Transcriptome-Wide Identification and Characterization of the JAZ Gene Family in Mentha canadensis L.
Abstract
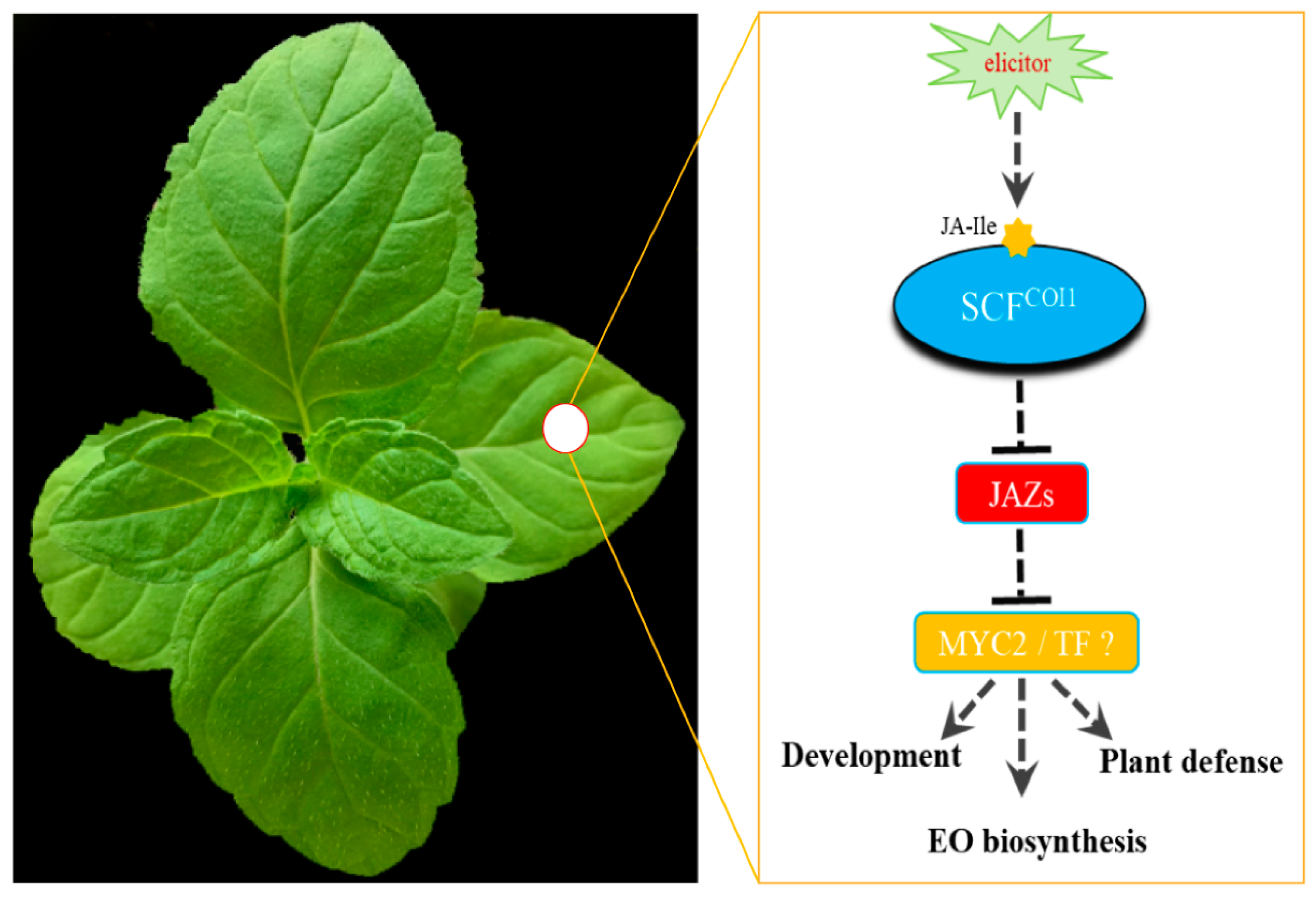
:1. Introduction
2. Results
2.1. Identification and Molecular Cloning of McJAZ Genes from M. canadensis
2.2. Sequence Alignment and Phylogenetic Analysis of McJAZ Genes
2.3. Tissue Expression Patterns of McJAZ Genes
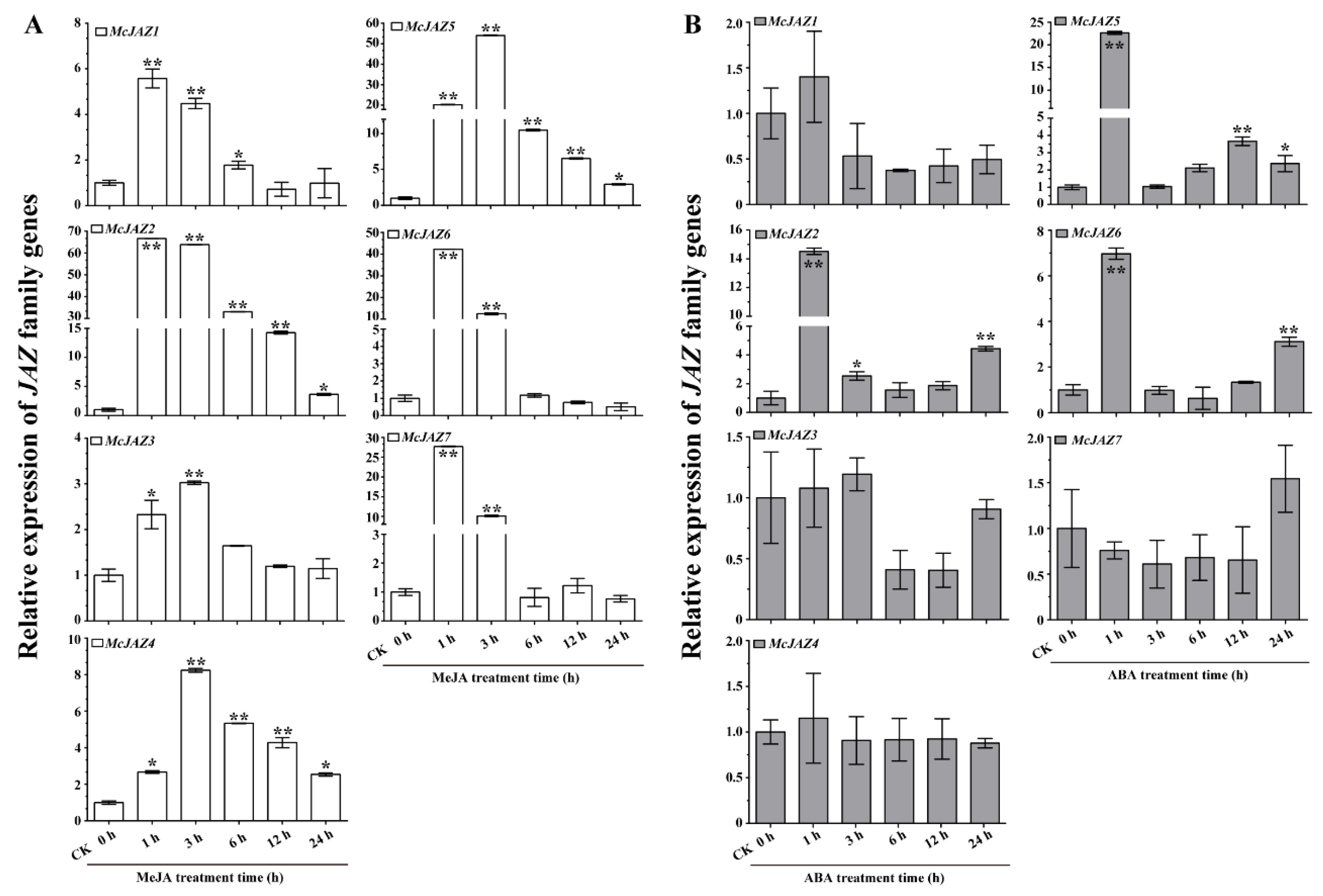
2.4. Expression Profiles of McJAZ Genes in Response to JA and ABA Treatments
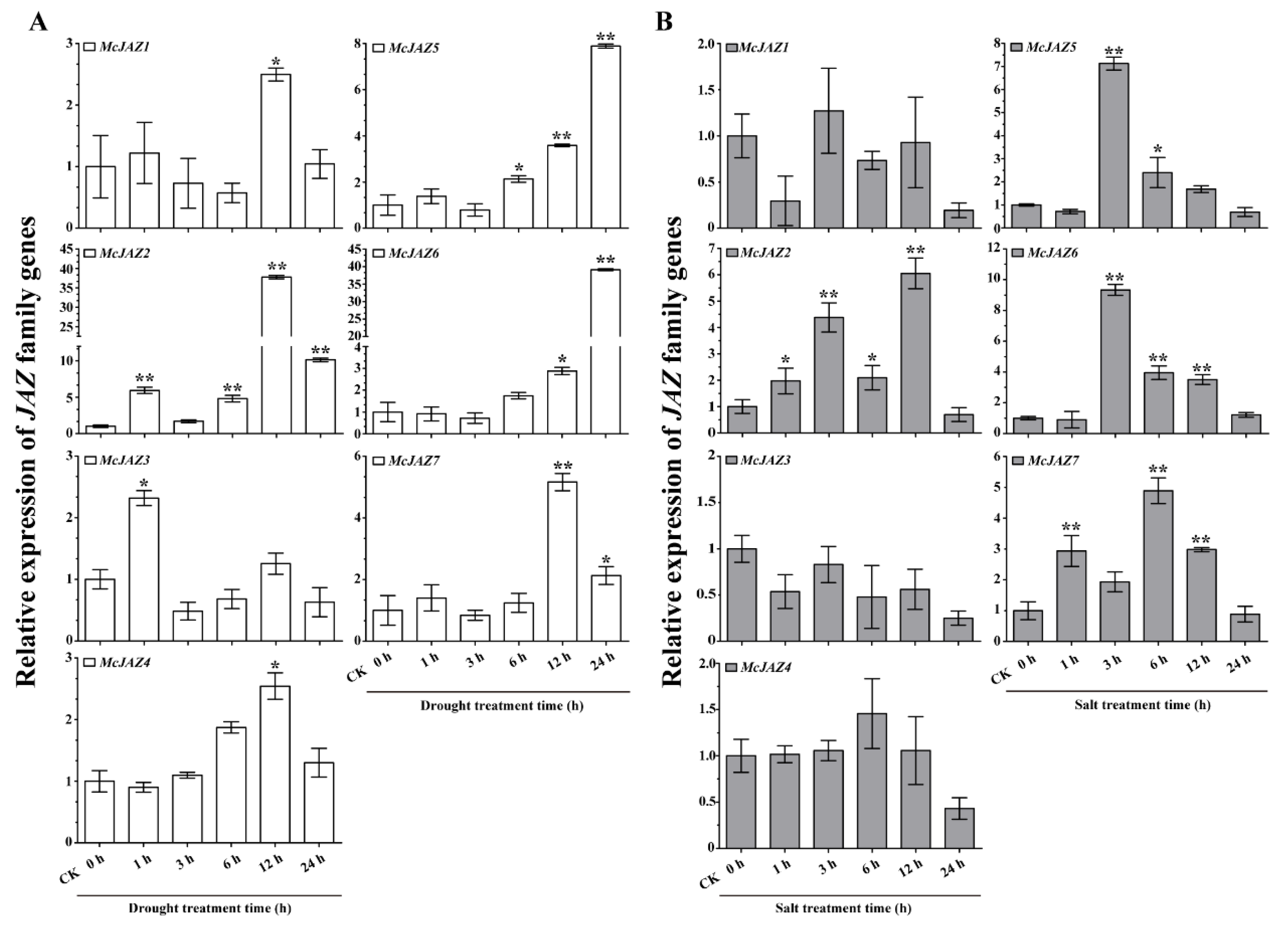
2.5. McJAZ Genes Are Involved in Response to Drought and Salt Stress
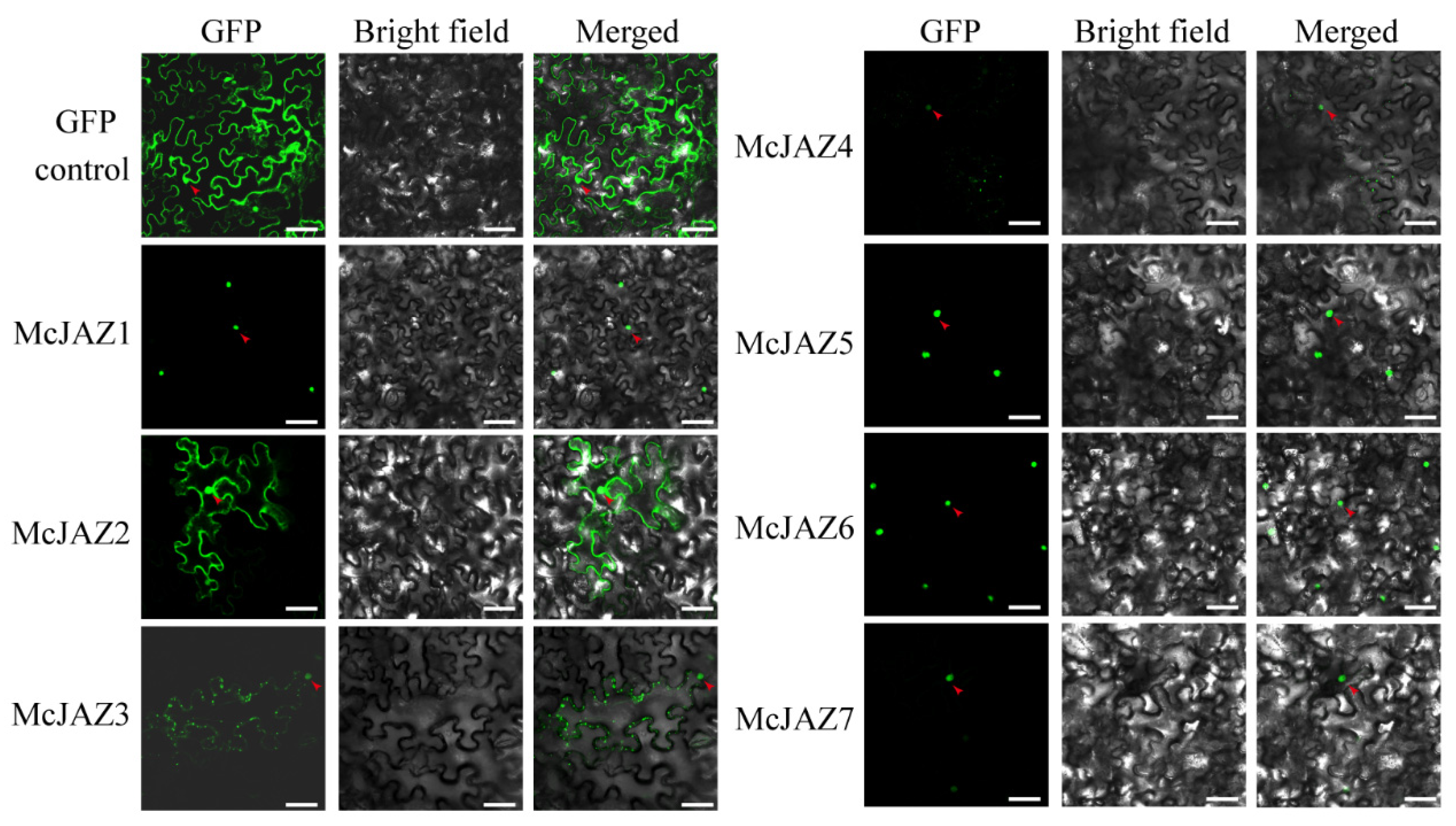
2.6. Subcellular Localization of McJAZ Proteins
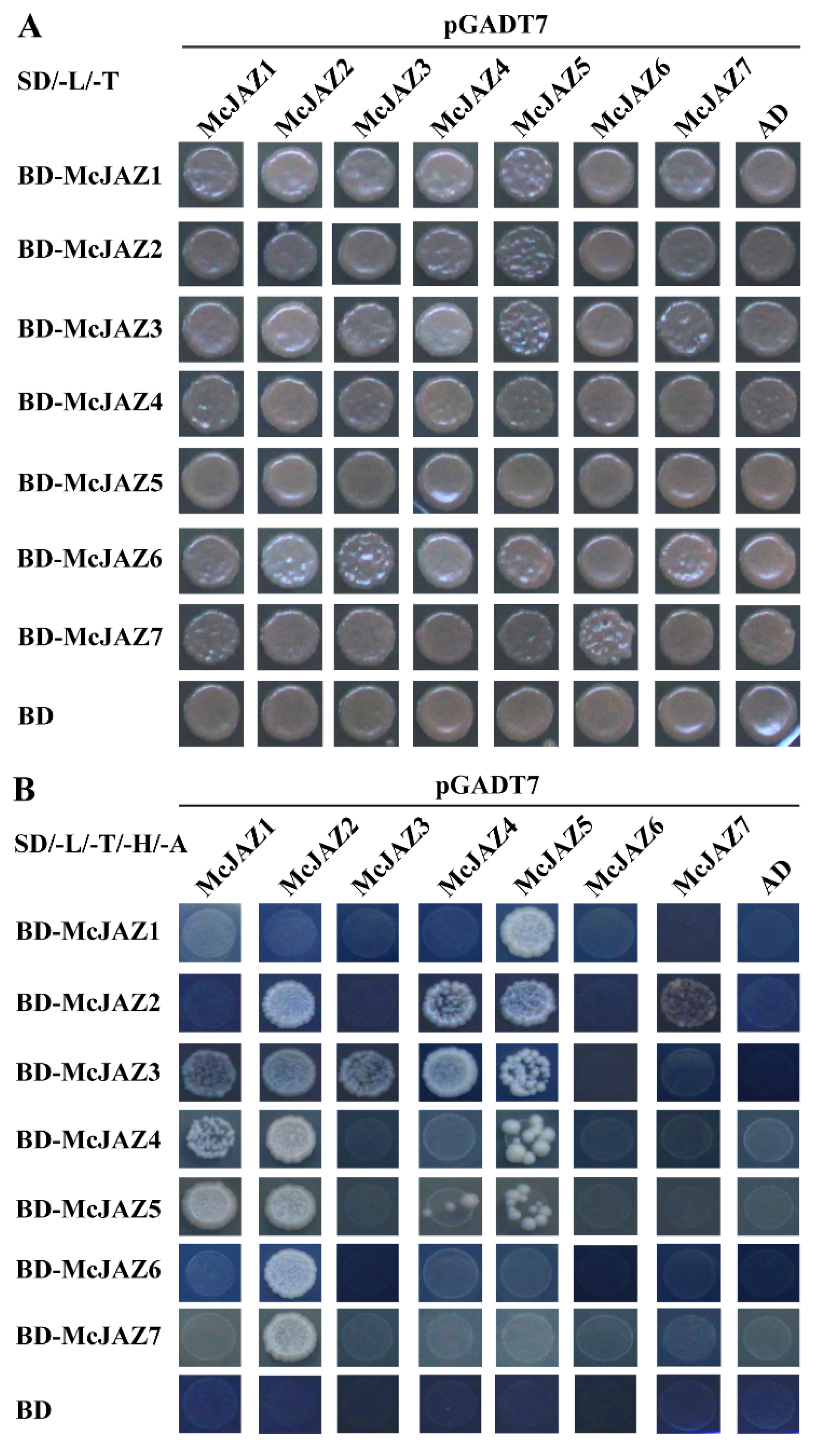
2.7. McJAZ Proteins Interact with McCOI1a
2.8. Homo- and Heterodimeric Interaction of McJAZ Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Isolation and Cloning of M. canadensis McJAZ Family Genes
4.3. Sequence Analysis and Phylogenetic Tree Analysis
4.4. Subcellular Localization of McJAZ Proteins
4.5. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
4.6. Yeast Two-Hybrid (Y2H) Experiments
4.7. Statistical Analysis
4.8. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Yuan, Z.; Zhang, D. Roles of jasmonate signaling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate: An oxylipin signal with many roles in plants. Vitam. Horm. 2005, 72, 431–456. [Google Scholar] [PubMed]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Zhang, M.; Yang, M.; Hu, Y. Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell 2020, 32, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Zhang, Y.; Turner, J.G. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 2008, 3, e3699. [Google Scholar] [CrossRef]
- Reinbothe, C.; Springer, A.; Samol, I.; Reinbothe, S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009, 276, 4666–4681. [Google Scholar] [CrossRef]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in jasmonate signaling for multistress resilience. Ann. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef] [Green Version]
- Basso, V.; Veneault-Fourrey, C. Role of jasmonates in beneficial microbe-root interactions. Methods Mol. Biol. 2020, 2085, 43–67. [Google Scholar]
