TF–RBP–AS Triplet Analysis Reveals the Mechanisms of Aberrant Alternative Splicing Events in Kidney Cancer: Implications for Their Possible Clinical Use as Prognostic and Therapeutic Biomarkers
Abstract
:1. Introduction
2. Results
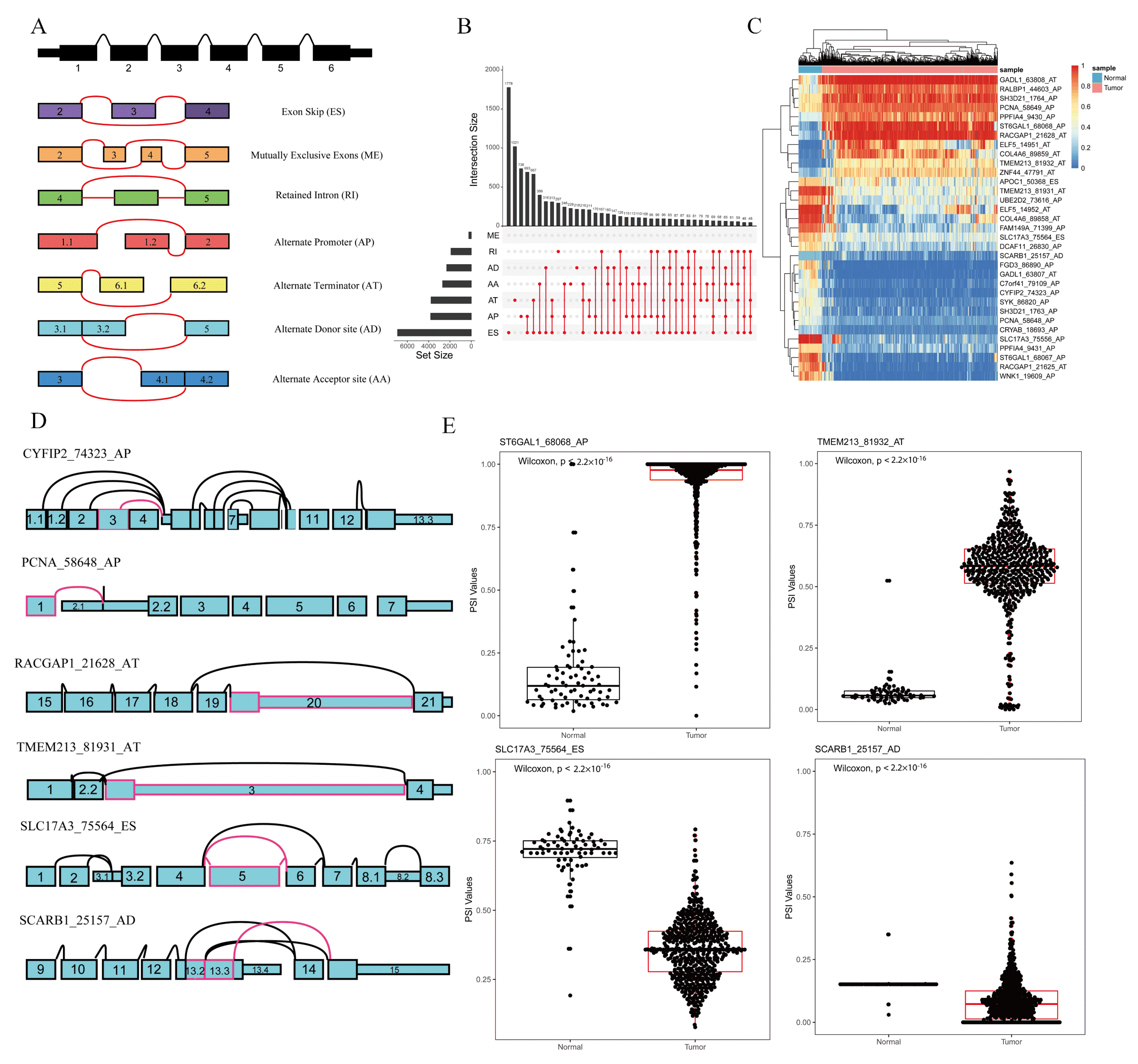
2.1. Screening of Key ASEs in Kidney Cancer
2.2. Identification of Differentially Expressed RBPs and TFs in Kidney Cancer
2.3. Category of TF Action
2.4. Detection of TF–RBP–AS Triplets in Kidney Cancer
2.5. Construction of Splicing-Regulatory Network for Triplets
2.6. Survival and Functional-Enrichment Analyses for Triplets
2.7. Analysis of Splicing Event of PCNA_58648_AP Influenced by Triplets in Kidney Cancer
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Processing
4.2. Identify Cancer-Specific Alternative Splicing Events
4.3. Identification of Differentially Expressed RBPs and TFs
4.4. Construction of TF–RBP–AS Triplets in KIRC
4.5. Protein–Protein Interaction Network Analysis
4.6. GO Functional and KEGG Pathway Enrichment Analyses of Genes
4.7. Establishment of Triplet Signature for KIRC Prognosis
4.8. Statistical Analysis and Software
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Li, W.; Wang, G.; Shi, H.; Wang, K.; Yang, H.; Peng, B. NR1B2 suppress kidney renal clear cell carcinoma (KIRC) progression by regulation of LATS 1/2-YAP signaling. J. Exp. Clin. Cancer Res. 2019, 38, 343. [Google Scholar] [CrossRef]
- Chen, J.; Cao, N.; Li, S.; Wang, Y. Identification of a Risk Stratification Model to Predict Overall Survival and Surgical Benefit in Clear Cell Renal Cell Carcinoma With Distant Metastasis. Front. Oncol. 2021, 11, 630842. [Google Scholar] [CrossRef]
- Capitanio, U.; Montorsi, F. Renal cancer. Lancet 2016, 387, 894–906. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Zhu, B.; Ma, L.; Xing, Q. A Novel Nine Apoptosis-Related Genes Signature Predicting Overall Survival for Kidney Renal Clear Cell Carcinoma and its Associations with Immune Infiltration. Front. Mol. Biosci. 2021, 8, 567730. [Google Scholar] [CrossRef] [PubMed]
- Orlandella, R.M.; Turbitt, W.J.; Gibson, J.T.; Boi, S.K.; Li, P.; Smith, D.L., Jr.; Norian, L.A. The Antidiabetic Agent Acarbose Improves Anti-PD-1 and Rapamycin Efficacy in Preclinical Renal Cancer. Cancers 2020, 12, 2872. [Google Scholar] [CrossRef]
- Roy, B.; Haupt, L.M.; Griffiths, L.R. Alternative splicing (AS) of genes as an approach for generating protein complexity. Curr. Genom. 2013, 14, 182–194. [Google Scholar] [CrossRef] [Green Version]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Le, K.Q.; Prabhakar, B.S.; Hong, W.J.; Li, L.C. Alternative splicing as a biomarker and potential target for drug discovery. Acta Pharmacol. Sin. 2015, 36, 1212–1218. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, C.; Li, L.; Yang, L.; Wang, S.; Ning, X.; Gao, S.; Ren, L.; Chaulagain, A.; Tang, J.; et al. Alternative splicing: An important regulatory mechanism in colorectal carcinoma. Mol. Carcinog. 2021, 60, 279–293. [Google Scholar] [CrossRef]
