LPEATs Tailor Plant Phospholipid Composition through Adjusting Substrate Preferences to Temperature
Abstract
:1. Introduction
2. Results
2.1. Temperature Alters Acyl-CoA Specificity of LPEAT Enzymes Present in Vegetative Tissues
2.2. Evolutionary Analyses Reveal Ancestral Origins of Camelina sativa LPEATs
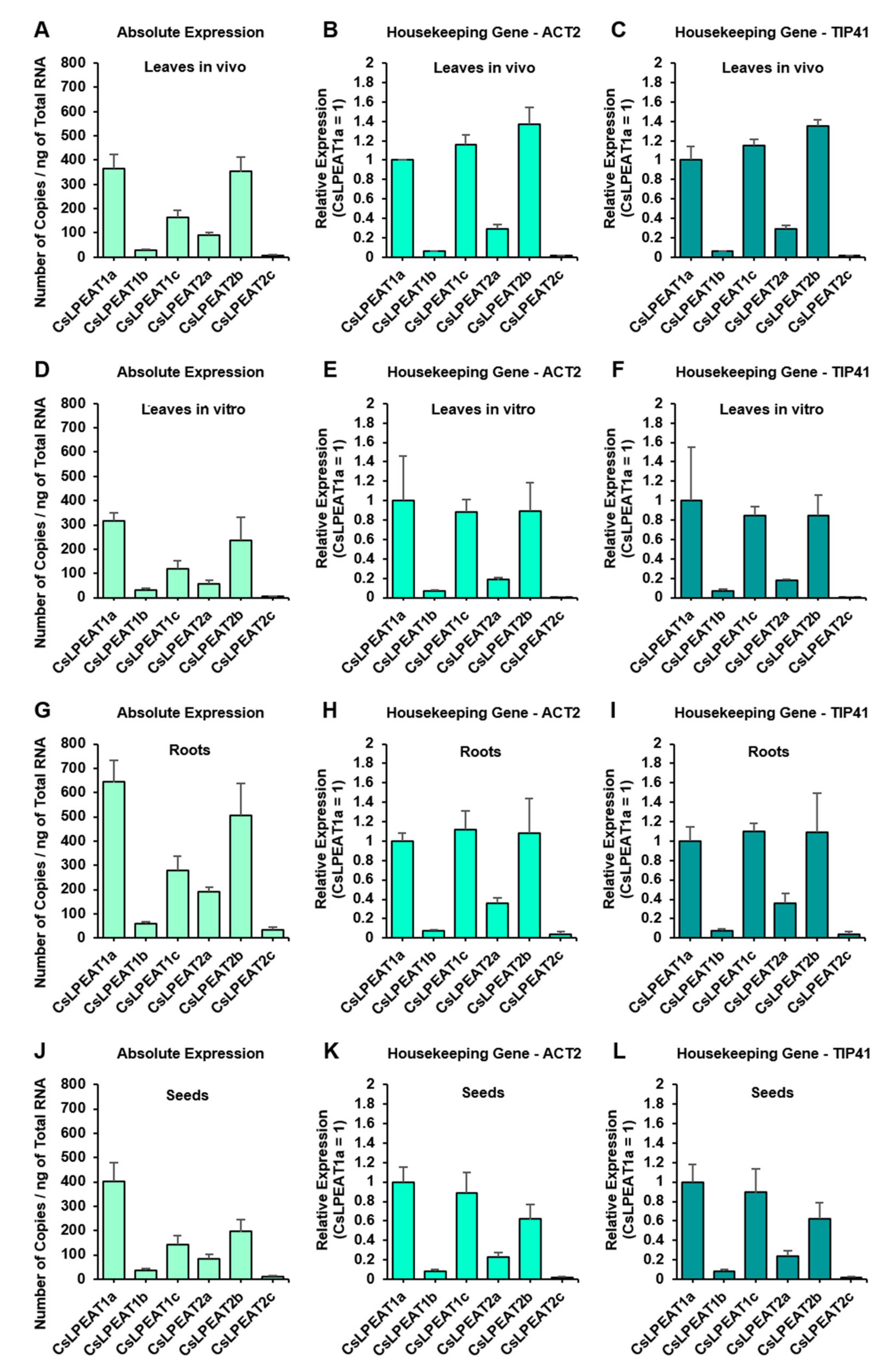
2.3. CsLPEAT1 and CsLPEAT2 Genes Expression Differs between Plant Tissues
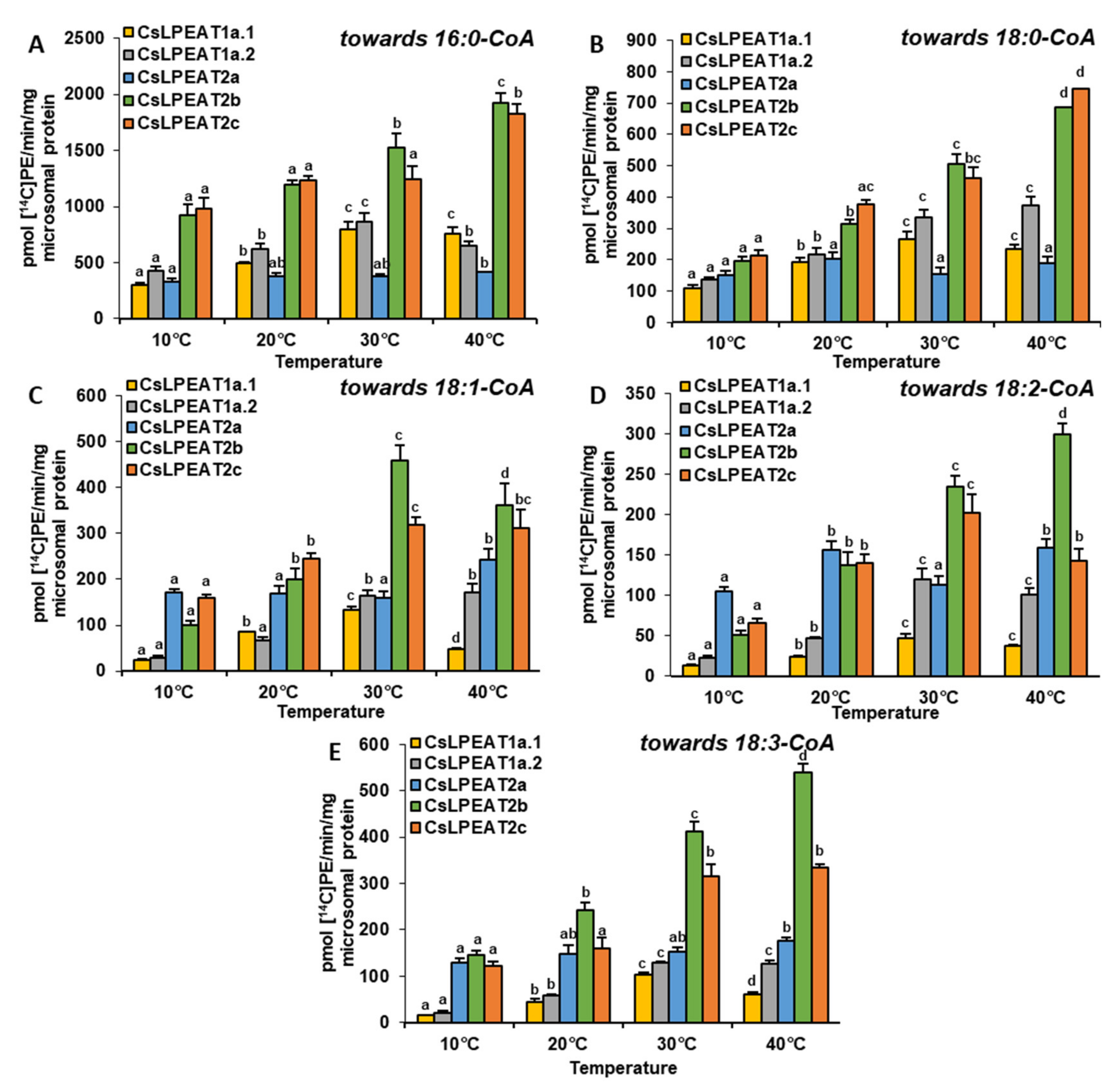
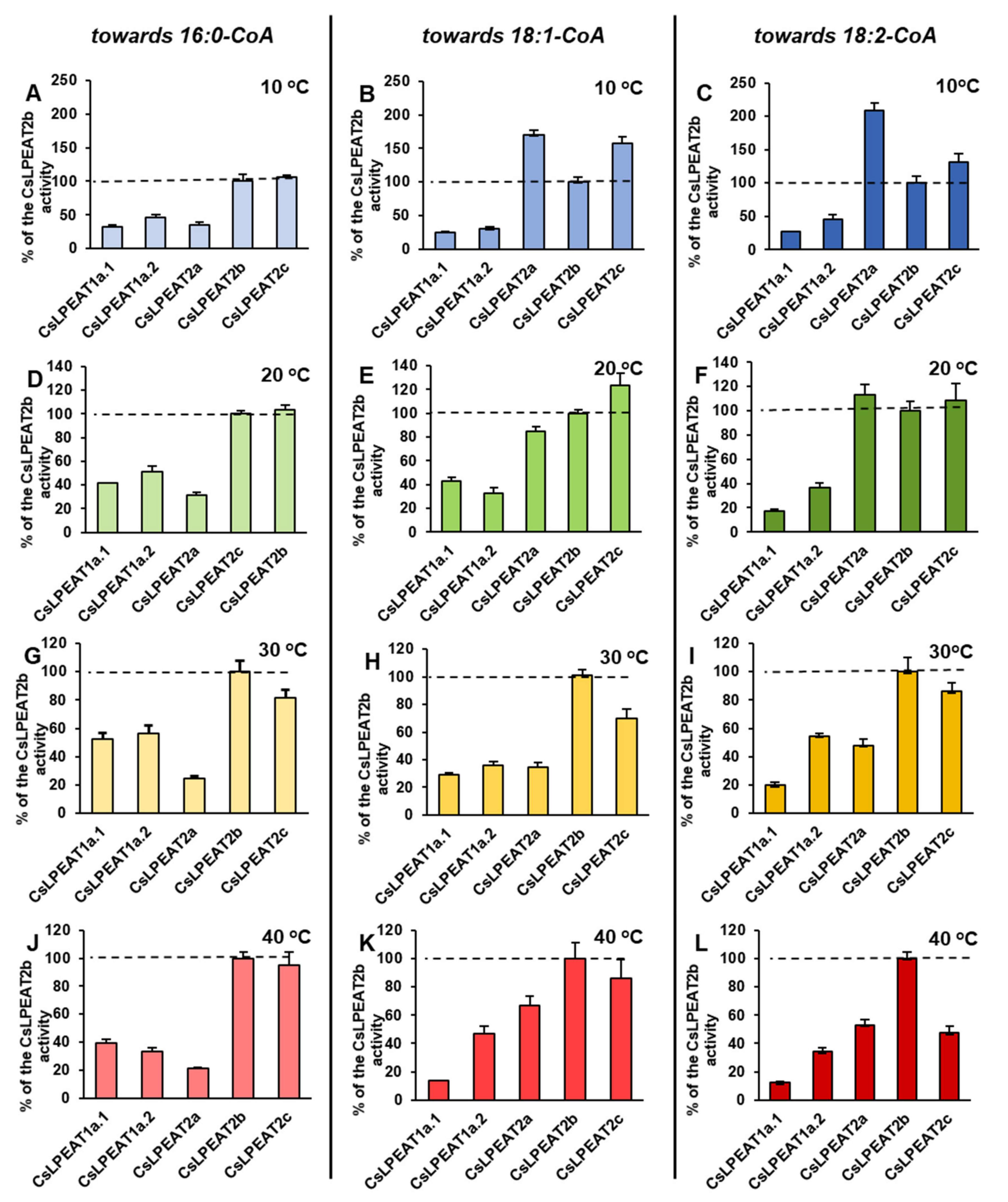
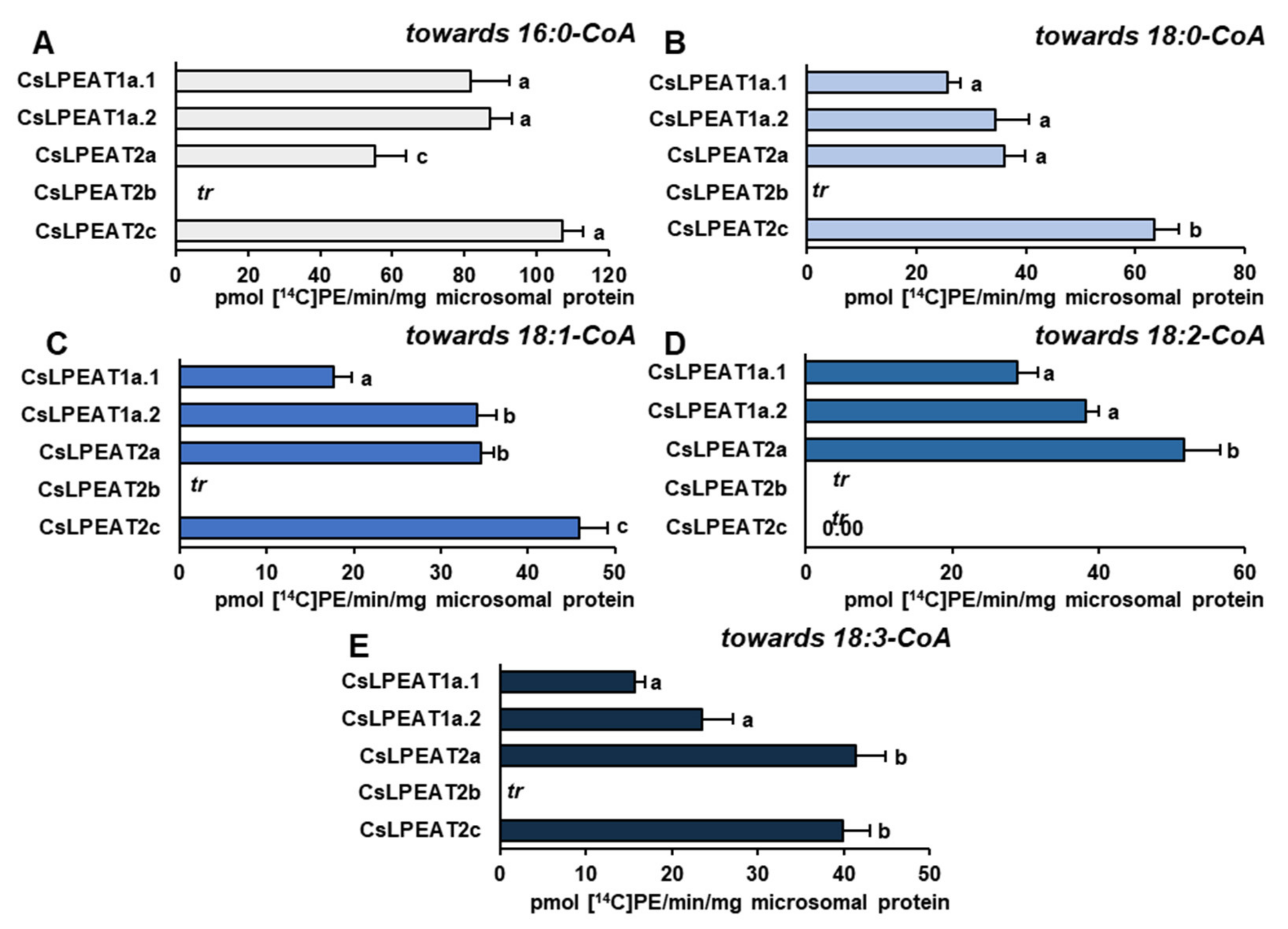
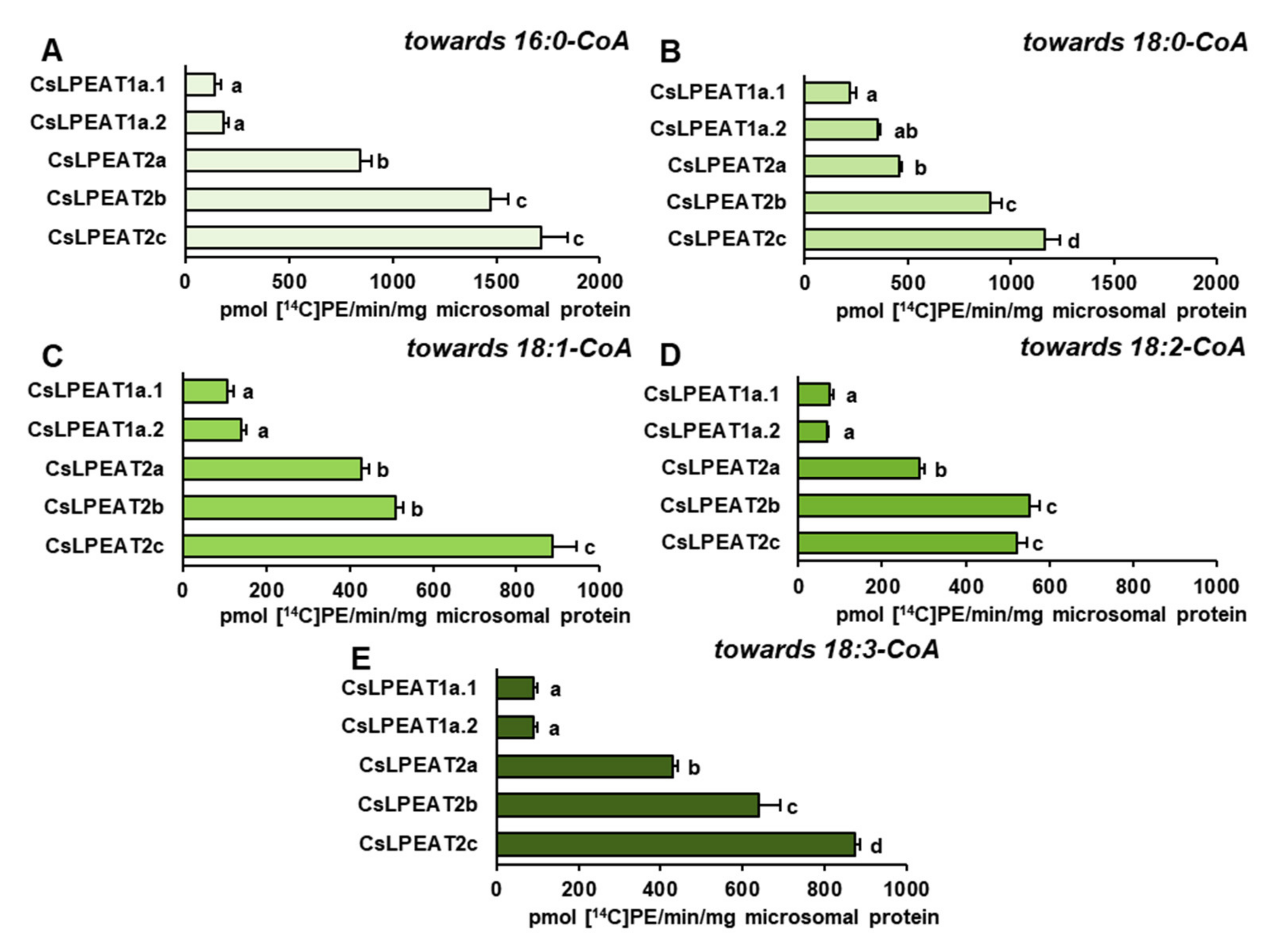
2.4. Diverse Acyl-CoA Utilization by CsLPEAT Enzymes at Various Temperatures
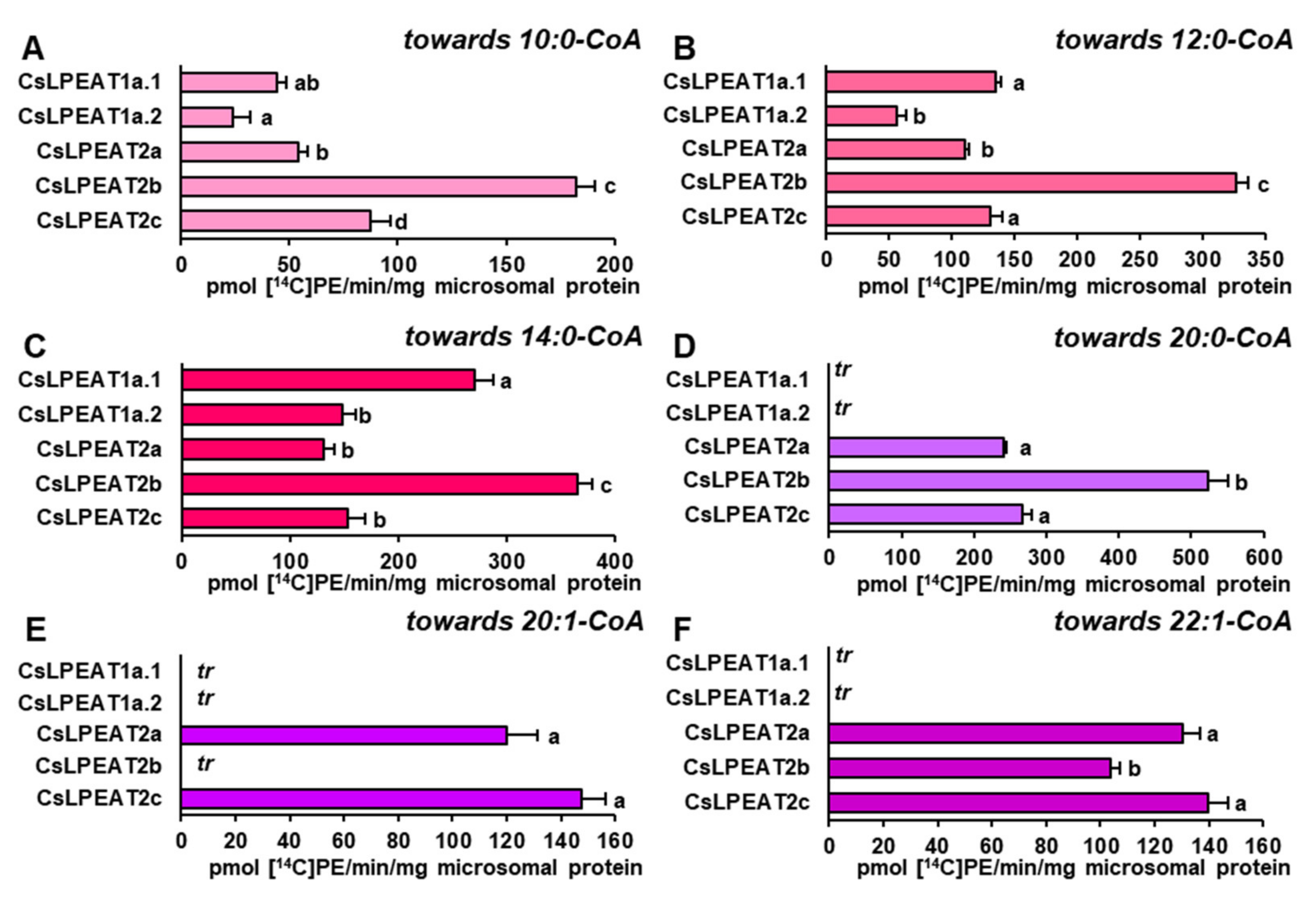
2.5. CsLPEAT2 Isoforms Are Specific towards Saturated and Very-Long-Chain Fatty Acids
2.6. Distinct Ability or Inability to Utilize Various Acyl Acceptors by CsLPEAT Enzymes
3. Discussion
3.1. Acyl-CoA: Lysophosphatidyethnolamine Acytransferases Present in Vegetative and Generative Organs of C. sativa Respond Directly to Temperature Changes and Shape Phospholipids’ Composition
3.2. The Origin of the CsLPEAT Isoforms, and Their Diverse Expression Pattern in C. sativa Organs
3.3. Biochemical CharacteristiCs and Temperature Susceptibility of CsLPEAT Enzymes
4. Materials and Methods
4.1. Chemicals
4.2. Plant Material and Growth Condition
4.3. Gene Cloning and Sequence Analysis
4.4. Vector Construction and Yeast Transformation
4.5. Cleavage of Phosphatidylethanolamine by Phospholipase A2
4.6. Plant Microsomal Fraction Isolation and Enzyme Assay
4.7. Yeast Microsomal Preparation and Assay of CsLPEAT Isoforms Variant
4.8. Expression Analysis
4.9. Sequence Comparison and Evolutionary Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ruelland, E.; Vaultier, M.N.; Zachowski, A.; Hurry, V. Cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar]
- Kates, M.; Pugh, E.L.; Ferrante, G. Regulation of membrane fluidity by lipid desaturases. Biomembranes 1984, 12, 379–395. [Google Scholar]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S. Effects of high temperature stress and traits associated with tolerance in wheat. Open Access J. Sci. 2018, 2, 177–186. [Google Scholar] [CrossRef]
