Electrochemical Sensors for Detection of Markers on Tumor Cells
Abstract
1. Introduction
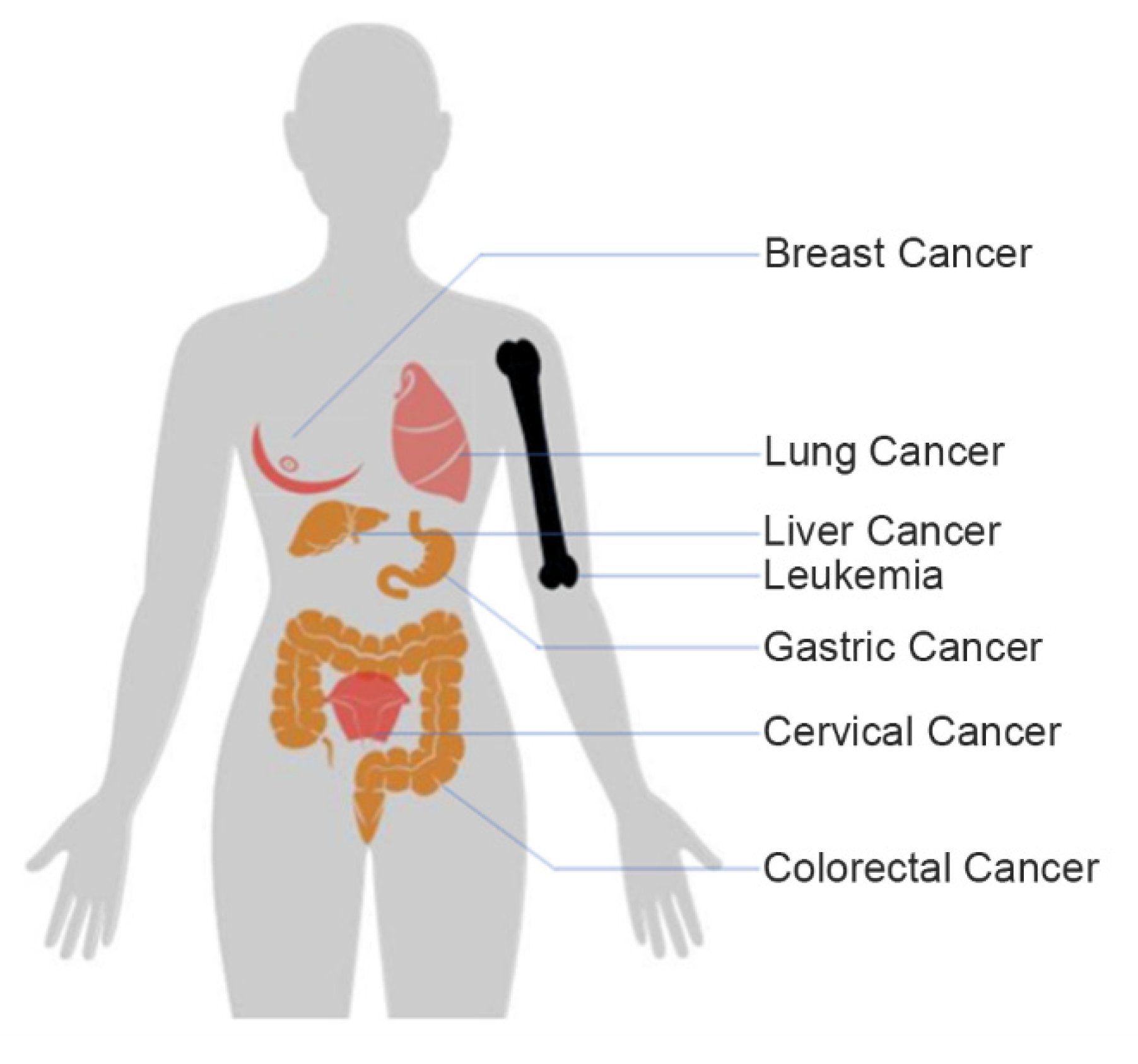
2. Electrochemical Detection of Markers on Tumor Cells
2.1. Breast Cancer
2.2. Lung Cancer
2.3. Cervical Cancer
2.4. Liver Cancer
| Cell Types | Cancer Markers | Modification Material | Linearity Range (Cells/mL) | Limit of Detection (Cells/mL) | Reference |
|---|---|---|---|---|---|
| HepG2 | MUC1 | pSC4 | 50–2 × 106 | 15 | [46] |
| HepG2 | FA receptors | FA-GAM-OA | 5–1 × 105 | 5 | [47] |
| HepG2 | EpCAM | — | 10–1 × 103 | 3 | [48] |
| HepG2 | Membrane surface | DNA nanotetrahedron | 10–1 × 106 | 5 | [49] |
| HepG2 | EpCAM | PAMAM | 1 × 104–1 × 106 | 2.1 × 103 | [54] |
| HepG2 | Membrane surface | ZnO@Au-Pd NPs | 1 × 102–1 × 107 | 10 | [55] |
| CSCs | CD44, CD90, CD133/2, and OV-6 | PDITC | 10–1 × 104 | 1 | [53] |
2.5. Leukemia Cells
2.6. Gastric Cancer and Colorectal Cancer
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. [Google Scholar] [CrossRef] [PubMed]
- Perumal, V.; Hashim, U. Advances in biosensors: Principle, architecture and applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Kell, D. Biosensors. Fundamentals and Applications.: A.P.F. Turner, I. Karube and G.S. Wilson, Oxford University Press, Oxford, 1987, ISBN 0-19-854724-2, xvi + 770 pp., £60.00. J. Electroanal. Chem. Interfacial Electrochem. 1988, 253, 589. [Google Scholar] [CrossRef][Green Version]
- Angelova, A.; Ollivon, M.; Campitelli, A.; Bourgaux, C. Lipid Cubic Phases as Stable Nanochannel Network Structures for Protein Biochip Development: X-ray Diffraction Study. Langmuir 2003, 19, 6928–6935. [Google Scholar] [CrossRef]
- Suhito, I.R.; Lee, W.; Baek, S.; Lee, D.; Min, J.; Kim, T.-H. Rapid and sensitive electrochemical detection of anticancer effects of curcumin on human glioblastoma cells. Sens. Actuators B Chem. 2019, 288, 527–534. [Google Scholar] [CrossRef]
- Corcoran, C.A.; Rechnitz, G.A. Cell-based biosensors. Trends Biotechnol. 1985, 3, 92–96. [Google Scholar] [CrossRef]
- Gupta, N.; Renugopalakrishnan, V.; Liepmann, D.; Paulmurugan, R.; Malhotra, B.D. Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron. 2019, 141, 111435. [Google Scholar] [CrossRef]
- Hong, H.; Koom, W.; Koh, W.-G. Cell Microarray Technologies for High-Throughput Cell-Based Biosensors. Sensors 2017, 17, 1293. [Google Scholar] [CrossRef]
- Kordasht, H.K.; Hasanzadeh, M. Aptamer based recognition of cancer cells: Recent progress and challenges in bioanalysis. Talanta 2020, 220, 121436. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, J.; Liu, J.; Wang, X.; Chen, B. Disease-Related Detection with Electrochemical Biosensors: A Review. Sensors 2017, 17, 2375. [Google Scholar] [CrossRef]
- Sun, D.; Lu, J.; Zhang, L.; Chen, Z. Aptamer-based electrochemical cytosensors for tumor cell detection in cancer diagnosis: A review. Anal. Chim. Acta 2019, 1082, 1–17. [Google Scholar] [CrossRef]