- Zuo, Y.; Zhang, L.; Tang, W.; Tang, W. Identification of prognosis-related alternative splicing events in kidney renal clear cell carcinoma. J. Cell. Mol. Med. 2019, 23, 7762–7772. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Zou, G.; Cheng, R.; Wang, P.; Ma, K.; Cao, H.; Zhou, W.; Jin, X.; Xu, Z.; Huang, Y.; et al. Alternative splicing associated with cancer stemness in kidney renal clear cell carcinoma. BMC Cancer 2021, 21, 703. [Google Scholar] [CrossRef]
- Gao, L.; He, R.Q.; Huang, Z.G.; Dang, Y.W.; Gu, Y.Y.; Yan, H.B.; Li, S.H.; Chen, G. Genome-wide Analysis of the Alternative Splicing Profiles Revealed Novel Prognostic Index for Kidney Renal Cell Clear Cell Carcinoma. J. Cancer 2020, 11, 1542–1554. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, S.X.; Rao, X.; Liu, Y. Modulator-Dependent RBPs Changes Alternative Splicing Outcomes in Kidney Cancer. Front. Genet. 2020, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.; Kazan, H.; Cook, K.B.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Gueroussov, S.; Albu, M.; Zheng, H.; Yang, A.; et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013, 499, 172–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.; Lee, Y.; Lee, J.S. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12, 2699. [Google Scholar] [CrossRef]
- Baker, K.E.; Parker, R. Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 2004, 16, 293–299. [Google Scholar] [CrossRef]
- Micale, L.; Muscarella, L.A.; Marzulli, M.; Augello, B.; Tritto, P.; D’Agruma, L.; Zelante, L.; Palumbo, G.; Merla, G. VHL frameshift mutation as target of nonsense-mediated mRNA decay in Drosophila melanogaster and human HEK293 cell line. J. Biomed. Biotechnol. 2009, 2009, 860761. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S. Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression. Int. J. Dev. Neurosci. 2016, 55, 102–108. [Google Scholar] [CrossRef]
- García-Moreno, J.F.; Romão, L. Perspective in Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay. Int. J. Mol. Sci. 2020, 21, 9424. [Google Scholar] [CrossRef]
- Hong, M.; Zhang, Z.; Chen, Q.; Lu, Y.; Zhang, J.; Lin, C.; Zhang, F.; Zhang, W.; Li, X.; Zhang, W.; et al. IRF1 inhibits the proliferation and metastasis of colorectal cancer by suppressing the RAS-RAC1 pathway. Cancer Manag. Res. 2019, 11, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morais, C.; Gobe, G.; Johnson, D.W.; Healy, H. The emerging role of nuclear factor kappa B in renal cell carcinoma. Int. J. Biochem. Cell Biol. 2011, 43, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. Ubiquitin- and ubiquitin-like proteins-conjugating enzymes (E2s) in breast cancer. Mol. Biol. Rep. 2013, 40, 2019–2034. [Google Scholar] [CrossRef] [Green Version]