- Mason, J.T. Investigation of phase transitions in bilayer membranes. Methods Enzymol. 1998, 295, 468–494. [Google Scholar] [PubMed]
- De Kroon, A.I.P.M. Metabolism of phosphatidylcholine and its implications for lipid acyl chain composition in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2007, 1771, 343–352. [Google Scholar] [CrossRef] [Green Version]
- Christie, W.W.; Han, X. Lipids: Their structures and occurrence. Lipid Anal. 2012, 1, 3–19. [Google Scholar]
- De Kruijff, B. Lipid polymorphism and biomembrane function. Curr. Opin. Chem. Biol. 1997, 1, 564–569. [Google Scholar] [CrossRef]
- Birner, R.; Bürgermeister, M.; Schneiter, R.; Daum, G. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 2001, 12, 997–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momchilova, A.; Markovska, T. Phosphatidylethanolamine and phosphatidylcholine are sources of diacylglycerol in ras-transformed NIH 3T3 fibroblasts. Int. J. Biochem. Cell Biol. 1999, 31, 311–318. [Google Scholar] [CrossRef]
- Rockenfeller, P.; Koska, M.; Pietrocola, F.; Minois, N.; Knittelfelder, O.; Sica, V.; Franz, J.; Carmona-Gutierrez, D.; Kroemer, G.; Madeo, F. Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death Differ. 2015, 22, 499–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Herde, K.; Krysko, D.V. Ferroptosis: Oxidized PEs trigger death. Nat. Chem. Biol. 2017, 13, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Eto, M.; Shindou, H.; Yamamoto, S.; Tamura-Nakano, M.; Shimizu, T. Lysophosphatidylethanolamine acyltransferase 2 (LPEAT2) incorporates DHA into phospholipids and has possible functions for fatty acid-induced cell death. Biochem. Biophys. Res. Commun. 2020, 526, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.P.; Weiss, S.B. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 1956, 222, 193–214. [Google Scholar] [CrossRef]