- Diaby, V.; Tawk, R.; Sanogo, V.; Xiao, H.; Montero, A.J. A review of systematic reviews of the cost-effectiveness of hormone therapy, chemotherapy, and targeted therapy for breast cancer. Breast Cancer Res. Treat. 2015, 151, 27–40. [Google Scholar] [CrossRef]
- Borghei, Y.-S.; Hosseini, M.; Dadmehr, M.; Hosseinkhani, S.; Ganjali, M.R.; Sheikhnejad, R. Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal. Chim. Acta 2016, 904, 92–97. [Google Scholar] [CrossRef]
- Nath, S.; Mukherjee, P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014, 20. [Google Scholar] [CrossRef]
- Ou, D.; Sun, D.; Liang, Z.; Chen, B.; Lin, X.; Chen, Z. A novel cytosensor for capture, detection and release of breast cancer cells based on metal organic framework PCN-224 and DNA tetrahedron linked dual-aptamer. Sens. Actuators B Chem. 2019, 285, 398–404. [Google Scholar] [CrossRef]
- Song, Y.; He, L.; Chen, K.; Wang, M.; Yang, L.; He, L.; Guo, C.; Jia, Q.; Zhang, Z. Quantification of EGFR and EGFR-overexpressed cancer cells based on carbon dots@bimetallic CuCo Prussian blue analogue. RSC Adv. 2020, 10, 28355–28364. [Google Scholar] [CrossRef]
- Yazdanparast, S.; Benvidi, A.; Banaei, M.; Nikukar, H.; Tezerjani, M.D.; Azimzadeh, M. Dual-aptamer based electrochemical sandwich biosensor for MCF-7 human breast cancer cells using silver nanoparticle labels and a poly(glutamic acid)/MWNT nanocomposite. Microchim. Acta 2018, 185. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.M.; Yin, X.H.; Gai, P.P.; Li, F. A label-free homogeneous electrochemical cytosensor for the ultrasensitive detection of cancer cells based on multiaptamer-functionalized DNA tetrahedral nanostructures. Chem. Commun. 2020, 56, 3883–3886. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Song, J.; Lu, Y.; Davis, J.J.; Gao, F.; Luo, X. Electrochemical Aptasensor for Ultralow Fouling Cancer Cell Quantification in Complex Biological Media Based on Designed Branched Peptides. Anal. Chem. 2019, 91, 8334–8340. [Google Scholar] [CrossRef]
- McFarlane, S.; Coulter, J.A.; Tibbits, P.; O’Grady, A.; McFarlane, C.; Montgomery, N.; Hill, A.; McCarthy, H.O.; Young, L.S.; Kay, E.W.; et al. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget 2015, 6, 11465–11476. [Google Scholar] [CrossRef]
- Safdar, H.; Hussain, Z.; Aborehab, M.; Hasan, H.; Afzal, S.; Thu, H. New Developments and Clinical Transition of Hyaluronic Acid-Based Nanotherapeutics for Treatment of Cancer: Reversing Multidrug Resistance, Tumor-Specific Targetability and Improved Anticancer Efficacy. Artif. Cells 2017, 46, 1967–1980. [Google Scholar] [CrossRef]
- Zhou, Y.; Wan, Y.; Yu, M.; Yuan, X.; Zhang, C. Hyaluronic acid-based label-free electrochemical impedance analysis for cancer cell quantification and CD44 expression. Microchem. J. 2021, 160, 105622. [Google Scholar] [CrossRef]