- Repana, D.; Nulsen, J.; Dressler, L.; Bortolomeazzi, M.; Venkata, S.K.; Tourna, A.; Yakovleva, A.; Palmieri, T.; Ciccarelli, F.D. The Network of Cancer Genes (NCG): A comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Kim, P.; Mitra, R.; Zhao, J.; Zhao, Z. TSGene 2.0: An updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016, 44, D1023–D1031. [Google Scholar] [CrossRef]
- Pramanik, S.; Sur, S.; Bankura, B.; Panda, C.K.; Pal, D.K. Expression of proliferating cell nuclear antigen and Ki-67 in renal cell carcinoma in eastern Indian patients. Int. Surg. J. 2019, 6. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Rao, X.; Wang, Y.; Feng, W.; Liang, H.; Liu, Y. Roles of alternative splicing in modulating transcriptional regulation. BMC Syst. Biol. 2017, 11, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, Z.; Wang, Y.; Zhao, Z.; Zhang, J.; Lu, J.; Xu, J.; Li, X. Identification and characterization of lncRNA mediated transcriptional dysregulation dictates lncRNA roles in glioblastoma. Oncotarget 2016, 7, 45027–45041. [Google Scholar] [CrossRef] [Green Version]
- Wrzesinski, T.; Szelag, M.; Cieslikowski, W.A.; Ida, A.; Giles, R.; Zodro, E.; Szumska, J.; Pozniak, J.; Kwias, Z.; Bluyssen, H.A.; et al. Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer 2015, 15, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapinskas, E.J.; Svobodova, S.; Davis, I.D.; Cebon, J.; Hertzog, P.J.; Pritchard, M.A. The Ets transcription factor ELF5 functions as a tumor suppressor in the kidney. Twin Res. Hum. Genet. 2011, 14, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Jinesh, G.G.; Kamat, A.M. RalBP1 and p19-VHL play an oncogenic role, and p30-VHL plays a tumor suppressor role during the blebbishield emergency program. Cell Death Discov. 2017, 3, 17023. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Hwang, K.H.; Eom, M.; Kim, M.; Park, E.Y.; Jeong, Y.; Park, K.S.; Cha, S.K. WNK1 promotes renal tumor progression by activating TRPC6-NFAT pathway. FASEB J. 2019, 33, 8588–8599. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Mitchell, T.J.; Braga, F.A.V.; Tran, M.G.; Stewart, B.J.; Ferdinand, J.R.; Collord, G.; Botting, R.A.; Popescu, D.M.; Loudon, K.W.; et al. Single- cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 2018, 599, 594–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Miao, C.; Hou, C.; Wang, Z.; Liu, B. Apolipoprotein C1 (APOC1): A Novel Diagnostic and Prognostic Biomarker for Clear Cell Renal Cell Carcinoma. Front. Oncol. 2020, 10, 1436. [Google Scholar] [CrossRef]