- Nerlich, A.; von Orlow, M.; Rontein, D.; Hanson, A.D.; Dörmann, P. Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol. 2007, 144, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Stålberg, K.; Ståhl, U.; Stymne, S.; Ohlrogge, J. Characterization of two Arabidopsis thaliana acyltransferases with preference for lysophosphatidylethanolamine. BMC Plant Biol. 2009, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Ståhl, U.; Stålberg, K.; Stymne, S.; Ronne, H. A family of eukaryotic lysophospholipid acyltransferases with broad specificity. FEBS Lett. 2008, 582, 305–309. [Google Scholar] [CrossRef] [Green Version]
- Bates, P.D.; Fatihi, A.; Snapp, A.R.; Carlsson, A.S.; Browse, J.; Lu, C. Acyl Editing and Headgroup Exchange Are the Major Mechanisms That Direct Polyunsaturated Fatty Acid Flux into Triacylglycerols. Plant Physiol. 2012, 160, 1530–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lager, I.; Yilmaz, J.L.; Zhou, X.R.; Jasieniecka, K.; Kazachkov, M.; Wang, P.; Zou, J.; Weselake, R.; Smith, M.A.; Bayon, S.; et al. Plant acyl-CoA:lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J. Biol. Chem. 2013, 288, 36902–36914. [Google Scholar] [CrossRef] [Green Version]
- Jasieniecka-Gazarkiewicz, K.; Demski, K.; Lager, I.; Stymne, S.; Banaś, A. Possible role of different yeast and plant lysophospholipid:acyl-CoA acyltransferases (LPLATs) in acyl remodelling of phospholipids. Lipids 2016, 51, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Lands, W.E. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 1958, 231, 883–888. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Ståhl, U.; Lenman, M.; Banaś, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acylCoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar]
- Tasseva, G.; Richard, L.; Zachowski, A. Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 2004, 566, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Jasieniecka-Gazarkiewicz, K.; Lager, I.; Carlsson, A.S.; Gutbrod, K.; Peisker, H.; Dörmann, P.; Stymne, S.; Banaś, A. Acyl-coa:Lysophosphatidylethanolamine acyltransferase activity regulates growth of Arabidopsis. Plant Physiol. 2017, 174, 986–998. [Google Scholar] [CrossRef] [Green Version]