- Vijayan, P.; Shockey, J.; Lévesque, C.A.; Cook, R.J.; Browse, J. A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 7209–7214. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.N.; Xu, D.B.; Yan, X.; Wu, Z.K.; Kayani, S.I.; Shen, Q.; Fu, X.Q.; Xie, L.H.; Hao, X.L.; Hassani, D.; et al. Jasmonate- and abscisic acid-activated AaGSW1-AaTCP15/AaORA transcriptional cascade promotes artemisinin biosynthesis in Artemisia annua. Plant Biotechnol. J. 2021, 19, 1412–1428. [Google Scholar] [CrossRef]
- Afrin, S.; Huang, J.-J.; Luo, Z.-Y. JA-mediated transcriptional regulation of secondary metabolism in medicinal plants. Sci. Bull. 2015, 60, 1062–1072. [Google Scholar] [CrossRef] [Green Version]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [Green Version]
- van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metab- olism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inzé, D.; Goossens, A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites -pathways, transcription factors and applied aspects—A brief review. N. Biotechnol. 2019, 48, 1–11. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Gfeller, A.; Liechti, R.; Farmer, E.E. Arabidopsis jasmonate signaling pathway. Sci. Signal. 2010, 3, cm4. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [Green Version]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.N.; Xu, D.B.; Li, L.; Zhang, F.; Fu, X.Q.; Shen, Q.; Lyu, X.Y.; Wu, Z.K.; Pan, Q.F.; Shi, P.; et al. Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv. 2018, 4, eaas9357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the Inducer of CBF expression-C-repeat Binding FAC TOR/DRE Binding Factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [Green Version]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef] [PubMed]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [Green Version]
- Cuéllar Pérez, A.; Nagels Durand, A.; Vanden Bossche, R.; De Clercq, R.; Persiau, G.; Van Wees, S.C.M.; Pieterse, C.M.J.; Gevaert, K.; De Jaeger, G.; Goossens, A.; et al. The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS ONE 2014, 9, e84891. [Google Scholar] [CrossRef] [Green Version]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernández-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interac- tions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef]