- Farzin, L.; Shamsipur, M.; Samandari, L.; Sheibani, S. Signalling probe displacement electrochemical aptasensor for malignant cell surface nucleolin as a breast cancer biomarker based on gold nanoparticle decorated hydroxyapatite nanorods and silver nanoparticle labels. Mikrochim. Acta 2018, 185, 154. [Google Scholar] [CrossRef] [PubMed]
- Zajchowski, D.A.; Bartholdi, M.F.; Gong, Y.; Webster, L.; Liu, H.L.; Munishkin, A.; Beauheim, C.; Harvey, S.; Ethier, S.P.; Johnson, P.H. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 2001, 61, 5168–5178. [Google Scholar]
- Bertrand, N.; Wu, J.; Xu, X.; Kamaly, N.; Farokhzad, O.C. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef]
- Zanghelini, F.; Frías, I.A.M.; Rêgo, M.J.B.M.; Pitta, M.G.R.; Sacilloti, M.; Oliveira, M.D.L.; Andrade, C.A.S. Biosensing breast cancer cells based on a three-dimensional TIO2 nanomembrane transducer. Biosens. Bioelectron. 2017, 92, 313–320. [Google Scholar] [CrossRef]
- Han, B.; Sha, L.; Yu, X.; Yang, M.; Cao, Y.; Zhao, J. Identification of dual therapeutic targets assisted by in situ automatous DNA assembly for combined therapy in breast cancer. Biosens. Bioelectron. 2021, 176, 112913. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Tang, H.; Gao, Z.; He, S.; Li, J.; Han, S. Ultrasensitive electrochemical detection of tumor cells based on multiple layer CdS quantum dots-functionalized polystyrene microspheres and graphene oxide—Polyaniline composite. Biosens. Bioelectron. 2018, 100, 1–7. [Google Scholar] [CrossRef]
- Rivera, M.P.; Mehta, A.C.; Wahidi, M.M. Establishing the Diagnosis of Lung Cancer Diagnosis and Management of Lung Cancer, 3rd ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest 2013, 143, E142–E165. [Google Scholar] [CrossRef]
- Bolat, G.; Vural, O.A.; Yaman, Y.T.; Abaci, S. Polydopamine nanoparticles-assisted impedimetric sensor towards label-free lung cancer cell detection. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 119, 111549. [Google Scholar] [CrossRef]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhang, Z.; Zheng, X.; Zhang, H.; Dong, D.; Zhang, Z.; Liu, M.; Zhou, J. An electrochemical biosensor for the detection of epithelial-mesenchymal transition. Nat. Commun. 2020, 11, 192. [Google Scholar] [CrossRef]
- Cheng, Y.; Zak, O.; Alsen, P.; Harrison, S.C.; Walz, T. Structure of the human transferrin receptor-transferrin complex. Cell 2004, 116, 565–576. [Google Scholar] [CrossRef]
- de Almeida, S.V.; Cancino-Bernardi, J.; de Andrade, J.K.; Felsner, M.L.; Zucolotto, V.; Galli, A. Cancer immunosensor based on apo and holo transferrin binding. Microchim. Acta 2020, 187. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Tu, W.; Wang, H.; Wang, Z.; Dai, Z. Formation of a Photoelectrochemical Z-Scheme Structure with Inorganic/Organic Hybrid Materials for Evaluation of Receptor Protein Expression on the Membrane of Cancer Cells. ACS Appl. Mater. Interfaces 2020, 12, 26905–26913. [Google Scholar] [CrossRef]
