Cytosine-Rich DNA Fragments Covalently Bound to Carbon Nanotube as Factors Triggering Doxorubicin Release at Acidic pH. A Molecular Dynamics Study
Abstract
:1. Introduction
2. Results and Discussion
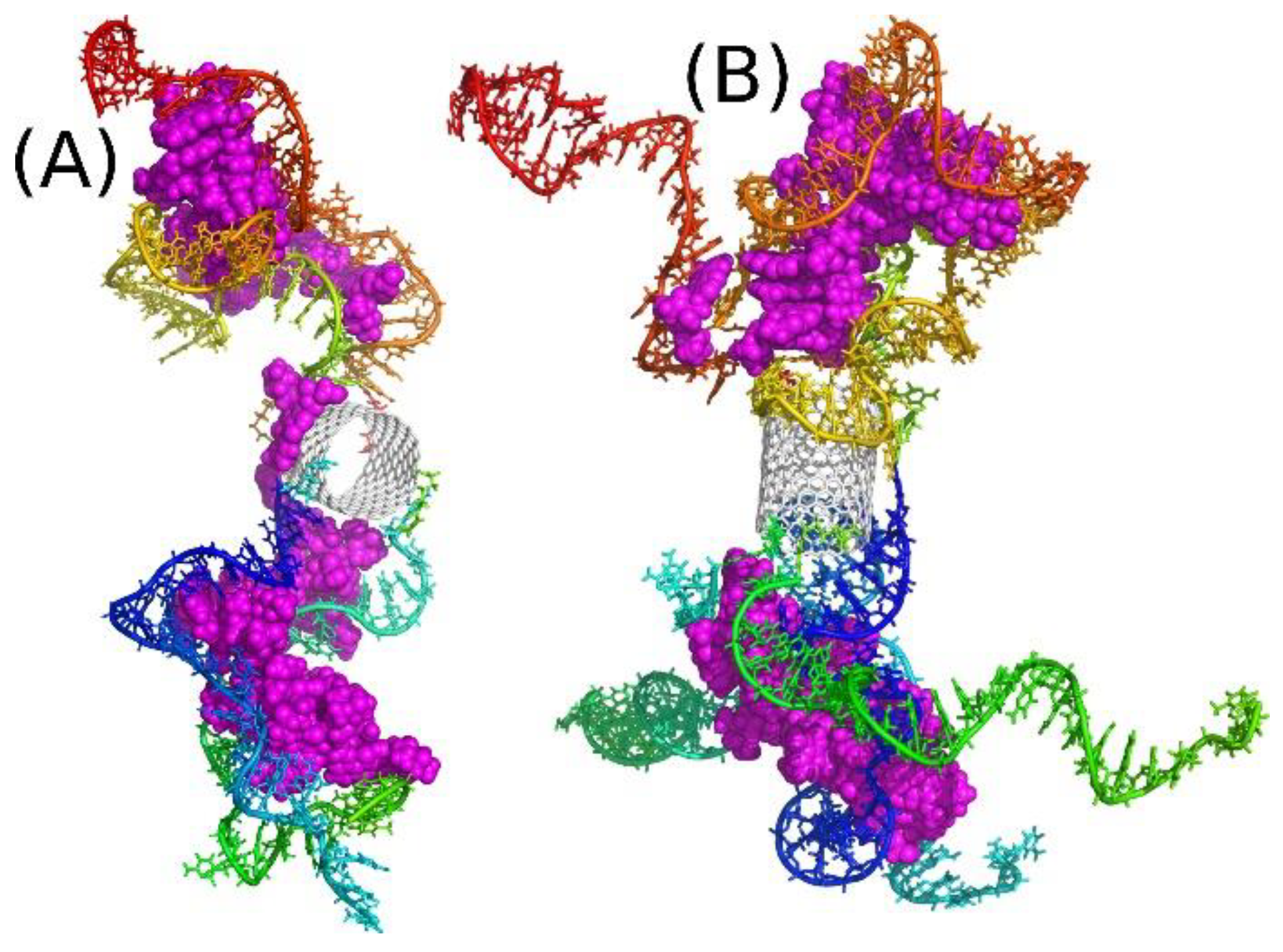
2.1. Definitions of the Analyzed Systems
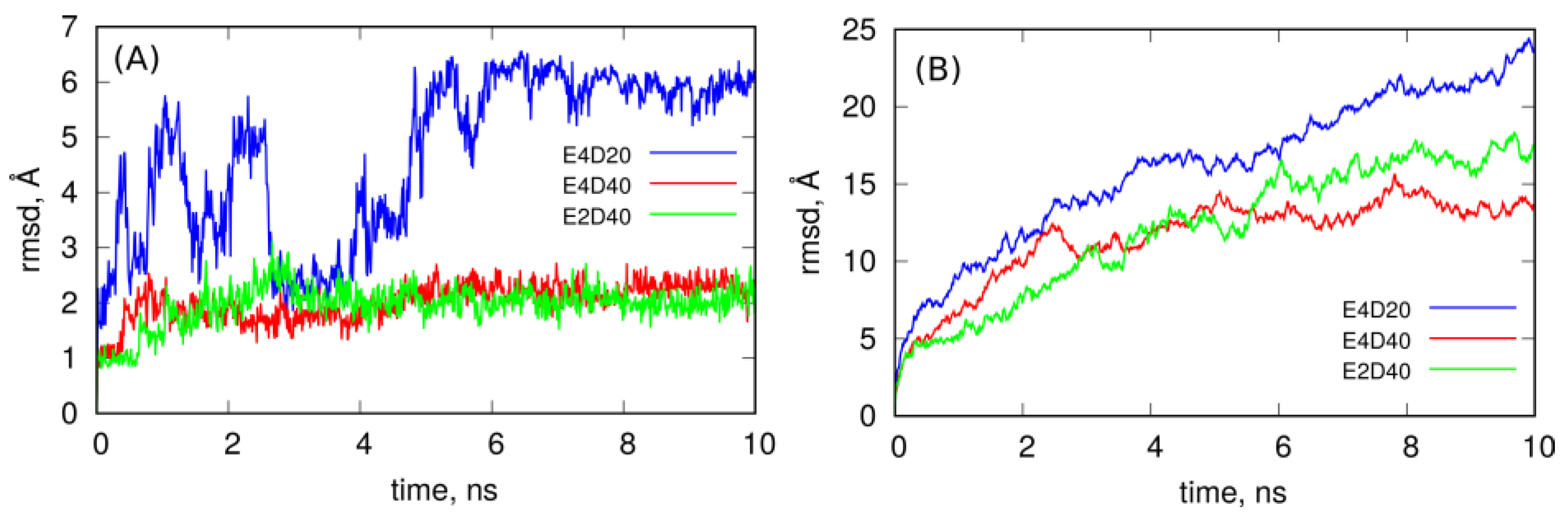
2.2. Covalent Functionalization of CNT by DNA Motifs
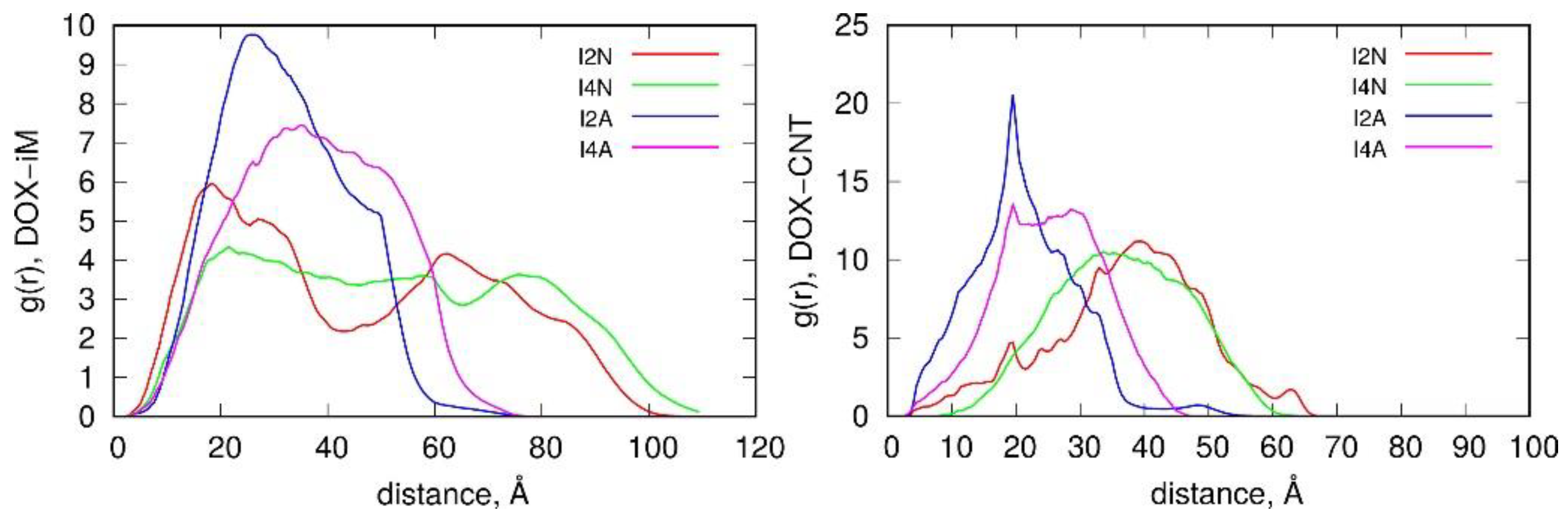
2.3. Encapsulation and Release of DOX from CNT Interior
2.4. Intercalation of DOX in DNA Chains
3. Summary and Conclusions
4. Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Battigelli, A.; Ménard-Moyon, C.; Da Ros, T.; Prato, M.; Bianco, A. Endowing carbon nanotubes with biological and biomedical properties by chemical modifications. Adv. Drug Deliv. Rev. 2013, 65, 1899–1920. [Google Scholar] [CrossRef]
- Wong, B.S.; Yoong, S.L.; Jagusiak, A.; Panczyk, T.; Ho, H.K.; Ang, W.H.; Pastorin, G. Pastorin, Carbon nanotubes for delivery of small molecule drugs. Adv. Drug Deliv. Rev. 2013, 65, 1964–2015. [Google Scholar] [CrossRef] [PubMed]
- Zare, H.; Ahmadi, S.; Ghasemi, A.; Ghanbari, M.; Rabiee, N.; Bagherzadeh, M.; Karimi, M.; Webster, T.J.; Hamblin, M.R.; Mostafavi, E. Carbon Nanotubes: Smart Drug/Gene Delivery Carriers. Int. J. Nanomed. 2021, 16, 1681–1706. [Google Scholar] [CrossRef]
- Karimi, M.; Solati, N.; Ghasemi, A.; Estiar, M.A.; Hashemkhani, M.; Kiani, P.; Mohamed, E.; Saeidi, A.; Taheri, M.; Avci, P.; et al. Carbon nanotubes part II: A remarkable carrier for drug and gene delivery. Expert Opin. Drug Deliv. 2015, 12, 1089–1105. [Google Scholar] [CrossRef]