- Pospiech, E.; Ligeza, J.; Wilk, W.; Golas, A.; Jaszczynski, J.; Stelmach, A.; Rys, J.; Blecharczyk, A.; Wojas-Pelc, A.; Jura, J.; et al. Variants of SCARB1 and VDR Involved in Complex Genetic Interactions May Be Implicated in the Genetic Susceptibility to Clear Cell Renal Cell Carcinoma. Biomed. Res. Int. 2015, 2015, 860405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; He, J.; Su, C.; Wang, H.; Chen, Y.; Guo, W.; Li, Y.; Ding, G. LINC00461 affects the survival of patients with renal cell carcinoma by acting as a competing endogenous RNA for microRNA942. Oncol. Rep. 2019, 42, 1924–1934. [Google Scholar] [CrossRef]
- Dimberg, A.; Rylova, S.; Dieterich, L.C.; Olsson, A.K.; Schiller, P.; Wikner, C.; Bohman, S.; Botling, J.; Lukinius, A.; Wawrousek, E.F.; et al. alphaB-crystallin promotes tumor angiogenesis by increasing vascular survival during tube morphogenesis. Blood 2008, 111, 2015–2023. [Google Scholar] [CrossRef] [Green Version]
- Murata, T.; Katayama, K.; Oohashi, T.; Jahnukainen, T.; Yonezawa, T.; Sado, Y.; Ishikawa, E.; Nomura, S.; Tryggvason, K.; Ito, M. COL4A6 is dispensable for autosomal recessive Alport syndrome. Sci. Rep. 2016, 6, 29450. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Zhao, S.; Xu, S.; Sun, Z.; Liang, Y.; Ding, X. RacGAP1 ameliorates acute kidney injury by promoting proliferation and suppressing apoptosis of renal tubular cells. Biochem. Biophys. Res. Commun. 2020, 527, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Lee, J.W.; Lee, N.; Lee, M.; Kim, H.S.; Chung, H.H.; Kim, J.W.; Park, N.H.; Song, Y.S.; Seo, J.S. LYL1 gene amplification predicts poor survival of patients with uterine corpus endometrial carcinoma: Analysis of the Cancer genome atlas data. BMC Cancer 2018, 18, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Shan, Z.; Yang, H.; Xu, J.; Li, W.; Guo, F. RelB plays an oncogenic role and conveys chemo-resistance to DLD-1 colon cancer cells. Cancer Cell Int. 2018, 18, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, B.; Qi, G.; Sun, X.; Tang, F.; Yuan, S.; Wang, Z.; Liang, X.; Li, B.; Yu, S.; Liu, J.; et al. HOXA7 plays a critical role in metastasis of liver cancer associated with activation of Snail. Mol. Cancer 2016, 15, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.D.; Feng, Q.C.; Qi, Y.; Cui, G.; Zhao, S. PPARGC1A is upregulated and facilitates lung cancer metastasis. Exp. Cell Res. 2017, 359, 356–360. [Google Scholar] [CrossRef]
- Ma, J.B.; Bai, J.Y.; Zhang, H.B.; Gu, L.; He, D.; Guo, P. Downregulation of Collagen COL4A6 Is Associated with Prostate Cancer Progression and Metastasis. Genet. Test. Mol. Biomarkers 2020, 24, 399–408. [Google Scholar] [CrossRef]
- Wu, T.N.; Chen, C.K.; Liu, I.C.; Wu, L.S.; Cheng, A.T. Effects of GADL1 overexpression on cell migration and the associated morphological changes. Sci. Rep. 2019, 9, 5298. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, M.J.; Stang, M.T.; Liu, Y.; Yan, J.; Pizzoferrato, E.; Yim, J.H. IRF-1 inhibits NF-kappaB activity, suppresses TRAF2 and cIAP1 and induces breast cancer cell specific growth inhibition. Cancer Biol. Ther. 2015, 16, 1029–1041. [Google Scholar] [CrossRef] [Green Version]
