Impact of Global Transcriptional Silencing on Cell Cycle Regulation and Chromosome Segregation in Early Mammalian Embryos
Abstract
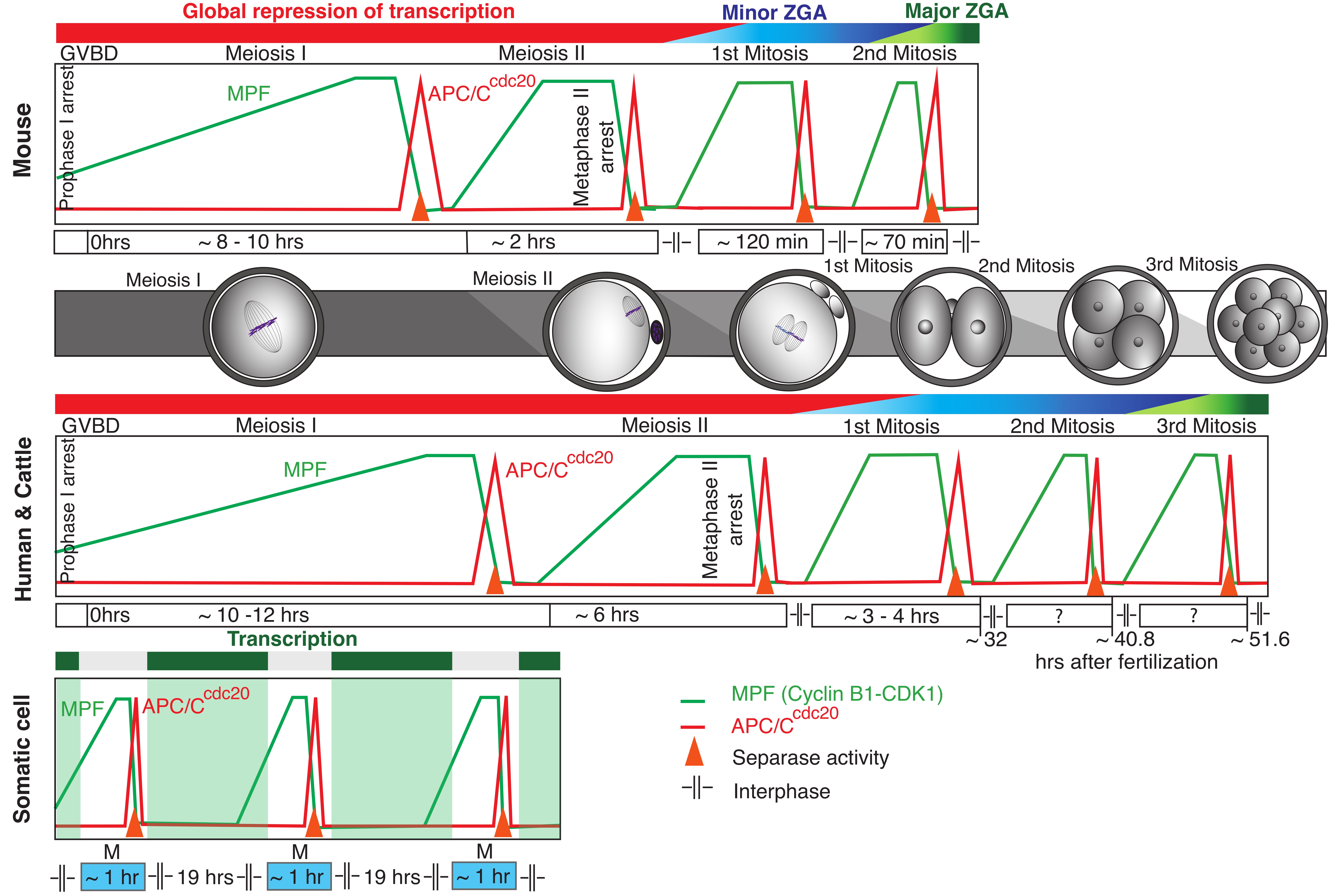
1. Peculiar Life of Mammalian Oocytes and Early Embryos
2. The Onset of Transcriptional Silencing during Female Meiosis
3. Activation of Zygotic Genome in Embryos
4. Transcription-Independent Regulation of Cell Cycle in Oocytes and Early Cleavage Embryos
5. The Control of Chromosome Separation in Oocytes and Early Embryos
6. The Control of Spindle Assembly in Oocytes and Early Embryos
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clarke, H.J. Regulation of germ cell development by intercellular signaling in the mammalian ovarian follicle. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e294. [Google Scholar] [CrossRef] [PubMed]
- Kalous, J.; Tetkova, A.; Kubelka, M.; Susor, A. Importance of ERK1/2 in Regulation of Protein Translation during Oocyte Meiosis. Int. J. Mol. Sci. 2018, 19, 698. [Google Scholar] [CrossRef] [PubMed]
- Jessus, C.; Munro, C.; Houliston, E. Managing the Oocyte Meiotic Arrest-Lessons from Frogs and Jellyfish. Cells 2020, 9, 1150. [Google Scholar] [CrossRef]
- Bhakta, H.H.; Refai, F.H.; Avella, M.A. The molecular mechanisms mediating mammalian fertilization. Development 2019, 146, dev176966. [Google Scholar] [CrossRef]
- Saunders, C.M.; Larman, M.G.; Parrington, J.; Cox, L.J.; Royse, J.; Blayney, L.M.; Swann, K.; Lai, F.A. PLC zeta: A sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development 2002, 129, 3533–3544. [Google Scholar] [CrossRef] [PubMed]
- Clift, D.; Schuh, M. Restarting life: Fertilization and the transition from meiosis to mitosis. Nat. Rev. Mol. Cell Biol. 2013, 14, 549–562. [Google Scholar] [CrossRef]
- Reichmann, J.; Nijmeijer, B.; Hossain, M.J.; Eguren, M.; Schneider, I.; Politi, A.Z.; Roberti, M.J.; Hufnagel, L.; Hiiragi, T.; Ellenberg, J. Dual-spindle formation in zygotes keeps parental genomes apart in early mammalian embryos. Science 2018, 361, 189–193. [Google Scholar] [CrossRef]
- Cavazza, T.; Takeda, Y.; Politi, A.Z.; Aushev, M.; Aldag, P.; Baker, C.; Choudhary, M.; Bucevičius, J.; Lukinavičius, G.; Elder, K.; et al. Parental genome unification is highly error-prone in mammalian embryos. Cell 2021, 184, 2860–2877.e22. [Google Scholar] [CrossRef]
- Schultz, R.M.; Stein, P.; Svoboda, P. The oocyte-to-embryo transition in mouse: Past, present, and future. Biol. Reprod. 2018, 99, 160–174. [Google Scholar] [CrossRef]