- Thireault, C.; Shyu, C.; Yoshida, Y.; Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef]
- Shyu, C.; Figueroa, P.; DePew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.E.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genom. 2017, 18, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Zhao, Y.; Jin, J.; Wu, S.; Deng, X.; Chen, Y.; Tian, W.M. Genome-wide identification and characterization of the JAZ gene family in Rubber Tree (Hevea brasiliensis). Front. Genet. 2019, 10, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chini, A.; Ben-Romdhane, W.; Hassairi, A.; Aboul-Soud, M.A.M. Identification of TIFY/JAZ family genes in Solanum lycoper- sicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381. [Google Scholar] [CrossRef]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef] [Green Version]
- Monte, I.; Franco-Zorrilla, J.M.; García-Casado, G.; Zamarreño, A.M.; García-Mina, J.M.; Nishihama, R.; Kohchi, T.; Solano, R. A single JAZ repressor controls the jasmonate pathway in Marchantia polymorpha. Mol. Plant 2019, 12, 185–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanif, M.; Rahman, M.U.; Gao, M.; Yang, J.; Ahmad, B.; Yan, X.; Wang, X. Heterologous expression of the Grapevine JAZ7 gene in Arabidopsis confers enhanced resistance to powdery mildew but not to Botrytis cinerea. Int. J. Mol. Sci. 2018, 19, 3889. [Google Scholar] [CrossRef] [Green Version]
- Jing, Y.; Liu, J.; Liu, P.; Ming, D.; Sun, J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 2019, 9, 5691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef]
- Shi, M.; Zhou, W.; Zhang, J.; Huang, S.; Wang, H.; Kai, G. Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 2016, 6, 20919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheljazkov, V.D.; Stewart, C.N.; Joyce, B.; Baxter, H.; Cantrell, C.L.; Astatkie, T.; Jeliazkova, E.A.; Poovaiah, C.R. Dual utilization of medicinal and aromatic crops as bioenergy feedstocks. J. Agric. Food Chem. 2018, 66, 8744–8752. [Google Scholar] [CrossRef]