- Keller, U.; Huber, J.; Nilsson, J.A.; Fallahi, M.; Hall, M.A.; Peschel, C.; Cleveland, J.L. Myc suppression of Nfkb2 accelerates lymphomagenesis. BMC Cancer 2010, 10, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, G.; Cui, K.; Zhang, Z.; Zhao, K. Division of labor between IRF1 and IRF2 in regulating different stages of transcriptional activation in cellular antiviral activities. Cell Biosci. 2015, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Brandt, M.; Hellmuth, S.K.; Ziosi, M.; Gokden, A.; Wolman, A.; Lam, N.; Recinos, Y.; Hornung, V.; Schumacher, J.; Lappalainen, T. An autoimmune disease risk variant has a trans master regulatory effect mediated by IRF1 under immune stimulation. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.F.; Jiang, G.; Xu, K.; Zheng, J.N. Function and mechanism by which interferon regulatory factor-1 inhibits oncogenesis. Oncol. Lett. 2013, 5, 417–423. [Google Scholar] [CrossRef] [Green Version]
- Hayden, M.S.; Ghosh, S. NF-kappaB in immunobiology. Cell. Res. 2011, 21, 223–244. [Google Scholar] [CrossRef] [Green Version]
- Kawai, T.; Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Attaix, D.; Taillandier, D. Role of E2-Ub-conjugating enzymes during skeletal muscle atrophy. Front. Physiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.; Wang, H.; Xia, L.; Lin, R.; Li, T.; Guan, C.; Liu, T. miR-30b-5p releases HMGB1 via UBE2D2/KAT2B/HMGB1 pathway to promote pro-inflammatory polarization and recruitment of macrophages. Atherosclerosis 2021, 324, 38–45. [Google Scholar] [CrossRef]
- Sacramento, L.A.; Benevides, L.; Maruyama, S.R.; Tavares, L.; Fukutani, K.F.; Francozo, M.; Sparwasser, T.; Cunha, F.Q.; Almeida, R.P.; da Silva, J.S.; et al. TLR4 abrogates the Th1 immune response through IRF1 and IFN-beta to prevent immunopathology during L. infantum infection. PLoS Pathog. 2020, 16, e1008435. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Pan, P.; Su, X.; Zhang, L.; Qin, Q.; Tan, H.; Huang, L.; Li, Y. Interferon Regulatory Factor-1 Mediates Alveolar Macrophage Pyroptosis During LPS-Induced Acute Lung Injury in Mice. Shock 2016, 46, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Liu, W.; Zhang, H.M.; Xie, G.Y.; Miao, Y.R.; Xia, M.; Guo, A.Y. hTFtarget: A Comprehensive Database for Regulations of Human Transcription Factors and Their Targets. Genom. Proteom. Bioinform. 2020, 18, 120–128. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Paz, I.; Kosti, I.; Ares, M., Jr.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef]
