfamily protein | 15.0 | 53.25 | 13.33 | 25.03 | 7.50 | 82 | 82 | [19] |
| AT5G53490.3 | TL17 PRP | Pentatricopeptide repeat superfamily protein | 18.8 | 215.00 | 26.37 | 22.75 | 12.89 | NF | 92 | [16,17] |
| AT1G12250.1 | TL20.3 PRP | Pentatricopeptide repeat superfamily protein | 20.4 | 9.25 | 3.20 | 14.00 | 3.83 | 91 | 101 | [19,21] |
| AT3G26060.1 | PrxQ | Thioredoxin superfamily protein | 16.7 | 45.00 | 3.92 | 20.50 | 4.51 | 68 | 68 | [23] |
| AT5G52970.1 | TLP15 | Thylakoid lumen 15.0 kDa protein | 16.5 | 29.75 | 6.65 | 3.50 | 2.65 | 76 | 76 | [16,17] |
| AT4G02530.1 | MPH2 (TLP16) | Thylakoid lumenal 16 kDa protein | 15.7 | 14.50 | 4.89 | 16.33 | 18.35 | 74 | 74 | [16,17] |
| AT4G24930.1 | TLP17.9 | Thylakoid lumenal 17.9 kDa protein | 18.0 | 27.25 | 4.19 | 6.25 | 2.63 | 64 | 64 | [16,17] |
| AT1G54780.1 | TLP18.3 | Thylakoid lumen 18.3 kDa protein, putative phosphatase | 22.2 | 12.00 | 4.08 | 5.00 | 2.16 | 85 | 85 | [16,17] |
| AT5G42765.1 | AT5G42765 | Uncharacterised protein | 18.3 | 21.25 | 3.59 | 12.50 | 1.00 | 65, 66, 67, 74 | 86 | [21] |
| AT2G26340.1 | AT2G26340 | Uncharacterised protein | 20.4 | 11.25 | 3.40 | 11.25 | 5.32 | 81 | none | current study |
| AT1G03600.1 | Psb27-1 | Photosystem II repair protein | 13.6 | 10.75 | 9.18 | 6.50 | 3.79 | 69 | 69 | [17] |
| AT1G05385.1 | Psb27-2 (LPA19) | Photosystem II D1 precursor processing protein | 14.9 | 2.25 | 0.96 | NF ** | NA | 68 | 150 | [21] |
| AT1G51400.1 | PsbTn2 | Photosystem II 5 kD protein | 11.4 | 0.25 ** | NA | NF ** | NA | NF | none | current study, below cut-off |
| AT5G45040.1 | Cyt c6a | Cytochrome c | 3.75 | 2.06 | 0.50 | 1.00 | 71 | none | current study | |
| AT2G23670.1 | AT2G23670 | YCF37-like protein | 10.0 | 3.50 | 0.58 | 2.75 | 0.96 | 72 | 72 | [21] |
| AT2G34860.1 | PSA2 PDI | Photosystem I assembly 2, DnaJ-like protein | 11.9 | 2.75 | 0.50 | 0.75 ** | NA | R_104 | none | [19] |
| AT2G36145.1 | AT2G36145 | Expressed protein, predicted lumenal | 12.3 | 0.50 ** | NA | 1.50 | 1.00 | K_62 | 75 | [21] |
| AT3G03630.1 | CS26 | Cysteine synthase 26 | 34.3 | 0.25 ** | NA | NF ** | NA | R_118 | none | current study, below cut-off |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gollan, P.J.; Trotta, A.; Bajwa, A.A.; Mancini, I.; Aro, E.-M. Characterization of the Free and Membrane-Associated Fractions of the Thylakoid Lumen Proteome in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 8126. https://doi.org/10.3390/ijms22158126
Gollan PJ, Trotta A, Bajwa AA, Mancini I, Aro E-M. Characterization of the Free and Membrane-Associated Fractions of the Thylakoid Lumen Proteome in Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(15):8126. https://doi.org/10.3390/ijms22158126
Chicago/Turabian StyleGollan, Peter J., Andrea Trotta, Azfar A. Bajwa, Ilaria Mancini, and Eva-Mari Aro. 2021. "Characterization of the Free and Membrane-Associated Fractions of the Thylakoid Lumen Proteome in Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 15: 8126. https://doi.org/10.3390/ijms22158126
APA StyleGollan, P. J., Trotta, A., Bajwa, A. A., Mancini, I., & Aro, E.-M. (2021). Characterization of the Free and Membrane-Associated Fractions of the Thylakoid Lumen Proteome in Arabidopsis thaliana. International Journal of Molecular Sciences, 22(15), 8126. https://doi.org/10.3390/ijms22158126