- Jasieniecka-Gazarkiewicz, K.; Demski, K.; Gidda, S.K.; Klińska, S.; Niedojadło, J.; Lager, I.; Carlsson, A.S.; Minina, E.A.; Mullen, R.T.; Bozhkov, P.V.; et al. Subcellular localization of acyl-CoA: Lysophosphatidylethanolamine acyltransferases (LPEATs) and the effects of knocking-out and overexpression of their genes on autophagy markers level and life span of A. thaliana. Int. J. Mol. Sci. 2021, 22, 3006. [Google Scholar] [CrossRef] [PubMed]
- Klińska, S.; Jasieniecka-Gazarkiewicz, K.; Demski, K.; Banaś, A. Editing of phosphatidic acid and phosphatidylethanolamine by acyl-CoA: Lysophospholipid acyltransferases in developing Camelina sativa seeds. Planta 2020, 10, 4. [Google Scholar] [CrossRef]
- Martinière, A.; Bassil, E.; Jublanc, E.; Alcon, C.; Reguera, M.; Sentenac, H.; Blumwald, E.; Paris, N. In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. Plant Cell 2013, 25, 4028–4043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klińska, S.; Jasieniecka-Gazarkiewicz, K.; Banaś, A. Acyl-CoA: Lysophosphatidylcholine acyltransferases (LPCATs) of Camelina sativa seeds: Biochemical properties and function. Planta 2019, 250, 1655–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandáková, T.; Pouch, M.; Brock, J.R.; Al-Shehbaz, I.A.; Lysak, M.A. Origin and Evolution of Diploid and Allopolyploid Camelina Genomes Were Accompanied by Chromosome Shattering. Plant Cell 2019, 31, 2596–2612. [Google Scholar]
- Search for Conserved Domains within a Protein or Coding Nucleotide Sequence. Available online: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 4 May 2021).
- Canvin, D.T. The effect of temperature on the oil content and fatty acid composition of oils from several oil seed crops. Can. J. Bot. 1965, 43, 63–69. [Google Scholar] [CrossRef]
- Wolf, R.B.; Cavins, J.F.; Kleiman, R.; Black, L.T. Effect of temperature on soybean seed constituents: Oil, protein, moisture, fatty acids, amino acids and sugars. J. Am. Oil Chem. Soc. 1982, 59, 230–232. [Google Scholar] [CrossRef]