- Parker, T.M.; Smith, E.M.; Ritchie, J.M.; Haugen, T.H.; Vonka, V.; Turek, L.P.; Hamsikova, E. Head and neck cancer associated with herpes simplex virus 1 and 2 and other risk factors. Oral Oncol. 2006, 42, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Stanziale, S.F.; Petrowsky, H.; Adusumilli, P.S.; Ben Porat, L.; Gonen, M.; Fong, Y. Infection with oncolytic herpes simplex virus-1 induces apoptosis in neighboring human cancer cells: A potential target to increase anticancer activity. Clin. Cancer Res. 2004, 10, 3225–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomasova, D.; Anders, H.J. Cell cycle control in the kidney. Nephrol. Dial. Transplant. 2015, 30, 1622–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hascoet, P.; Chesnel, F.; Le Goff, C.; Le Goff, X.; Arlot-Bonnemains, Y. Unconventional Functions of Mitotic Kinases in Kidney Tumorigenesis. Front. Oncol. 2015, 5, 241. [Google Scholar] [CrossRef] [Green Version]
- Tun, H.W.; Marlow, L.A.; von Roemeling, C.A.; Cooper, S.J.; Kreinest, P.; Wu, K.; Luxon, B.A.; Sinha, M.; Anastasiadis, P.Z.; Copland, J.A. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS ONE 2010, 5, e10696. [Google Scholar] [CrossRef] [PubMed]
- Ruijtenberg, S.; van den Heuvel, S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016, 15, 196–212. [Google Scholar] [CrossRef] [Green Version]
- Ryan, M.; Wong, W.C.; Brown, R.; Akbani, R.; Su, X.; Broom, B.; Melott, J.; Weinstein, J. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016, 44, D1018–D1022. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, W.; Wang, X.; Xiao, W.; Zhu, H. A Generic Sure Independence Screening Procedure. J. Am. Stat. Assoc. 2019, 114, 928–937. [Google Scholar] [CrossRef] [PubMed]





| Symbol | Gene | AS Type | Exons | From Exon | To Exon | Mean_T | Mean_N | Mi Value |
|---|---|---|---|---|---|---|---|---|
| Upregulated | ||||||||
| PPFIA4_9430_AP | PPFIA4 | AP | 19.1 | 0.816 | 0.345 | 0.354 | ||
| ST6GAL1_68068_AP | ST6GAL1 | AP | 1 | 0.932 | 0.161 | 0.347 | ||
| RACGAP1_21628_AT | RACGAP1 | AT | 20 | 0.944 | 0.216 | 0.341 | ||
| TMEM213_81932_AT | TMEM213 | AT | 4 | 0.565 | 0.070 | 0.340 | ||
| SH3D21_1764_AP | SH3D21 | AP | 4.1 | 0.924 | 0.600 | 0.338 | ||
| GADL1_63808_AT | GADL1 | AT | 12.2 | 0.986 | 0.549 | 0.338 | ||
| ELF5_14951_AT | ELF5 | AT | 4.2 | 0.648 | 0.026 | 0.336 | ||
| PCNA_58649_AP | PC | AP | 2.1 | 0.884 | 0.631 | 0.333 | ||
| ZNF44_47791_AT | ZNF44 | AT | 6.2 | 0.652 | 0.177 | 0.328 | ||
| RALBP1_44603_AP | RALBP1 | AP | 2 | 0.878 | 0.455 | 0.325 | ||
| COL4A6_89859_AT | COL4A6 | AT | 52 | 0.708 | 0.054 | 0.322 | ||
| Downregulated | ||||||||
| PPFIA4_9431_AP | PPFIA4 | AP | 1 | 0.180 | 0.655 | 0.354 | ||
| ST6GAL1_68067_AP | ST6GAL1 | AP | 3 | 0.068 | 0.839 | 0.347 | ||
| C7orf41_79109_AP | C7orf41 | AP | 2 | 0.047 | 0.470 | 0.346 | ||
| FGD3_86890_AP | FGD3 | AP | 3 | 0.019 | 0.589 | 0.342 | ||
| RACGAP1_21625_AT | RACGAP1 | AT | 21 | 0.056 | 0.784 | 0.341 | ||
| TMEM213_81931_AT | TMEM213 | AT | 3 | 0.435 | 0.931 | 0.340 | ||
| WNK1_19609_AP | WNK1 | AP | 5 | 0.036 | 0.736 | 0.340 | ||