- CLay, C.L.; Liu, J.; Liu, Y. Functionalized carbon nanotubes for anticancer drug delivery. Expert Rev. Med. Devices 2011, 8, 561–566. [Google Scholar]
- Li, Z.; de Barros, A.L.B.; Soares, D.C.F.; Moss, S.N.; Alisaraie, L. Functionalized single-walled carbon nanotubes: Cellular uptake, biodistribution and applications in drug delivery. Int. J. Pharm. 2017, 524, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Alidori, S.; Asqiriba, K.; Londero, P.; Bergkvist, M.; Leona, M.; Scheinberg, D.A.; McDevitt, M.R. Deploying RNA and DNA with Functionalized Carbon Nanotubes. J. Phys. Chem. C 2013, 117, 5982–5992. [Google Scholar] [CrossRef] [Green Version]
- Roxbury, D.; Jagota, A.; Mittal, J. Structural Characteristics of Oligomeric DNA Strands Adsorbed onto Single-Walled Carbon Nanotubes. J. Phys. Chem. B 2013, 117, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Roxbury, D.; Zhang, S.Q.; Mittal, J.; DeGrado, W.F.; Jagota, A. Structural Stability and Binding Strength of a Designed Peptide–Carbon Nanotube Hybrid. J. Phys. Chem. C 2013, 117, 26255–26261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadi, M.; Salmasi, Z.; Hashemi, M.; Mosaffa, F.; Abnous, K.; Ramezani, M. Single-walled carbon nanotubes functionalized with aptamer and piperazine–polyethylenimine derivative for targeted siRNA delivery into breast cancer cells. Int. J. Pharm. 