- Zhao, D.; Xu, Y.W.; Yang, G.L.; Husaini, A.M.; Wu, W. Variation of essential oil of Mentha haplocalyx Briq. and Mentha spicata L. from China. Ind. Crops Prod. 2013, 42, 251–260. [Google Scholar] [CrossRef]
- Lange, B.M.; Ahkami, A. Metabolic engineering of plant monoterpenes, sesquiterpenes and diterpenes-current status and fu- ture opportunities. Plant Biotechnol. J. 2013, 11, 169–196. [Google Scholar] [CrossRef]
- Heydari, M.; Zanfardino, A.; Taleei, A.; Bushehri, A.A.S.; Hadian, J.; Maresca, V.; Sorbo, S.; Napoli, M.D.; Varcamonti, M.; Basile, A.; et al. Effect of heat stress on yield, monoterpene content and antibacterial activity of essential oils of Mentha x piperita var. Mitcham and Mentha arvensis var. piperascens. Molecules 2018, 23, 1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liu, Y.; Ma, A.; Bao, Y.; Wang, M.; Sun, Z. In vitro antiviral, anti-inflammatory, and antioxidant activities of the ethanol extract of Mentha piperita L. Food Sci. Biotechnol. 2017, 26, 1675–1683. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; Rahier, A.; Camara, B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 2005, 44, 357–429. [Google Scholar] [CrossRef] [PubMed]
- Sukegawa, S.; Shiojiri, K.; Higami, T.; Suzuki, S.; Arimura, G.I. Pest management using mint volatiles to elicit resistance in soy: Mechanism and application potential. Plant J. 2018, 96, 910–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueroa-Pérez, M.G.; Reynoso-Camacho, R.; Garcia-Ortega, L.F.; Guevara-González, R.G. Transcriptome profiling of pep- permint (Mentha piperita) with improved antioxidant properties in response to salicylic acid elicitation. J. Plant Biochem. Biot. 2019, 28, 301–311. [Google Scholar] [CrossRef]
- Joo-Sun, S.; Yung-Jin, C.; Yang-Do, C.; Soo-Un, K. Role of jasmonic acid in biotransformation of (--)-isopiperitenone in suspen- sion cell culture of Mentha piperita. Mol. Cells 1998, 8, 366–369. [Google Scholar]