- Schulte, L.R.; Ballard, T.; Samarakoon, T.; Yao, L.; Vadlani, P.; Staggenborg, S.; Rezac, M. Increased growing temperature reduces content of polyunsaturated fatty acids in four oilseed crops. Industrial Crops Prod. 2013, 51, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Welti, R.; Li, W.; Li, M.; Sang, Y.; Biesiada, H.; Zhou, H.E.; Rajashekar, C.B.; Williams, T.D.; Wang, X. Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef] [Green Version]
- Falcone, D.L.; Ogas, J.P.; Somerville, C.R. Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol. 2004, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Saidi, Y.; Peter, M.; Finka, A.; Cicekli, C.; Vigh, L.; Goloubinoff, P. Membrane lipid composition affects plant heat sensing and modulates Ca2+-dependent heat shock response. Plant Signal. Behav. 2010, 5, 1530–1533. [Google Scholar] [CrossRef] [Green Version]
- Qin, F.; Lin, L.; Jia, Y.; Li, W.; Yu, B. Quantitative Profiling of Arabidopsis Polar Glycerolipids under Two Types of Heat Stress. Plants 2020, 9, 693. [Google Scholar] [CrossRef]
- Williams, J.P.; Khan, M.U.; Mitchell, K.; Johnson, G. The Effect of Temperature on the Level and Biosynthesis of Unsaturated Fatty Acids in Diacylglycerols of Brassica napus Leaves. Plant Physiol. 1988, 87, 904–910. [Google Scholar] [CrossRef] [Green Version]
- Murakami, Y.; Tsuyama, M.; Kobayashi, Y.; Kodama, H.; Iba, K. Trienoic Fatty Acids and Plant Tolerance of High Temperature. Science 2000, 287, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Mao, X.; Zhao, K.; Ji, X.; Ji, C.; Xue, J.; Li, R. Characterisation of phospholipid: Diacylglycerol acyltransferases (PDATs) from Camelina sativa and their roles in stress responses. Biol. Open. 2017, 6, 1024–1034. [Google Scholar] [CrossRef] [Green Version]
- Stymne, S.; Stobart, A.K. The biosynthesis of triacylglycerols in microsomal preparations of developing cotyledons of sunfower (Helianthus annuus L.). Biochem. J. 1984, 220, 481–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engeseth, N.J.; Pacovskya, R.S.; Newman, T.; Ohlrogge, J.B. Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch. Biochem. Biophys. 