2015, 485, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Son, K.H.; Hong, J.H.; Lee, J.W. Carbon nanotubes as cancer therapeutic carriers and mediators. Int. J. Nanomed. 2016, 11, 5163–5185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohajeri, M.; Behnam, B.; Sahebkar, A. Biomedical applications of carbon nanomaterials: Drug and gene delivery potentials. J. Cell Physiol. 2019, 234, 298–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Qu, K.; Zhao, C.; Wu, L.; Ren, J.; Wang, J.; Qu, X. Insights into the biomedical effects of carboxylated single-wall carbon nanotubes on telomerase and telomeres. Nat. Commun. 2012, 3, 1074. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Peng, Y.; Ren, J.; Qu, X. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl. Acad. Sci. USA 2006, 103, 19658–19663. [Google Scholar] [CrossRef] [Green Version]
- Wolski, P.; Wojton, P.; Nieszporek, K.; Panczyk, T. Interaction of Human Telomeric i-Motif DNA with Single-Walled Carbon Nanotubes: Insights from Molecular Dynamics Simulations. J. Phys. Chem. B 2019, 123, 10343–10353. [Google Scholar] [CrossRef]
- Chen, C.; Pu, F.; Huang, Z.; Liu, Z.; Ren, J.; Qu, X. Stimuli-responsive controlled-release system using quadruplex DNA-capped silica nanocontainers. Nucleic Acids Res. 2011, 39, 1638–1644. [Google Scholar] [CrossRef]
- Chen, L.; Di, J.; Cao, C.; Zhao, Y.; Ma, Y.; Luo, J.; Wen, Y.; Song, W.; Song, Y.; Jiang, L. A pH-driven DNA nanoswitch for responsive controlled release. Chem. Commun. 2011, 47, 2850–2852. [Google Scholar] [CrossRef]