- Zhang, H.; Ke, H.; Wang, Y.; Li, P.; Huang, C.; Jia, N. 3D carbon nanosphere and gold nanoparticle-based voltammetric cytosensor for cell line A549 and for early diagnosis of non-small cell lung cancer cells. Microchim. Acta 2018, 186. [Google Scholar] [CrossRef]
- Ma, S.S.; Hu, S.S.; Wang, Q.; Liu, Y.H.; Zhao, G.Y.; Zhang, Q.C.; Mao, C.; Zhao, B. Evaluation of sialic acid based on electrochemical cytosensor with 3D micro/nanostructured sensing interface. Anal. Methods 2017, 9, 6171–6176. [Google Scholar] [CrossRef]
- Zhang, R.; Gu, Y.; Wang, Z.; Li, Y.; Fan, Q.; Jia, Y. Aptamer cell sensor based on porous graphene oxide decorated ion-selective-electrode: Double sensing platform for cell and ion. Biosens. Bioelectron. 2018, 117, 303–311. [Google Scholar] [CrossRef]
- Saslow, D.; Solomon, D.; Lawson, H.W.; Killackey, M.; Kulasingam, S.L.; Cain, J.; Garcia, F.A.R.; Moriarty, A.T.; Waxman, A.G.; Wilbur, D.C.; et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. CA Cancer J. Clin. 2012, 62, 147–172. [Google Scholar] [CrossRef]
- Fan, G.-C.; Li, Z.; Lu, Y.; Ma, L.; Zhao, H.; Luo, X. Robust photoelectrochemical cytosensor in biological media using antifouling property of zwitterionic peptide. Sens. Actuators B Chem. 2019, 299, 126996. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Wu, H.; Huang, W.; Ju, H.; Ding, S. A amperometric immunosensor for sensitive detection of circulating tumor cells using a tyramide signal amplification-based signal enhancement system. Biosens. Bioelectron. 2019, 130, 88–94. [Google Scholar] [CrossRef]
- Dutta Chowdhury, A.; Ganganboina, A.B.; Tsai, Y.C.; Chiu, H.C.; Doong, R.A. Multifunctional GQDs-Concanavalin A@Fe3O4 nanocomposites for cancer cells detection and targeted drug delivery. Anal Chim, Acta 2018, 1027, 109–120. [Google Scholar] [CrossRef]
- Motaghed Mazhabi, R.; Ge, L.; Jiang, H.; Wang, X. A label-free aptamer-based cytosensor for specific cervical cancer HeLa cell recognition through a g-C3N4-AgI/ITO photoelectrode. J. Mater. Chem. B 2018, 6, 5039–5049. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Shen, Y.; Liu, Y.H.; Hou, Z.Y.; Gu, Y.Q.; Zhao, W.B. An electrochemiluminescence cytosensor for sensitive detection of HeLa cells based on a signal amplification strategy of Au-NaYF4:Yb, Er nanocomposites. Analyst 2018, 143, 4199–4205. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. CURRENT CONCEPTS Hepatocellular Carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Han, B.; Wang, Y.; Zhao, J.; Cao, Y. Simple and universal signal labeling of cell surface for amplified detection of cancer cells via mild reduction. Biosens. Bioelectron. 2019, 145, 111714. [Google Scholar] [CrossRef] [PubMed]