- Bose, S.K.; Yadav, R.K.; Mishra, S.; Sangwan, R.S.; Singh, A.K.; Mishra, B.; Srivastava, A.K.; Sangwan, N.S. Effect of gibber- ellic acid and calliterpenone on plant growth attributes, trichomes, essential oil biosynthesis and pathway gene expression in differential manner in Mentha arvensis L. Plant Physiol. Biochem. 2013, 66, 150–158. [Google Scholar] [CrossRef]
- Chrysargyris, A.; Papakyriakou, E.; Petropoulos, S.A.; Tzortzakis, N. The combined and single effect of salinity and copper stress on growth and quality of Mentha spicata plants. J. Hazard. Mater. 2019, 368, 584–593. [Google Scholar] [CrossRef]
- Ahkami, A.; Johnson, S.R.; Srividya, N.; Lange, B.M. Multiple levels of regulation determine monoterpenoid essential oil compositional variation in the mint family. Mol. Plant 2015, 8, 188–191. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.A.; Wang, Q.; Dhar, N.; Kumar, N.; Venkatesh, P.N.; Rajan, C.; Panicker, D.; Sridhar, V.; Mao, H.-Z.; Sarojam, R. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol. J. 2017, 15, 1105–1119. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Reddy, V.A.; Panicker, D.; Mao, H.-Z.; Kumar, N.; Rajan, C.; Venkatesh, P.N.; Chua, N.H.; Sarojam, R. Metabolic engineering of terpene biosynthesis in plants using a trichome-specific transcription factor MsYABBY5 from spearmint (Mentha spicata). Plant Biotechnol. J. 2016, 14, 1619–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tafrihi, M.; Imran, M.; Tufail, T.; Gondal, T.A.; Caruso, G.; Sharma, S.; Sharma, R.; Atanassova, M.; Atanassov, L.; Valere Tsouh Fokou, P.; et al. The wonderful activities of the genus Mentha: Not only antioxidant properties. Molecules 2021, 26, 1118. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liang, C.; Chen, J.; Qi, X.; Liu, Y.; Li, W. The effects of salinity stress on morphological characteristics, mineral nutrient accumulation and essential oil yield and composition in Mentha canadensis L. Sci. Hortic. 2015, 197, 579–583. [Google Scholar] [CrossRef]
- Wang, H.T.; Yu, X.; Liu, Y.; Liang, C.Y.; Li, W.L. Analysis of genetic variability and relationships among Mentha L. using the limonene synthase gene, LS. Gene 2013, 524, 246–252. [Google Scholar] [CrossRef]