- Song, L.; Ho, V.H.; Chen, C.; Yang, Z.; Liu, D.; Chen, R.; Zhou, D. Efficient, pH-Triggered Drug Delivery Using a pH-Responsive DNA-Conjugated Gold Nanoparticle. Adv. Healthc. Mater. 2013, 2, 275–280. [Google Scholar] [CrossRef]
- Xu, C.; Zhao, C.; Ren, J.; Qu, X. pH-controlled reversible drug binding and release using a cytosine-rich hairpin DNA. Chem. Commun. 2011, 47, 8043–8045. [Google Scholar] [CrossRef]
- Kim, J.Y.; Song, J.; Jung, H.; Mok, H. I-motif-coated exosomes as a pH-sensitive carrier for anticancer drugs. Appl. Biol. Chem. 2018, 61, 599–606. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Kim, J.; Jung, S.; Kim, W.J. DNA-Au Nanomachine Equipped with i-Motif and G-Quadruplex for Triple Combinatorial Anti-Tumor Therapy. Adv. Funct. Mater. 2018, 28, 1705416. [Google Scholar] [CrossRef]
- Wolski, P.; Nieszporek, K.; Panczyk, T. Carbon Nanotubes and Short Cytosine-Rich Telomeric DNA Oligomeres as Platforms for Controlled Release of Doxorubicin—A Molecular Dynamics Study. Int. J. Mol. Sci. 2020, 21, 3619. [Google Scholar] [CrossRef]
- Phan, A.T.; Mergny, J.L. Mergny, Human telomeric DNA: G-quadruplex, i-motif and Watson—Crick double helix. Nucleic Acids Res. 2002, 30, 4618–4625. [Google Scholar] [CrossRef]
- Dembska, A. The analytical and biomedical potential of cytosine-rich oligonucleotides: A review. Anal. Chim. Acta 2016, 930, 1–12. [Google Scholar] [CrossRef]