1996, 331, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Lopez, N.; Broughton, R.; Usher, S.; Salas, J.J.; Haslam, R.P.; Napier, J.A.; Beaudoin, F. Tailoring the composition of novel wax esters in the seeds of transgenic Camelina sativa through systematic metabolic engineering. Plant Biotechnol. J. 2017, 15, 837–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagale, S.; Koh, C.; Nixon, J.; Bollina, V.; Clarke, W.E.; Tuteja, R.; Spillane, C.; Robinson, S.J.; Links, M.G.; Clarke, C.; et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014, 5, 3706. [Google Scholar] [CrossRef] [Green Version]
- Roberts, J.A.; Digby, H.R.; Kara, M.; El Ajouz, S.; Sutcliffe, M.J.; Evans, R.J. Cysteine substitution mutagenesis and the effects of methanethiosulfonate reagents at P2X2 and P2X4 receptors support a core common mode of ATP action at P2X receptors. J. Biol. Chem. 2008, 283, 20126–20136. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, H.M.; Akbari, P.; Paulose, B.; Schnell, D.; Qi, W.; Park, Y.; Pareek, A.; Dhankher, O.P. Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds. Biotechnol. Biofuels. 2016, 9, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marmon, S.; Sturtevant, D.; Herrfurth, C.; Chapman, K.; Stymne, S.; Feussner, I. Two Acyltransferases Contribute Differently to Linolenic Acid Levels in Seed Oil. Plant Physiol. 2017, 173, 2081–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdullah, H.M.; Chhikara, S.; Akbari, P.; Schnell, D.J.; Pareek, A.; Dhankher, O.P. Comparative transcriptome and metabolome analysis suggests bottlenecks that limit seed and oil yields in transgenic Camelina sativa expressing diacylglycerol acyltransferase 1 and glycerol-3-phosphate dehydrogenase. Biotechnol. Biofuels. 2018, 11, 335. [Google Scholar] [CrossRef] [Green Version]
- Lightner, J.; Wu, J.; Browse, J. A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiol. 1994, 106, 1443–1451. [Google Scholar] [CrossRef] [Green Version]
- Maatta, S.; Scheu, B.; Roth, M.R.; Tamura, P.; Li, M.; Williams, T.D.; Wang, X.; Welti, R. Levels of Arabidopsis thaliana Leaf Phosphatidic Acids, Phosphatidylserines, and Most Trienoate-Containing Polar Lipid Molecular Species Increase during the Dark Period of the Diurnal Cycle. Front. Plant Sci. 2012, 3, 49. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.R.; Callahan, D.L.; Shrestha, P.; Liu, Q.; Petrie, J.R.; Singh, S.P. Lipidomic analysis of Arabidopsis seed genetically engineered to contain DHA. Front. Plant Sci. 2014, 5, 419. [Google Scholar] [CrossRef] [Green Version]