- Qi, X.; Fang, H.; Yu, X.; Xu, D.; Li, L.; Liang, C.; Lu, H.; Li, W.; Chen, Y.; Chen, Z. Transcriptome analysis of JA signal trans- duction, transcription factors, and monoterpene biosynthesis pathway in response to methyl jasmonate elicitation in Mentha canadensis L. Int. J. Mol. Sci. 2018, 19, 2364. [Google Scholar] [CrossRef] [Green Version]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Yu, S.; Han, X.; Xu, J.; He, Q.; Xu, S.; Wang, R. Identification, molecular characterization and expression of JAZ genes in Lycoris aurea. PLoS ONE 2020, 15, e0230177. [Google Scholar] [CrossRef]
- Garrido-Bigotes, A.; Valenzuela-Riffo, F.; Torrejón, M.; Solano, R.; Morales-Quintana, L.; Figueroa, C.R. A new functional JAZ degron sequence in strawberry JAZ1 revealed by structural and interaction studies on the COI1-JA-Ile/COR-JAZs complexes. Sci. Rep. 2020, 10, 11310. [Google Scholar] [CrossRef] [PubMed]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Xie, D. Jasmonate in plant defence: Sentinel or double agent? Plant Biotechnol. J. 2015, 13, 1233–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Zou, Z.; Xing, H.; Duan, Y.; Zhu, X.; Ma, Y.; Wang, Y.; Fang, W. Genome-wide analysis reveals stress and hormone responsive patterns of JAZ family genes in Camellia Sinensis. Int. J. Mol. Sci. 2020, 21, 2433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunewald, W.; Vanholme, B.; Pauwels, L.; Plovie, E.; Inzé, D.; Gheysen, G.; Goossens, A. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009, 10, 923–928. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.S.; Cooke, T.F.; DePew, C.L.; Patel, L.C.; Ogawa, N.; Kobayashi, Y.; Howe, G.A. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 2010, 63, 613–622. [Google Scholar] [CrossRef] [Green Version]
- Saha, G.; Park, J.I.; Kayum, M.A.; Nou, I.S. A Genome-wide analysis reveals stress and hormone responsive patterns of TIFY family genes in Brassica rapa. Front. Plant Sci. 2016, 7, 936. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, F.; Fernández-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef]
- Hsouna, A.B.; Touj, N.; Hammami, I.; Dridi, K.; Al-Ayed, A.S.; Hamdi, N. Chemical composition and in vivo efficacy of the essential oil of Mentha piperita L. in the suppression of crown gall disease on tomato plants. J. Oleo Sci. 2019, 68, 419–426. [Google Scholar] [CrossRef]