- Koltai, T. Cancer: Fundamentals behind pH targeting and the double-edged approach. OncoTargets Ther. 2016, 9, 6343–6360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilon-Thomas, S.; Kodumudi, K.N.; El-Kenawi, A.E.; Russell, S.; Weber, A.M.; Luddy, K.; Damaghi, M.; Wojtkowiak, J.W.; Mulé, J.J.; Ibrahim-Hashim, A.; et al. Neutralization of Tumor Acidity Improves Antitumor Responses to Immunotherapy. Cancer Res. 2016, 76, 1381–1390. [Google Scholar] [CrossRef] [Green Version]
- Ulbrich, K. Polymeric anticancer drugs with pH-controlled activation. Adv. Drug Deliv. Rev. 2004, 56, 1023–1050. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Niemeyer, C.M. Designer DNA–silica/carbon nanotube nanocomposites for traceable and targeted drug delivery. J. Mater. Chem. B 2020, 8, 2250–2255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastorin, G. Crucial Functionalizations of Carbon Nanotubes for Improved Drug Delivery: A Valuable Option? Pharm. Res. 2009, 26, 746–769. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Pomales, G.; Santiago-Rodríguez, L.; Cabrera, C.R. DNA-Functionalized Carbon Nanotubes for Biosensing Applications. J. Nanosci. Nanotech. 2009, 9, 2175–2188. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Cheatham, T.E., III; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [Green Version]
- Panczyk, T.; Wolski, P.; Lajtar, L. Coadsorption of Doxorubicin and Selected Dyes on Carbon Nanotubes. Theoretical Investigation of Potential Application as a pH-Controlled Drug Delivery System. Langmuir 2016, 32, 4719–4728. [Google Scholar] [CrossRef] [PubMed]