| SH3D21_1763_AP | SH3D21 | AP | 1 | 0.076 | 0.408 | 0.339 | ||
| GADL1_63807_AT | GADL1 | AT | 15 | 0.014 | 0.451 | 0.338 | ||
| ELF5_14952_AT | ELF5 | AT | 8 | 0.352 | 0.974 | 0.336 | ||
| PCNA_58648_AP | PC | AP | 1 | 0.116 | 0.369 | 0.333 | ||
| SLC17A3_75564_ES | SLC17A3 | ES | 5 | 4 | 6 | 0.358 | 0.707 | 0.331 |
| APOC1_50368_ES | APOC1 | ES | 4:5.1 | 3.2 | 7 | 0.424 | 0.692 | 0.331 |
| SCARB1_25157_AD | SCARB1 | AD | 13.3 | 13.2 | 15 | 0.089 | 0.151 | 0.326 |
| SYK_86820_AP | SYK | AP | 2 | 0.079 | 0.456 | 0.324 | ||
| SLC17A3_75556_AP | SLC17A3 | AP | 2 | 0.199 | 0.970 | 0.323 | ||
| UBE2D2_73616_AP | UBE2D2 | AP | 2 | 0.395 | 0.780 | 0.322 | ||
| COL4A6_89858_AT | COL4A6 | AT | 51.2 | 0.292 | 0.946 | 0.322 | ||
| DCAF11_26830_AP | DCAF11 | AP | 2.1 | 0.262 | 0.621 | 0.321 | ||
| CYFIP2_74323_AP | CYFIP2 | AP | 3 | 0.030 | 0.371 | 0.319 | ||
| CRYAB_18693_AP | CRYAB | AP | 5.1 | 0.060 | 0.224 | 0.318 | ||
| FAM149A_71399_AP | FAM149A | AP | 3 | 0.288 | 0.867 | 0.318 |
| Pattern | PCClow | PCChigh | ΔPCC | Subtype Mode |
|---|---|---|---|---|
| Enhances | – | –– | |PCClow| < |PCChigh| | Strengthens attenuation interaction (SAI) |
| Attenuates | –– | – | |PCClow| > |PCChigh| | Weakens attenuation interaction (WAI) |
| Inverts | + | – | Inverts positive to negative (IPN) | |
| Inverts | – | + | Inverts negative to positive (INP) | |
| Enhances | + | ++ | |PCClow| < |PCChigh| | Strengthens enhancement interaction (SEI) |
| Attenuates | ++ | + | |PCClow| > |PCChigh| | Weakens enhancement interaction (WEI) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, M.; Hu, F. TF–RBP–AS Triplet Analysis Reveals the Mechanisms of Aberrant Alternative Splicing Events in Kidney Cancer: Implications for Their Possible Clinical Use as Prognostic and Therapeutic Biomarkers. Int. J. Mol. Sci. 2021, 22, 8789. https://doi.org/10.3390/ijms22168789
He M, Hu F. TF–RBP–AS Triplet Analysis Reveals the Mechanisms of Aberrant Alternative Splicing Events in Kidney Cancer: Implications for Their Possible Clinical Use as Prognostic and Therapeutic Biomarkers. International Journal of Molecular Sciences. 2021; 22(16):8789. https://doi.org/10.3390/ijms22168789
Chicago/Turabian StyleHe, Meng, and Fuyan Hu. 2021. "TF–RBP–AS Triplet Analysis Reveals the Mechanisms of Aberrant Alternative Splicing Events in Kidney Cancer: Implications for Their Possible Clinical Use as Prognostic and Therapeutic Biomarkers" International Journal of Molecular Sciences 22, no. 16: 8789. https://doi.org/10.3390/ijms22168789
APA StyleHe, M., & Hu, F. (2021). TF–RBP–AS Triplet Analysis Reveals the Mechanisms of Aberrant Alternative Splicing Events in Kidney Cancer: Implications for Their Possible Clinical Use as Prognostic and Therapeutic Biomarkers. International Journal of Molecular Sciences, 22(16), 8789. https://doi.org/10.3390/ijms22168789