- Lager, I.; Jeppson, S.; Gippert, A.L.; Feussner, I.; Stymne, S.; Marmon, S. Acyltransferases Regulate Oil Quality in Camelina sativa Through Both Acyl Donor and Acyl Acceptor Specificities. Front. Plant Sci. 2020, 11, 1144. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rodríguez, M.F.; Sánchez-García, A.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Characterization of the morphological changes and fatty acid profile of developing Camelina sativa seeds. Ind. Crops Prod. 2013, 50, 673–679. [Google Scholar] [CrossRef]
- Rhodes, D.; Hanson, A.D. Quaternary ammonium and tertiary sulfonium compouns in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 357–384. [Google Scholar] [CrossRef]
- Banaś, A.; Dahlqvist, A.; Ståhl, U.; Lenman, M.; Stymne, S. The involvement of phospholipid:diacylglycerol acyltransferases in triacylglycerol production. Biochem. Soc. Trans. 2000, 28, 703–705. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.; Yu, S.I.; Lee, H.; Yim, J.H.; Zhu, J.K.; Lee, B.H. Arabidopsis serine decarboxylase mutants implicate the roles of ethanolamine in plant growth and development. Int. J. Mol. Sci. 2012, 13, 3176–3188. [Google Scholar] [CrossRef] [Green Version]
- Hishikawa, D.; Shindou, H.; Kobayashi, S.; Nakanishi, H.; Taguchi, R.; Shimizu, T. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA 2008, 105, 2830–2835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oo, K.C.; Huang, A.H. Lysophosphatidate acyltransferase activities in the microsomes from palm endosperm, maize scutellum, and rapeseed cotyledon of maturing seeds. Plant Physiol. 