- Ruiyi, L.; Fangchao, C.; Haiyan, Z.; Xiulan, S.; Zaijun, L. Electrochemical sensor for detection of cancer cell based on folic acid and octadecylamine-functionalized graphene aerogel microspheres. Biosens. Bioelectron. 2018, 119, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Shi, H.; Wang, M.; Duan, C.; Huang, Y.; Li, C.; Xiang, Y.; Li, G. Homogenous Electrochemical Method for Ultrasensitive Detection of Tumor Cells Designed by Introduction of Poly(A) Tails onto Cell Membranes. Anal. Chem. 2020, 92, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Lu, J.; Chen, D.; Jiang, Y.; Wang, Z.; Qin, W.; Yu, Y.; Chen, Z.; Zhang, Y. Label-free electrochemical detection of HepG2 tumor cells with a self-assembled DNA nanostructure-based aptasensor. Sens. Actuators B Chem. 2018, 268, 359–367. [Google Scholar] [CrossRef]
- Sun, D.; Lu, J.; Luo, Z.; Zhang, L.; Liu, P.; Chen, Z. Competitive electrochemical platform for ultrasensitive cytosensing of liver cancer cells by using nanotetrahedra structure with rolling circle amplification. Biosens. Bioelectron. 2018, 120, 8–14. [Google Scholar] [CrossRef]
- Chen, D.; Sun, D.; Wang, Z.; Qin, W.; Chen, L.; Zhou, L.; Zhang, Y. A DNA nanostructured aptasensor for the sensitive electrochemical detection of HepG2 cells based on multibranched hybridization chain reaction amplification strategy. Biosens. Bioelectron. 2018, 117, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Wang, X.W. Cancer stem cells in the development of liver cancer. J. Clin. Investig. 2013, 123, 1911–1918. [Google Scholar] [CrossRef]
- Eissa, S.; Chinnappan, R.; Zourob, M. Ultrasensitive Label-free Electrochemical Immunosensors for Multiple Cell Surface Biomarkers on Liver Cancer Stem Cells. Electroanalysis 2017, 29, 1994–2000. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.; Yan, C.; Chen, W. A Polyamidoamine Dendrimer-Based Electrochemical Immunosensor for Label-Free Determination of Epithelial Cell Adhesion Molecule- Expressing Cancer Cells. Sensors 2019, 19, 1879. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Lu, J.; Wang, X.; Zhang, Y.; Chen, Z. Voltammetric aptamer based detection of HepG2 tumor cells by using an indium tin oxide electrode array and multifunctional nanoprobes. Microchim. Acta 2017, 184, 3487–3496. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Shinohara, H.; Kuramitz, H.; Sugawara, K. Design of an electroactive peptide probe for sensing of a protein. Anal. Chim. Acta 2015, 890, 143–149. [Google Scholar] [CrossRef]
- Sugawara, K.; Kuramitz, H.; Kadoya, T. Label-free cytosensing of cancer cells based on the interaction between protein and an electron-transfer carbohydrate-mimetic peptide. Anal. Chim. Acta 2018, 1040, 166–176. [Google Scholar] [CrossRef]
- Sugawara, K.; Ishizaki, S.; Kodaira, K.; Kuramitz, H.; Kadoya, T. Fabrication of a cell-recognition/electron-transfer/cross-linker, peptide-immobilized electrode for the sensing of K562 cells. Anal. Chim. Acta 2020, 1116, 53–61. [Google Scholar] [CrossRef]