- Pang, X.; Feng, Y.X.; Qi, X.J.; Wang, Y.; Almaz, B.; Xi, C.; Du, S.S. Toxicity and repellent activity of essential oil from Mentha piperita Linn. leaves and its major monoterpenoids against three stored product insects. Environ. Sci. Pollut. Res. Int. 2020, 27, 7618–7627. [Google Scholar] [CrossRef]
- Liu, X.J.; An, X.H.; Liu, X.; Hu, D.G.; Wang, X.F.; You, C.X.; Hao, Y.J. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J. Exp. Bot. 2017, 68, 2977–2990. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.N.; Chen, M.; Xu, D.B.; Fang, G.N.; Wang, E.H.; Gao, S.Q.; Xu, Z.S.; Li, L.C.; Zhang, X.H.; Min, D.H.; et al. G-protein β subunit AGB1 positively regulates salt stress tolerance in Arabidopsis. J. Integr. Agr. 2015, 14, 314–325. [Google Scholar] [CrossRef]
- Xu, D.B.; Chen, M.; Ma, Y.N.; Xu, Z.S.; Li, L.C.; Chen, Y.F.; Ma, Y.Z. A G-protein β subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis. PLoS ONE 2015, 10, e0116385. [Google Scholar] [CrossRef]
- Tian, S.; Liu, S.; Wang, Y.; Wang, K.; Yin, C.; Yue, Y.; Hu, H. Genome-wide identification and characterization of JAZ protein family in two Petunia Progenitors. Plants 2019, 8, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Q.; Zhang, L.; Liao, Z.; Wang, S.; Yan, T.; Shi, P.; Liu, M.; Fu, X.; Pan, Q.; Wang, Y.; et al. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant 2018, 11, 776–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strat- egies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.B.; Gao, S.Q.; Ma, Y.N.; Wang, X.T.; Feng, L.; Li, L.C.; Xu, Z.S.; Chen, Y.F.; Chen, M.; Ma, Y.Z. The G-protein β subunit AGB1 promotes hypocotyl elongation through inhibiting transcription activation function of BBX21 in Arabidopsis. Mol. Plant 2017, 10, 1206–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bender, C.L. Chlorosis-inducing phytotoxins produced by Pseudomonas syringae. Eur. J. Plant Pathol. 1999, 105, 1–12. [Google Scholar] [CrossRef]
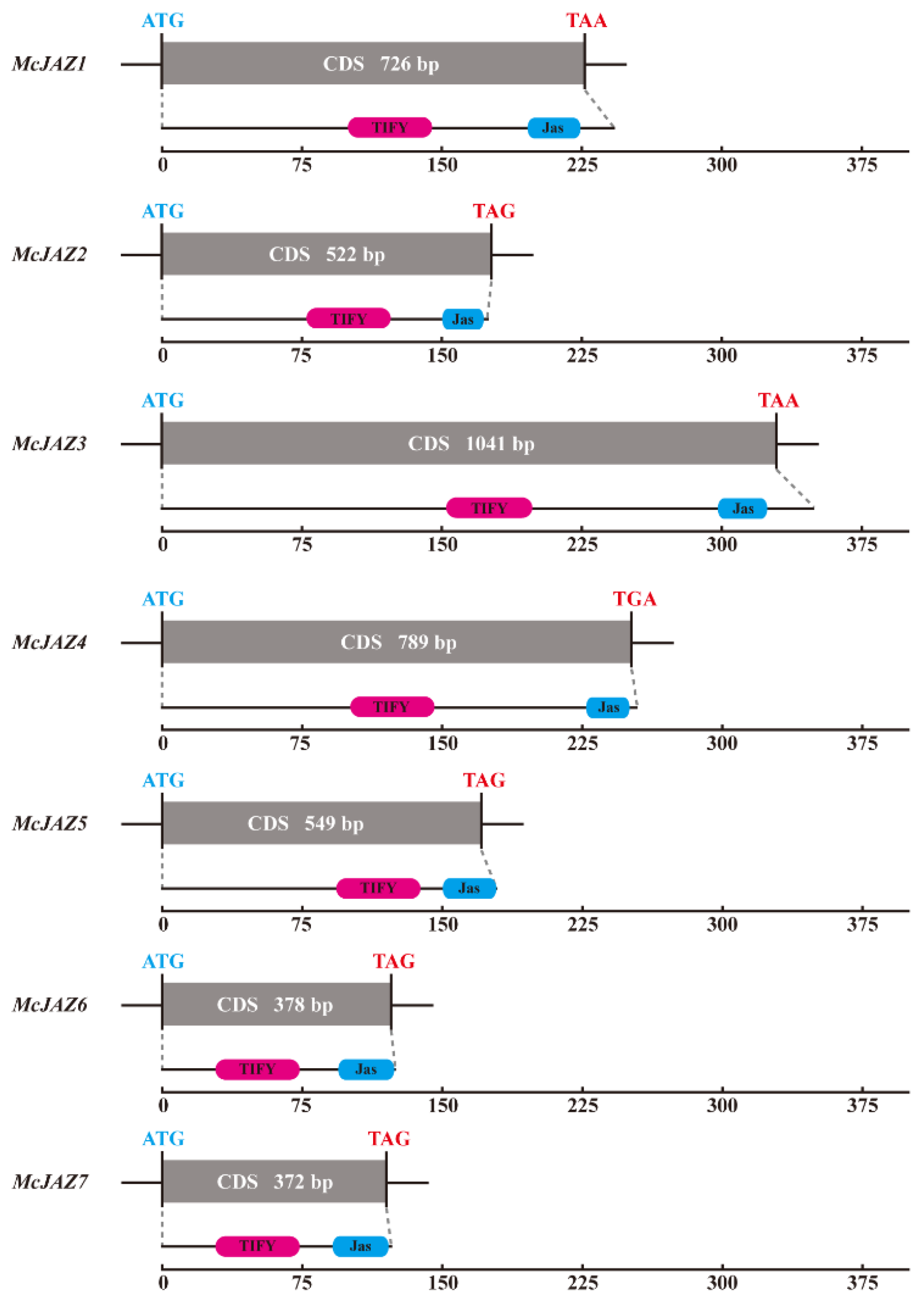
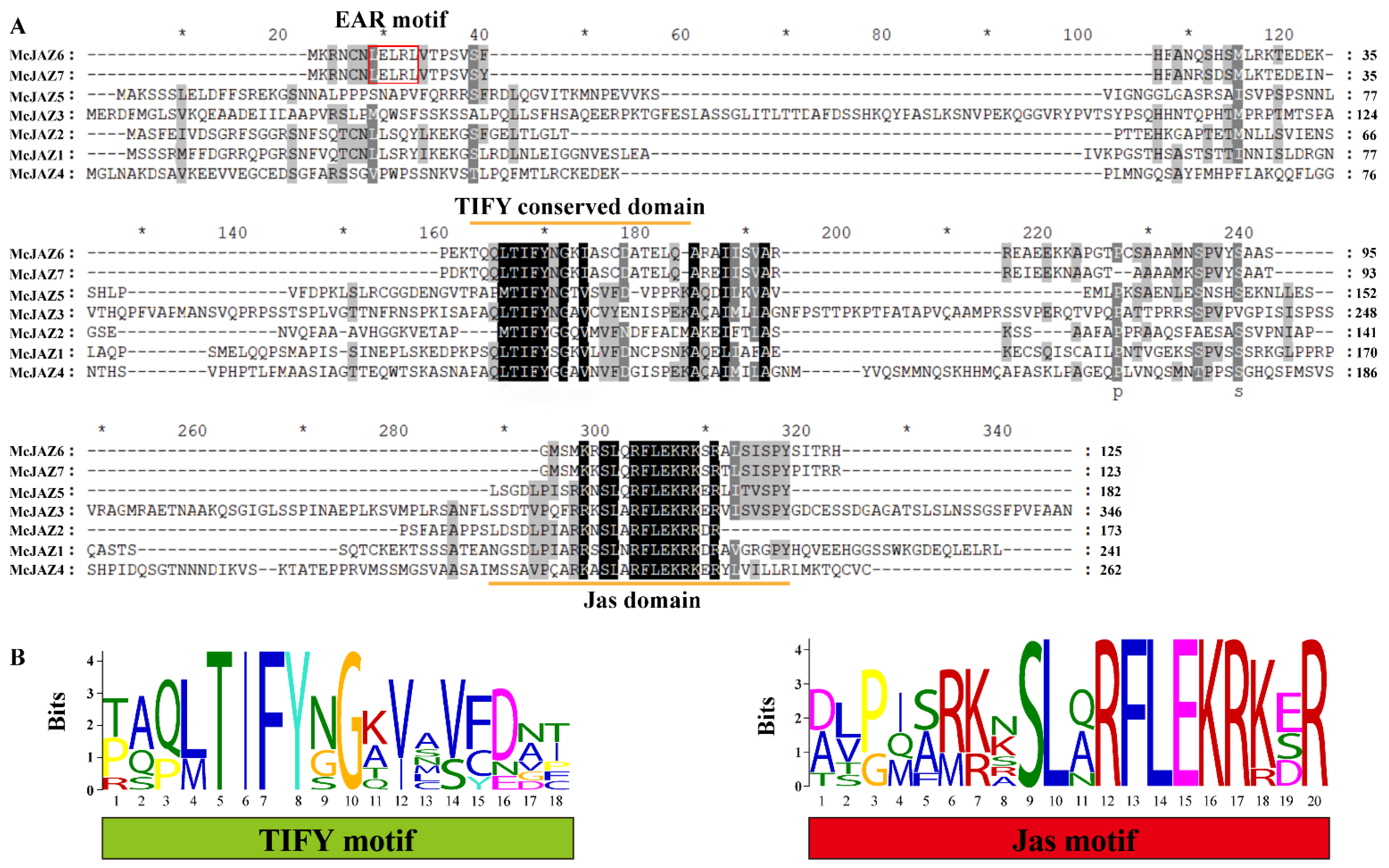