- Wolski, P.; Nieszporek, K.; Panczyk, T. Pegylated and folic acid functionalized carbon nanotubes as pH controlled carriers of doxorubicin. Molecular dynamics analysis of the stability and drug release mechanism. Phys. Chem. Chem. Phys. 2017, 19, 9300–9312. [Google Scholar] [CrossRef]
- Jagusiak, A.; Chlopas, K.; Zemanek, G.; Wolski, P.; Panczyk, T. Controlled Release of Doxorubicin from the Drug Delivery Formulation Composed of Single-Walled Carbon Nanotubes and Congo Red: A Molecular Dynamics Study and Dynamic Light Scattering Analysis. Pharmaceutics 2020, 12, 622. [Google Scholar] [CrossRef] [PubMed]
- Weiner, S.J.; Kollman, P.A.; Nguyen, D.T.; Case, D.A. Case, An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 1986, 7, 230–252. [Google Scholar] [CrossRef] [PubMed]
- Ivani, I.; Dans, P.D.; Noy, A.; Pérez, A.; Faustino, I.; Hospital, A.; Walther, J.; Andrio, P.; Goñi, R.; Balaceanu, A.; et al. Parmbsc1: A refined force field for DNA simulations. Nat. Methods 2016, 13, 55–58. [Google Scholar] [CrossRef] [Green Version]
- Dupradeau, F.Y.; Pigache, A.; Zaffran, T.; Savineau, C.; Lelong, R.; Grivel, N.; Lelong, D.; Rosanski, W.; Cieplak, P. The R.E.D. tools: Advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010, 12, 7821–7839. [Google Scholar] [CrossRef] [Green Version]