- Li, J.; Lin, X.; Zhang, Z.; Tu, W.; Dai, Z. Red light-driven photoelectrochemical biosensing for ultrasensitive and scatheless assay of tumor cells based on hypotoxic AgInS2 nanoparticles. Biosens. Bioelectron. 2019, 126, 332–338. [Google Scholar] [CrossRef]
- Khoshroo, A.; Hosseinzadeh, L.; Adib, K.; Rahimi-Nasrabadi, M.; Ahmadi, F. Earlier diagnoses of acute leukemia by a sandwich type of electrochemical aptasensor based on copper sulfide-graphene composite. Anal. Chim. Acta 2021, 1146, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Zhao, X.P.; Liu, F.F.; Younis, M.R.; Xia, X.H.; Wang, C. Direct Plasmon-Enhanced Electrochemistry for Enabling Ultrasensitive and Label-Free Detection of Circulating Tumor Cells in Blood. Anal. Chem. 2019, 91, 4413–4420. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Gulati, P.; Kaur, P.; Rajam, M.V.; Srivastava, T.; Mishra, P.; Islam, S.S. Vertically aligned multi-walled carbon nanotubes based flexible immunosensor for extreme low level detection of multidrug resistant leukemia cells. Sens. Actuators B Chem. 2019, 301, 127047. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, H.; Huang, Z.; Luo, Z.; Huang, N.; Ding, S.; Feng, W. A Simple Electrochemical Aptamer Cytosensor for Direct Detection of Chronic Myelogenous Leukemia K562 Cells. Electroanalysis 2017, 29, 828–834. [Google Scholar] [CrossRef]
- Sun, P.; Xiong, W.W.; Zhu, D.; Dong, Z.; Jin, X.; Liu, B.; Zhang, Y.; Bao, B.; Yao, W.; Zhang, L.; et al. An ultrasensitive electrochemical cytosensor for highly specific detection of HL-60 cancer cells based on metal ion functionalized titanium phosphate nanospheres. Analyst 2018, 143, 5170–5175. [Google Scholar] [CrossRef]
- Amouzadeh Tabrizi, M.; Shamsipur, M.; Saber, R.; Sarkar, S. Flow injection amperometric sandwich-type aptasensor for the determination of human leukemic lymphoblast cancer cells using MWCNTs-Pdnano/PTCA/aptamer as labeled aptamer for the signal amplification. Anal. Chim Acta 2017, 985, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.D.; Cheng, H.; Chen, W.H.; Cheng, S.X.; Zhuo, R.X.; Zhang, X.Z. In situ recognition of cell-surface glycans and targeted imaging of cancer cells. Sci. Rep. 2013, 3, 2679. [Google Scholar] [CrossRef]
- Dervisevic, M.; Senel, M.; Sagir, T.; Isik, S. Highly sensitive detection of cancer cells with an electrochemical cytosensor based on boronic acid functional polythiophene. Biosens. Bioelectron. 2017, 90, 6–12. [Google Scholar] [CrossRef]
- Dervisevic, M.; Senel, M.; Sagir, T.; Isik, S. Boronic Acid vs. Folic Acid: A Comparison of the bio-recognition performances by Impedimetric Cytosensors based on Ferrocene cored dendrimer. Biosens. Bioelectron. 2017, 91, 680–686. [Google Scholar] [CrossRef]