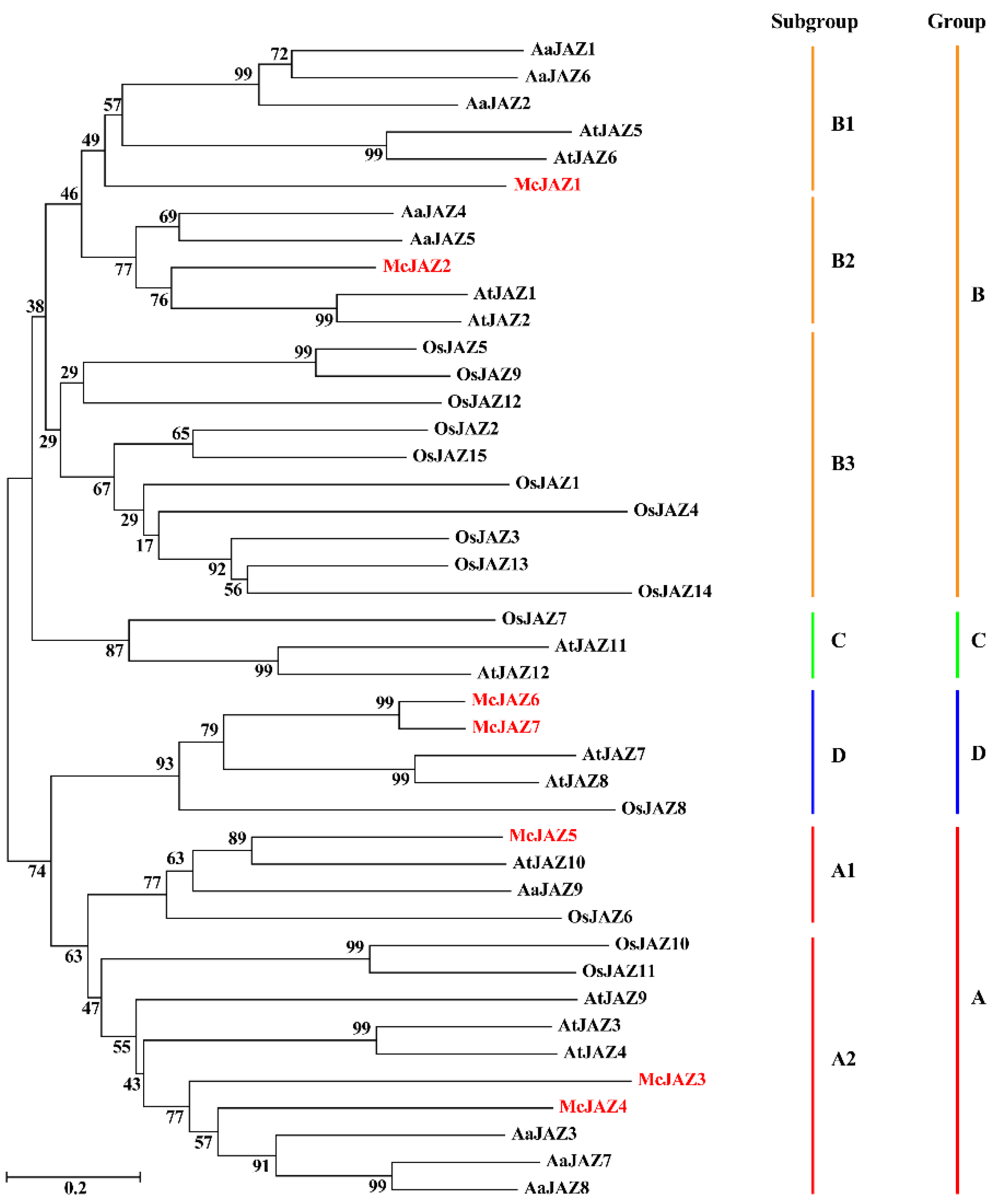
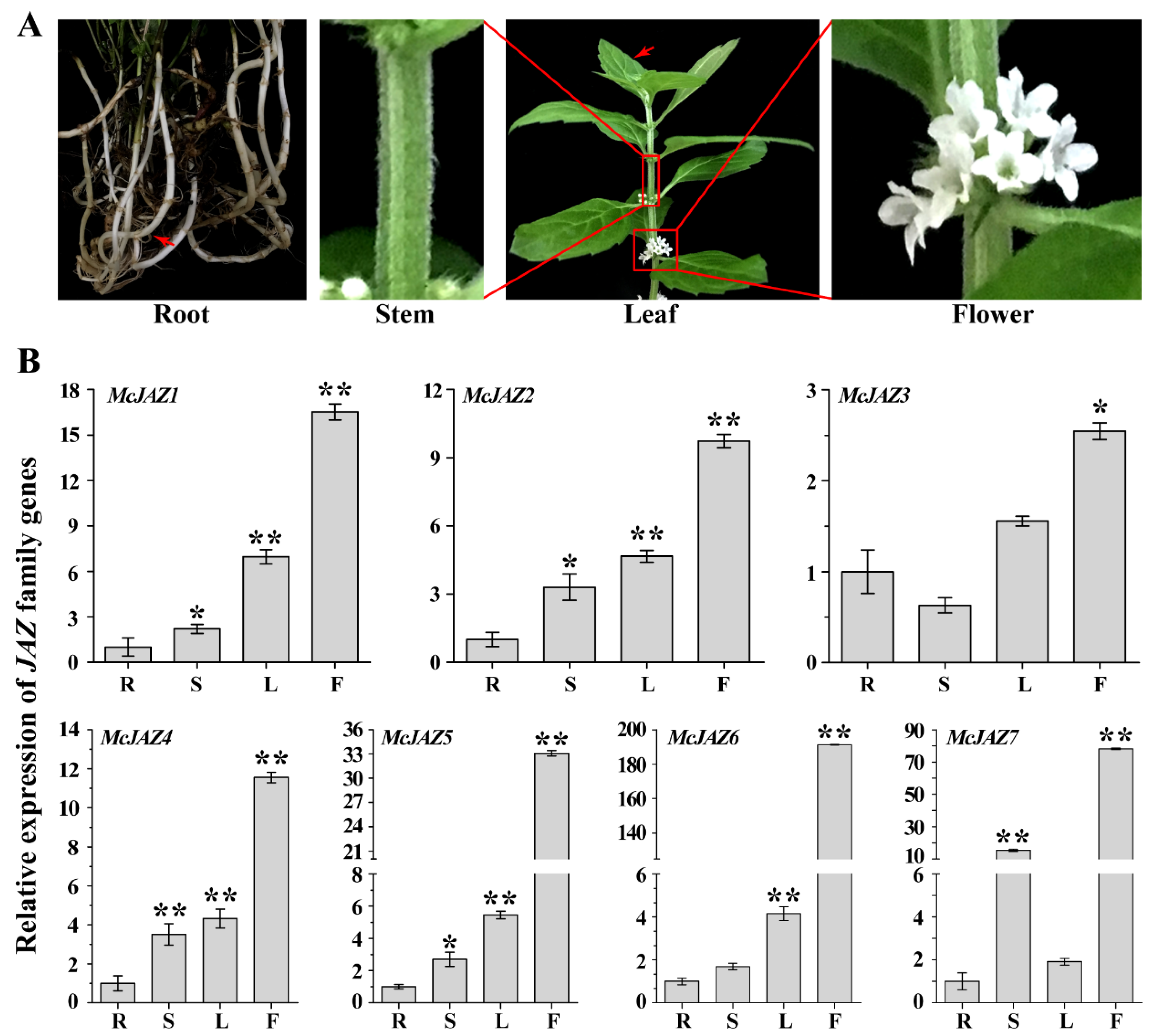

| Gene Name | Genbank Accession No. 1 | Length CDs 2 | Amino Acids 3 | MW (kDa) 4 | pI 5 | TIFY Motif 6 | Jas Motif (aa) 7 | Protein Localization |
|---|---|---|---|---|---|---|---|---|
| McJAZ1 | MZ229995 | 726 | 241 | 26.30 | 8.98 | TIFYSG | 196–221 | Nuclei |
| McJAZ2 | MZ229996 | 522 | 173 | 18.35 | 6.12 | TIFYGG | 156–173 | Nuclei |
| McJAZ3 | MZ229997 | 1041 | 346 | 36.89 | 9.75 | TIFYNG | 294–319 | Nuclei |
| McJAZ4 | MZ229998 | 789 | 262 | 28.09 | 9.17 | TIFYGG | 230–249 | Nuclei |
| McJAZ5 | MZ229999 | 549 | 182 | 19.93 | 9.76 | TIFYNG | 158–182 | Nuclei |
| McJAZ6 | MZ230000 | 378 | 125 | 14.02 | 9.96 | TIFYNG | 93–121 | Nuclei |
| McJAZ7 | MZ230001 | 372 | 123 | 13.87 | 9.77 | TIFYNG | 91–119 | Nuclei |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.-B.; Ma, Y.-N.; Qin, T.-F.; Tang, W.-L.; Qi, X.-W.; Wang, X.; Liu, R.-C.; Fang, H.-L.; Chen, Z.-Q.; Liang, C.-Y.; et al. Transcriptome-Wide Identification and Characterization of the JAZ Gene Family in Mentha canadensis L. Int. J. Mol. Sci. 2021, 22, 8859. https://doi.org/10.3390/ijms22168859
Xu D-B, Ma Y-N, Qin T-F, Tang W-L, Qi X-W, Wang X, Liu R-C, Fang H-L, Chen Z-Q, Liang C-Y, et al. Transcriptome-Wide Identification and Characterization of the JAZ Gene Family in Mentha canadensis L. International Journal of Molecular Sciences. 2021; 22(16):8859. https://doi.org/10.3390/ijms22168859
Chicago/Turabian StyleXu, Dong-Bei, Ya-Nan Ma, Teng-Fei Qin, Wei-Lin Tang, Xi-Wu Qi, Xia Wang, Rui-Cen Liu, Hai-Ling Fang, Ze-Qun Chen, Cheng-Yuan Liang, and et al. 2021. "Transcriptome-Wide Identification and Characterization of the JAZ Gene Family in Mentha canadensis L." International Journal of Molecular Sciences 22, no. 16: 8859. https://doi.org/10.3390/ijms22168859
APA StyleXu, D.-B., Ma, Y.-N., Qin, T.-F., Tang, W.-L., Qi, X.-W., Wang, X., Liu, R.-C., Fang, H.-L., Chen, Z.-Q., Liang, C.-Y., & Wu, W. (2021). Transcriptome-Wide Identification and Characterization of the JAZ Gene Family in Mentha canadensis L. International Journal of Molecular Sciences, 22(16), 8859. https://doi.org/10.3390/ijms22168859