- Vanquelef, E.; Simon, S.; Marquant, G.; Garcia, E.; Klimerak, G.; Delepine, J.C.; Cieplak, P.; Dupradeau, F.Y. Server: A web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011, 39, W511–W517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berendsen, H.J.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Fülöp, Z.; Gref, R.; Loftsson, T. A permeation method for detection of self-aggregation of doxorubicin in aqueous environment. Int. J. Pharm. 2013, 454, 559–561. [Google Scholar] [CrossRef]
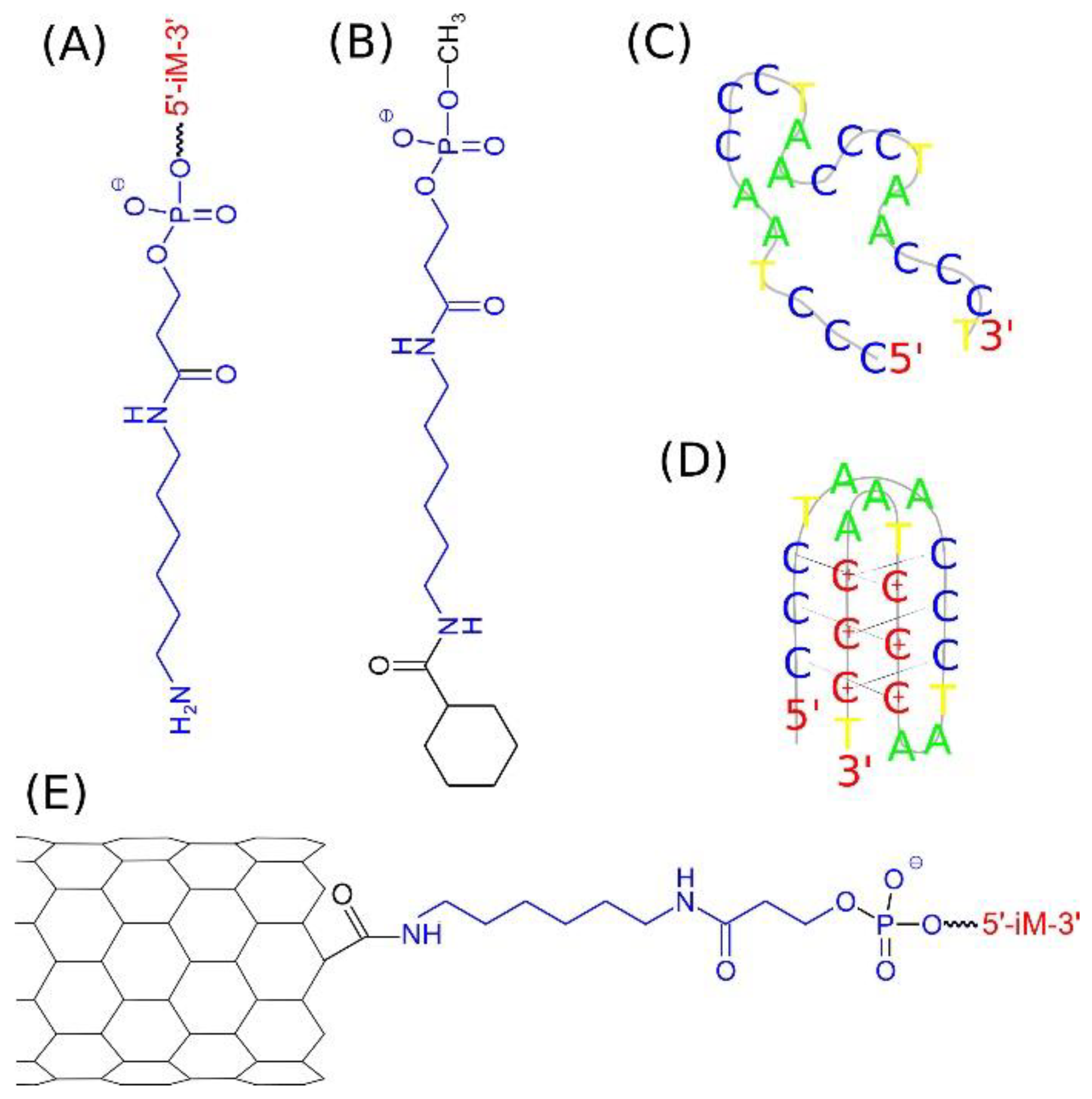
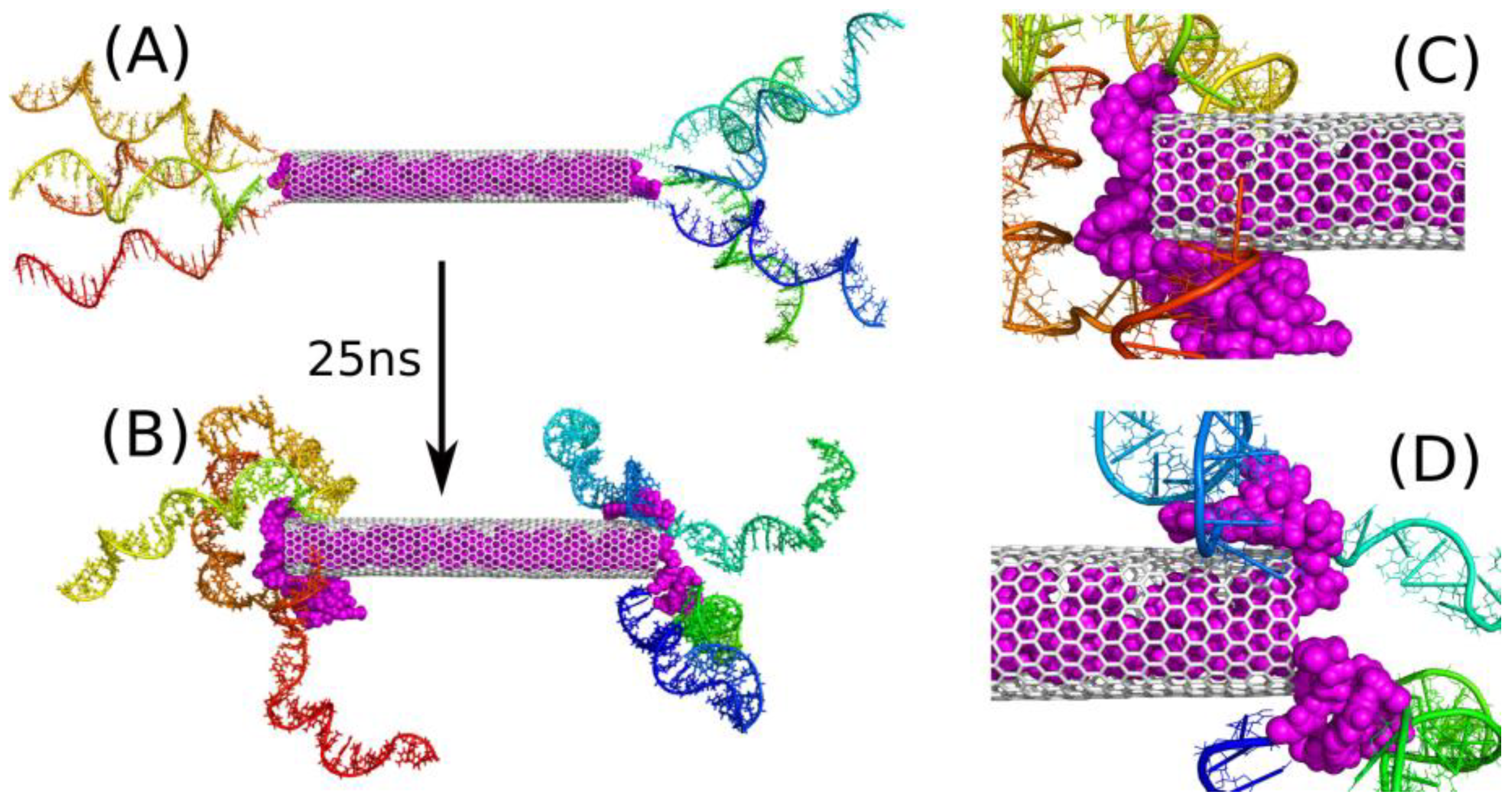
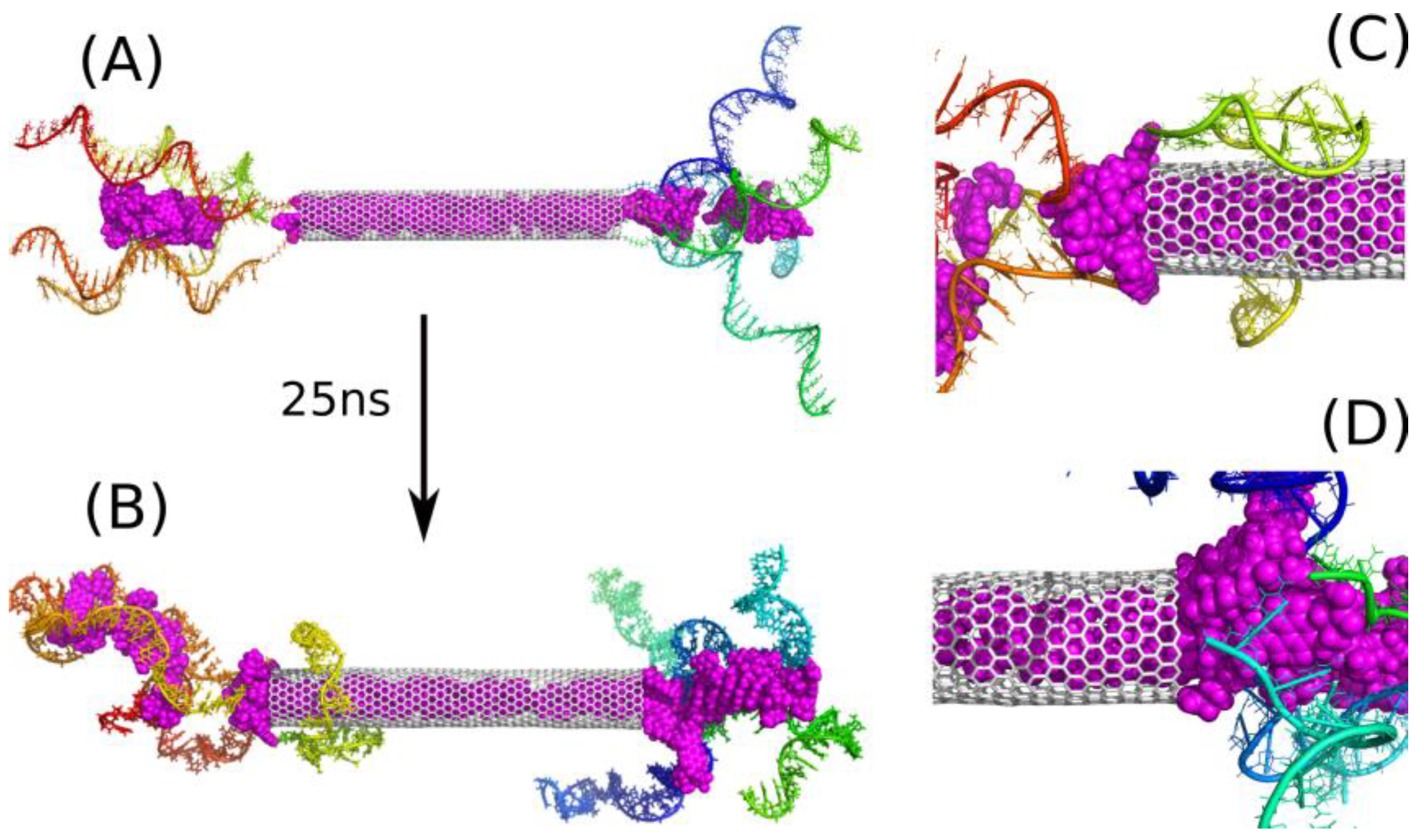
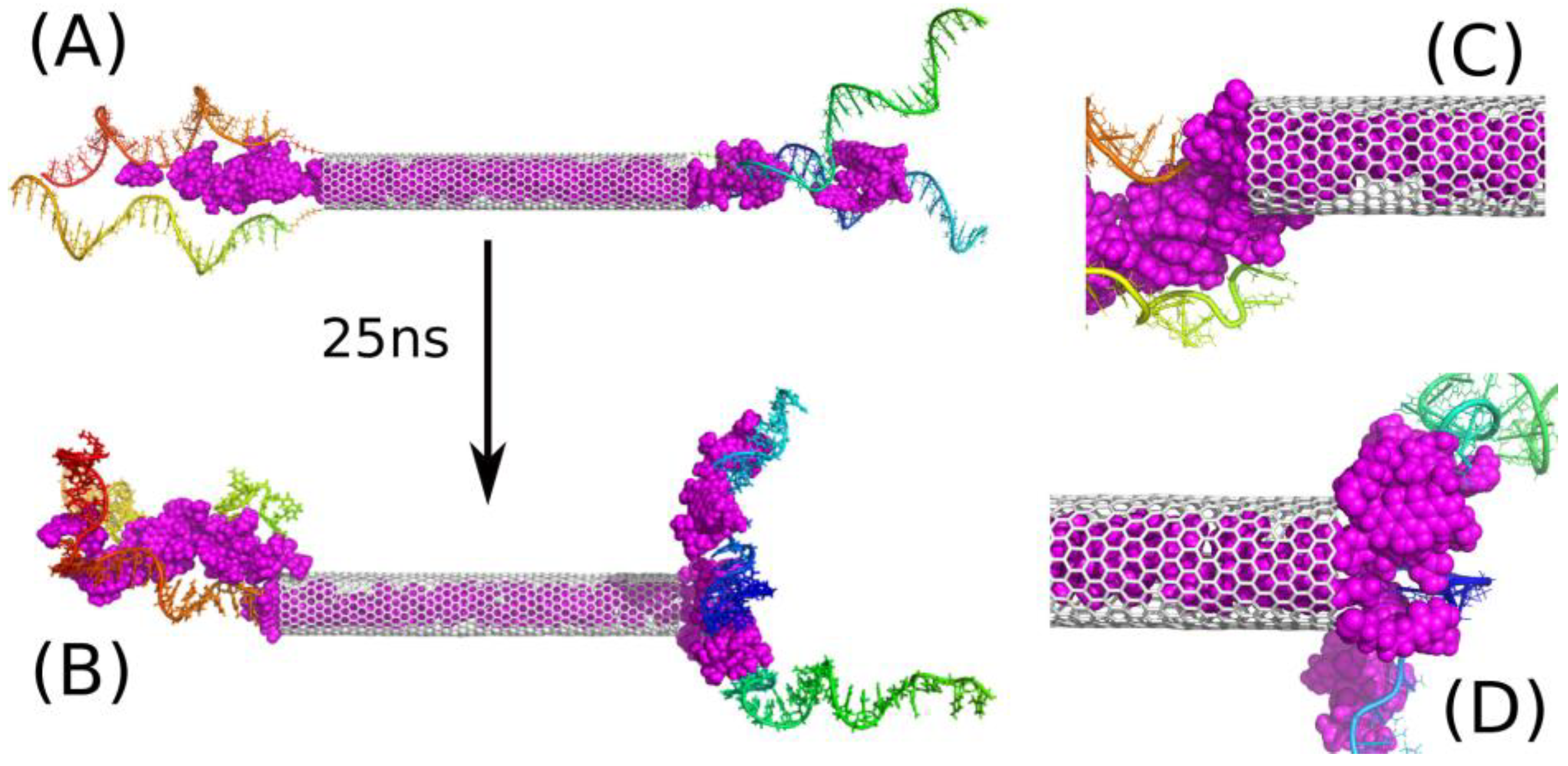

| Name | CNT Length, Å | Number of DNA Chains on Each Side | pH | Number of DOX Molecules | Number of Water Molecules |
|---|---|---|---|---|---|
| E4D20 | 100 | 4 | neutral | 20 | 248,339 |
| E4D40 | 100 | 4 | neutral | 40 | 246,544 |
| E2D40 | 100 | 2 | neutral | 40 | 247,719 |
| I2N | 20 | 2 | neutral | 30 | 101,423 |
| I4N | 20 | 4 | neutral | 30 | 100,296 |
| I2A | 20 | 2 | acidic | 30 | 37,255 |
| I4A | 20 | 4 | acidic | 30 | 36,242 |
| System | EDOX-iM | EDOX-CNT | EDOX-CNT/iM | EDOX-DOX | EDOX-Sol | DOX SASA (Å2) | Nmax |
|---|---|---|---|---|---|---|---|
| I2N | −6169.14 ± 368.39 −4304.12 ± 279.19 * | −332.20 ± 24.05 | −6501.34 ± 369.20 | −13,613.11 ± 169.29 | −9007.70 ± 306.19 | 7900.33 ± 252.70 | 15 ± 0.8 |
| I4N | −7160.94 ± 313.70 −5205.18 ± 247.75 * | −7.36 ± 6.92 | −7168.29 ± 315.23 | −13,880.85 ± 158.42 | −7783.71 ± 335.71 | 6913.92 ± 286.23 | 15 ± 0.1 |