- Amouzadeh Tabrizi, M.; Shamsipur, M.; Saber, R.; Sarkar, S.; Sherkatkhameneh, N. Flow injection amperometric sandwich-type electrochemical aptasensor for the determination of adenocarcinoma gastric cancer cell using aptamer-Au@Ag nanoparticles as labeled aptamer. Electrochim. Acta 2017, 246, 1147–1154. [Google Scholar] [CrossRef]
- Duan, F.; Hu, M.; Guo, C.; Song, Y.; Wang, M.; He, L.; Zhang, Z.; Pettinari, R.; Zhou, L. Chromium-based metal-organic framework embedded with cobalt phthalocyanine for the sensitively impedimetric cytosensing of colorectal cancer (CT26) cells and cell imaging. Chem. Eng. J. 2020, 398, 125452. [Google Scholar] [CrossRef]
- Akbal, O.; Bolat, G.; Yaman, Y.T.; Abaci, S. Folic acid conjugated Prussian blue nanoparticles: Synthesis, physicochemical characterization and targeted cancer cell sensing. Colloids Surf. B Biointerfaces 2020, 187, 110655. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, J.; Hasanzadeh, M.; Somi, M.H.; Shadjou, N.; Jouyban, A. Highly sensitive and specific cytosensing of HT 29 colorectal cancer cells using folic acid functionalized-KCC-1 nanoparticles. Biosens. Bioelectron. 2019, 132, 122–131. [Google Scholar] [CrossRef]
- An, Y. Sensitive Electrochemical Cytosensor Based on Biocompatible Au@BSA Conductive Architecture and Lectin-Modified Nanoprobe. J. Electrochem. Soc. 2016, 163, B242–B247. [Google Scholar] [CrossRef]
- Geng, P.; Feng, C.; Zhu, L.; Zhang, J.; Wang, F.; Liu, K.; Xu, Z.; Zhang, W. Evaluation of Sialic Acid Expression on Cancer Cells via an Electrochemical Assay Based on Biocompatible Au@BSA Architecture and Lectin-modified Nanoprobes. Electroanalysis 2016, 28, 1331–1339. [Google Scholar] [CrossRef]





| Cell Types | Cancer Markers | Modification Material | Linearity Range (Cells/mL) | Limit of Detection (Cells/mL) | Reference |
|---|---|---|---|---|---|
| MCF7 | MUC1 and Nucleoli | TDNs | 20–1 × 107 | 6 | [15] |
| MCF7 | EGFR | CD@CuCoPBA | 5 × 102–1 × 105 | 80 | [16] |
| MCF7 | MUC1 | Apt-DTNs | 50–1 × 106 | 5 | [18] |
| MCF7 | MUC1 | PANI Films | 50–1 × 106 | 20 | [19] |
| MCF7 | Nucleoli | IL/HAp-Au NPs | 10–1 × 106 | 8 ± 2 | [23] |
| MCF7 | MUC1 | MWCNT-PGA | 1 × 102–1 × 107 | 25 | [17] |
| MDA-MB-231 | CD44 | GNPs | 2 × 102–3 × 105 | 128 | [22] |
| T47D | Carbohydrate profile | TiO2-MN | 10–1 × 106 | 10 | [26] |
| Cell Types | Cancer Markers | Modification Material | Linearity Range (Cells/mL) | Limit of Detection (Cells/mL) | Reference |
|---|---|---|---|---|---|
| A549 | — | PDA NPs | 1 × 102–1 × 105 | 25 | [30] |
| A549 | E-cadherin | CNT-Au NPs | 75–5.5 × 104 | 75 | [32] |
| A549 | CD44 | FePc/Cys AZIS QDs | 2 × 102–4.5 × 106 | 15 | [35] |
| A549 | Carcinoembryonic antigen | CNS@AuNP | 42–4.2 × 106 | 14 | [36] |
| A549 | Sialic acid | CS-Au/hPPy | 10–1 × 107 | 2 | [37] |
| A549 | Nucleoli | PGO Film | — | 10 | [38] |