1989, 91, 1288–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, M.; David, G.N.; David, N.B. The relationship between palmitoyl-coenzyme A synthetase activity and esterification of sn-glycerol 3-phosphate in rat liver mitochondria. Biochem. J. 1973, 132, 697–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary GenetiCs Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winer, J.; Jung, C.K.; Shackel, I.; Williams, P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999, 270, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. BioinformatiCs 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
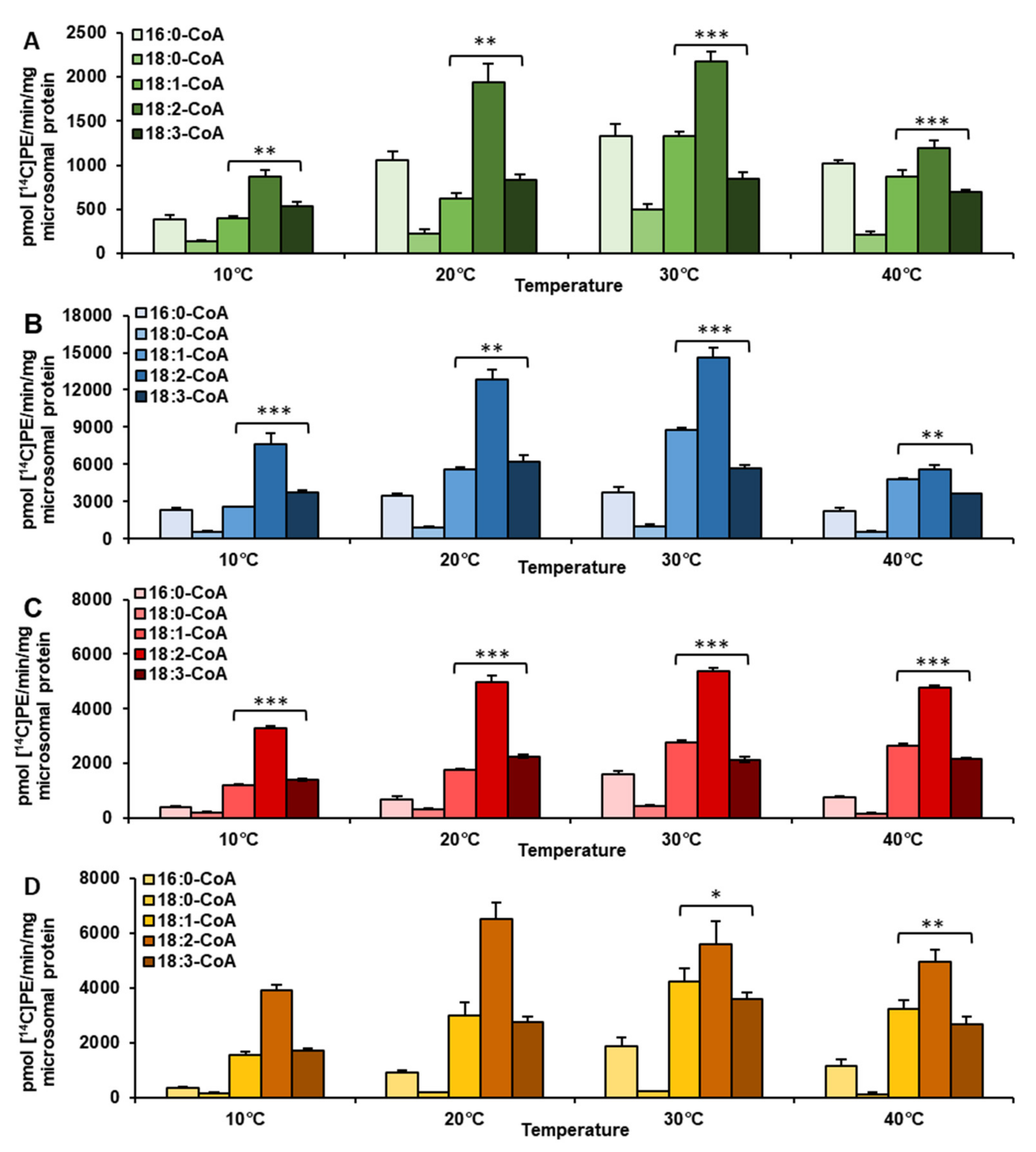
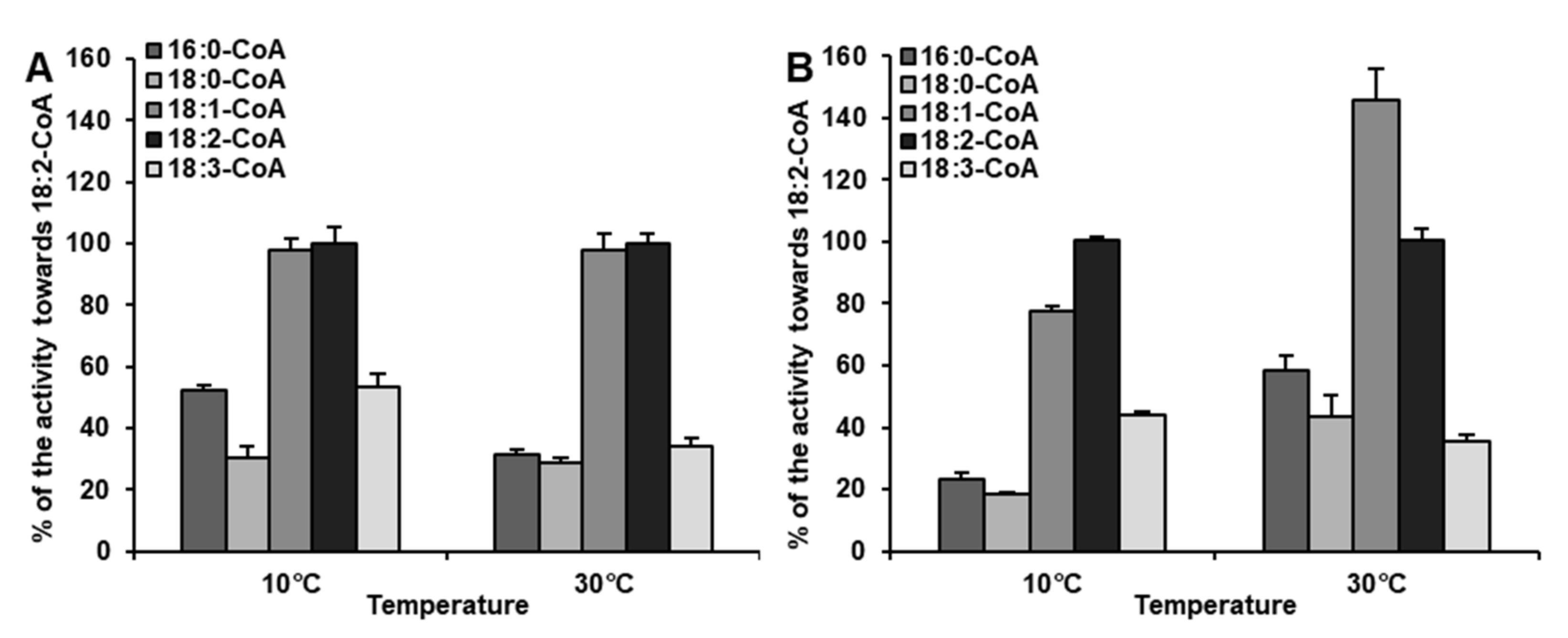
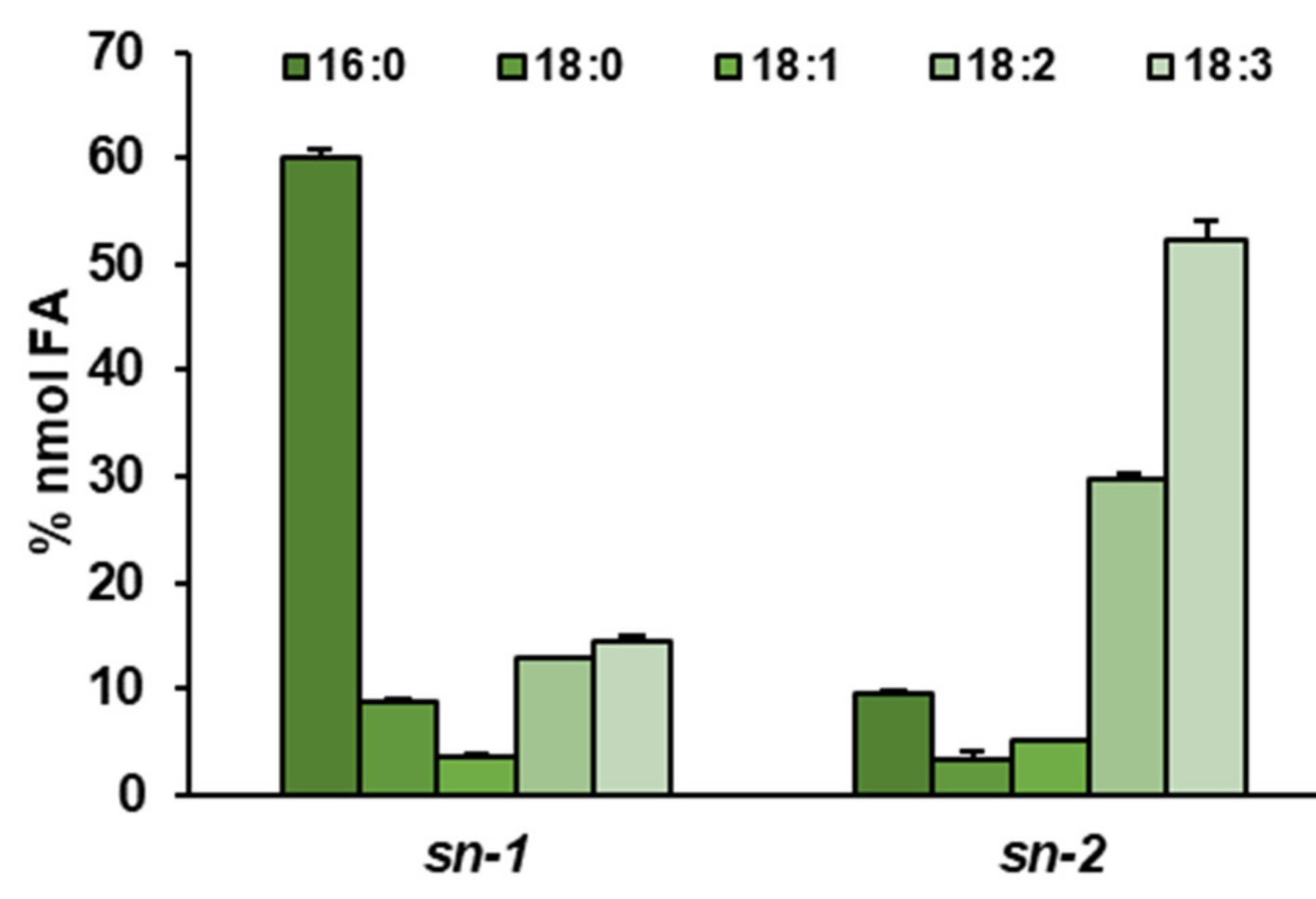
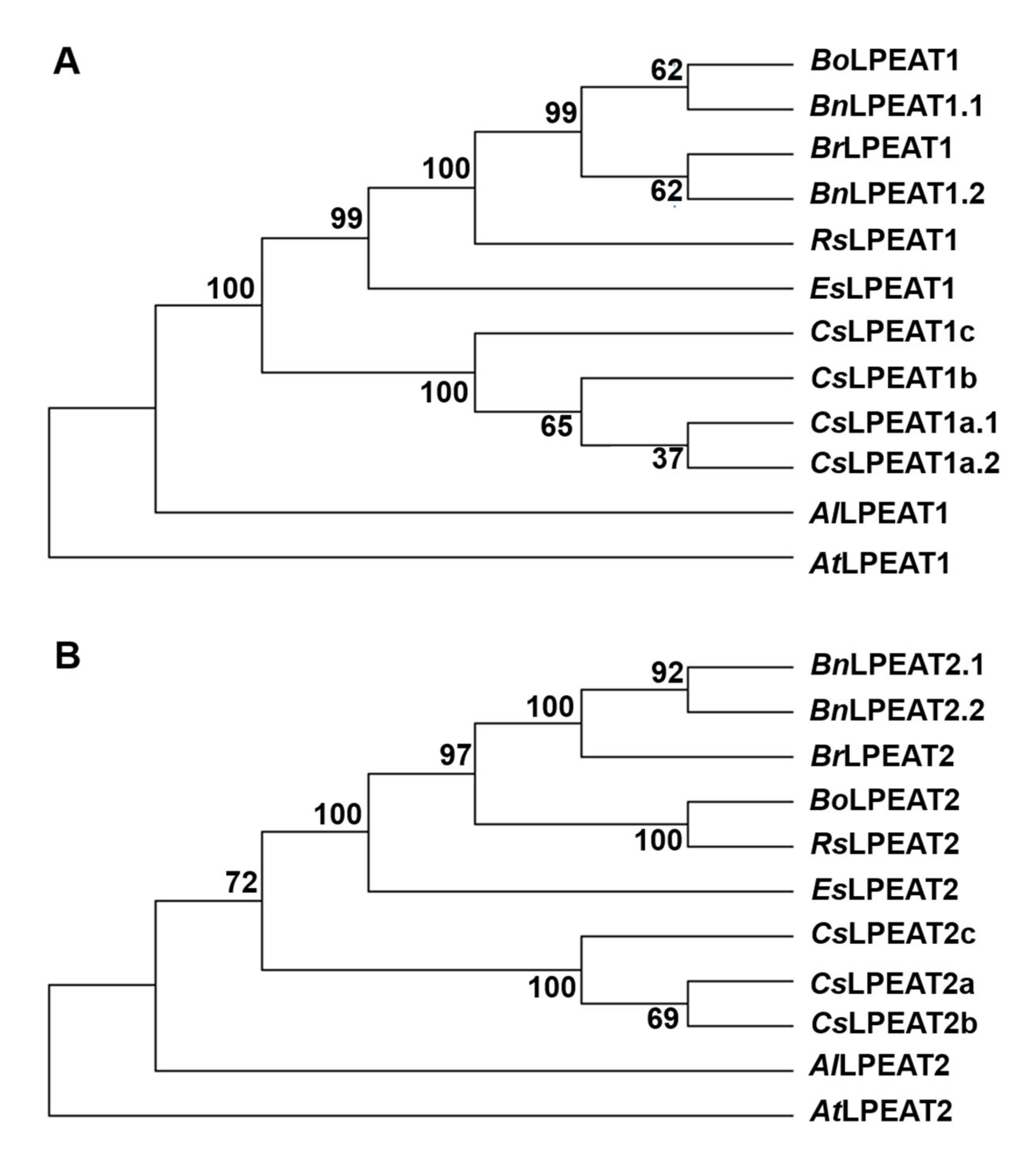
| Temperature | Acyl-CoA Used in the Assay | pmol [ 14C]PE/nmol Microsomal PE/min] | Time of Exchange of All PE Fatty Acids (FA) for FA from Acyl-CoA Pool [Days] |
|---|---|---|---|
| 10 °C | [14C]18:1-CoA | 0.194 ± 0.03 | 7.2 |
| [14C]18:2-CoA | 0.155 ± 0.004 | 8.9 | |
| [14C]18:3-CoA | 0.101 ± 0.005 | 13.8 | |
| 40 °C | [14C]18:1-CoA | 0.711 ± 0.03 | 2.0 |
| [14C]18:2-CoA | 0.763 ± 0.007 | 1.8 | |
| [14C]18:3-CoA | 0.295 ± 0.02 | 5.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klińska, S.; Demski, K.; Jasieniecka-Gazarkiewicz, K.; Banaś, A. LPEATs Tailor Plant Phospholipid Composition through Adjusting Substrate Preferences to Temperature. Int. J. Mol. Sci. 2021, 22, 8137. https://doi.org/10.3390/ijms22158137
Klińska S, Demski K, Jasieniecka-Gazarkiewicz K, Banaś A. LPEATs Tailor Plant Phospholipid Composition through Adjusting Substrate Preferences to Temperature. International Journal of Molecular Sciences. 2021; 22(15):8137. https://doi.org/10.3390/ijms22158137
Chicago/Turabian StyleKlińska, Sylwia, Kamil Demski, Katarzyna Jasieniecka-Gazarkiewicz, and Antoni Banaś. 2021. "LPEATs Tailor Plant Phospholipid Composition through Adjusting Substrate Preferences to Temperature" International Journal of Molecular Sciences 22, no. 15: 8137. https://doi.org/10.3390/ijms22158137
APA StyleKlińska, S., Demski, K., Jasieniecka-Gazarkiewicz, K., & Banaś, A. (2021). LPEATs Tailor Plant Phospholipid Composition through Adjusting Substrate Preferences to Temperature. International Journal of Molecular Sciences, 22(15), 8137. https://doi.org/10.3390/ijms22158137