| I2A | −4031.98 ± 323.11 −3396.64 ± 301.49 * | −1581.86 ± 103.39 | −5613.83 ± 328.15 | −13,211.97 ± 173.55 | −10,031.52 ± 259.08 | 8551.25 ± 210.79 | 28 ± 0.2 |
| I4A | −5833.06 ± 238.00 −4427.44 ± 212.68 * | −709.01± 49.42 | −6542.12 ± 236.75 | −13,067.73 ± 158.67 | −8908.34 ± 292.37 | 8443.85 ± 204.07 | 21 ± 8.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolski, P.; Nieszporek, K.; Panczyk, T. Cytosine-Rich DNA Fragments Covalently Bound to Carbon Nanotube as Factors Triggering Doxorubicin Release at Acidic pH. A Molecular Dynamics Study. Int. J. Mol. Sci. 2021, 22, 8466. https://doi.org/10.3390/ijms22168466
Wolski P, Nieszporek K, Panczyk T. Cytosine-Rich DNA Fragments Covalently Bound to Carbon Nanotube as Factors Triggering Doxorubicin Release at Acidic pH. A Molecular Dynamics Study. International Journal of Molecular Sciences. 2021; 22(16):8466. https://doi.org/10.3390/ijms22168466
Chicago/Turabian StyleWolski, Pawel, Krzysztof Nieszporek, and Tomasz Panczyk. 2021. "Cytosine-Rich DNA Fragments Covalently Bound to Carbon Nanotube as Factors Triggering Doxorubicin Release at Acidic pH. A Molecular Dynamics Study" International Journal of Molecular Sciences 22, no. 16: 8466. https://doi.org/10.3390/ijms22168466
APA StyleWolski, P., Nieszporek, K., & Panczyk, T. (2021). Cytosine-Rich DNA Fragments Covalently Bound to Carbon Nanotube as Factors Triggering Doxorubicin Release at Acidic pH. A Molecular Dynamics Study. International Journal of Molecular Sciences, 22(16), 8466. https://doi.org/10.3390/ijms22168466