| Cell Types | Cancer Markers | Modification Material | Linearity Range (Cells/mL) | Limit of Detection (Cells/mL) | Reference |
|---|---|---|---|---|---|
| HeLa | Nucleoli | ITO/TiO2/ZnIn2S4 | 1×102–1 × 106 | 34 | [40] |
| HeLa | Nucleoli | ICPs@Tyr | 2–2 × 104 | 2 | [41] |
| HeLa | Carbohydrate profile | GQDs-Con A@Fe3O4 | 5 × 102–1 × 105 | 274 | [42] |
| HeLa | CEM/PTK7 | WDg-C3N4-AgI | 10–1 × 106 | 5 | [43] |
| HeLa | Folate receptors | Au-NaYF4:Yb.Er | 4.25 × 102–4.25 × 105 | 326 | [44] |
| Cell Types | Cancer Markers | Modification Material | Linearity Range (Cells/mL) | Limit of Detection (Cells/mL) | Reference |
|---|---|---|---|---|---|
| K562 | ASGP receptors | — | 1 × 102–5 × 103 | — | [58] |
| K562 | CD44 | Collagen and Peptide | 27–2 × 103 | 8 | [59] |
| K562 | P-glycoprotein | PET and MWNT | 1.5 × 102–1.5 × 107 | 10 | [64] |
| K562 | Carbohydrate profile | GO-PANI-GA-Con A | 10–1 × 107 | 3 | [28] |
| K562 | Carbohydrate profile | — | 1 × 102–1 × 107 | 79 | [65] |
| HL-60 | — | C-MWNT | 1 × 102–1 × 107 | 35 | [66] |
| CCRF-CEM | PTK-7 | AgInS2 | 1.5 × 102–3 × 105 | 16 | [60] |
| CCRF-CEM | PTK-7 | Au-GR NPs | 50–1 × 106 | 18 | [61] |
| CCRF-CEM | PTK-7 | AuNSs | 5–1 × 105 | 5 | [62] |
| CCRF-CEM | PTK-7 | MWNT-Pd NPs/PTCA | 10–5 × 105 | 8 | [67] |
| Cell Types | Cancer Markers | Modification Material | Linearity Range (Cells/mL) | Limit of Detection (Cells/mL) | Reference |
|---|---|---|---|---|---|
| AGS | Sialic acid | p(TBA0.5Th0.5) | 10–1 × 106 | 10 | [69] |
| AGS | Folate receptors and Sialic acid | Au/Fc-PAMAM(G2)/FA and Au/Fc-PAMAM(G2)/BA | 1 × 102–1 × 106 | 20/28 | [70] |
| AGS | Carbohydrate profile | MWCNT-Au NPs | 10–5 × 105 | 6 | [71] |
| BGC-823 | Mannosyl groups and Sialic acid | TGA-Au@BSA microspheres-APBA | 1 × 102–1 × 106 | 40 | [75] |
| BGC-823 | Mannosyl groups and Sialic acid | Au@BSA microspheres-Con A | 5 × 102–1 × 106 | 120 | [76] |
| CT26 | — | Cr-MOF@CoPc | 50–1 × 107 | 36/8 | [72] |
| DLD-1 | FA receptors | FA-PB NPs | 5 × 102–1 × 105 | 48 | [73] |
| HT29 | FA receptors | KCC-1-NH2-FA | 50–1.2 × 104 | 50 | [74] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Du, X.; Zhang, Z. Electrochemical Sensors for Detection of Markers on Tumor Cells. Int. J. Mol. Sci. 2021, 22, 8184. https://doi.org/10.3390/ijms22158184
Zhou H, Du X, Zhang Z. Electrochemical Sensors for Detection of Markers on Tumor Cells. International Journal of Molecular Sciences. 2021; 22(15):8184. https://doi.org/10.3390/ijms22158184
Chicago/Turabian StyleZhou, Han, Xin Du, and Zhenguo Zhang. 2021. "Electrochemical Sensors for Detection of Markers on Tumor Cells" International Journal of Molecular Sciences 22, no. 15: 8184. https://doi.org/10.3390/ijms22158184
APA StyleZhou, H., Du, X., & Zhang, Z. (2021). Electrochemical Sensors for Detection of Markers on Tumor Cells. International Journal of Molecular Sciences, 22(15), 8184. https://doi.org/10.3390/ijms22158184