- Debey, P.; Szöllösi, M.S.; Szöllösi, D.; Vautier, D.; Girousse, A.; Besombes, D. Competent mouse oocytes isolated from antral follicles exhibit different chromatin organization and follow different maturation dynamics. Mol. Reprod. Dev. 1993, 36, 59–74. [Google Scholar] [CrossRef]
- Longo, F.; Garagna, S.; Merico, V.; Orlandini, G.; Gatti, R.; Scandroglio, R.; Redi, C.A.; Zuccotti, M. Nuclear localization of NORs and centromeres in mouse oocytes during folliculogenesis. Mol. Reprod. Dev. 2003, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.H.; Wang, H.L.; Sun, X.S.; Liu, Y.; Sui, H.S.; Zhang, J. Chromatin configurations in the germinal vesicle of mammalian oocytes. Mol. Hum. Reprod. 2009, 15, 1–9. [Google Scholar] [CrossRef]
- Turner, S.; Wong, H.P.; Rai, J.; Hartshorne, G.M. Telomere lengths in human oocytes, cleavage stage embryos and blastocysts. Mol. Hum. Reprod. 2010, 16, 685–694. [Google Scholar] [CrossRef]
- Bonnet-Garnier, A.; Feuerstein, P.; Chebrout, M.; Fleurot, R.; Jan, H.U.; Debey, P.; Beaujean, N. Genome organization and epigenetic marks in mouse germinal vesicle oocytes. Int. J. Dev. Biol. 2012, 56, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef]
- Vastenhouw, N.L.; Cao, W.X.; Lipshitz, H.D. The maternal-to-zygotic transition revisited. Development 2019, 146, dev161471. [Google Scholar] [CrossRef]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.Y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E.; et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef]
- Abe, K.I.; Funaya, S.; Tsukioka, D.; Kawamura, M.; Suzuki, Y.; Suzuki, M.G.; Schultz, R.M.; Aoki, F. Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. USA 2018, 115, E6780–E6788. [Google Scholar] [CrossRef] [PubMed]
- Sha, Q.Q.; Zhu, Y.Z.; Li, S.; Jiang, Y.; Chen, L.; Sun, X.H.; Shen, L.; Ou, X.H.; Fan, H.Y. Characterization of zygotic genome activation-dependent maternal mRNA clearance in mouse. Nucleic Acids Res. 2020, 48, 879–894. [Google Scholar] [CrossRef]
- Sha, Q.Q.; Zheng, W.; Wu, Y.W.; Li, S.; Guo, L.; Zhang, S.; Lin, G.; Ou, X.H.; Fan, H.Y. Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat. Commun. 2020, 11, 4917. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Brandão, H.B.; Ulianov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.A.; Tachibana-Konwalski, K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Halstead, M.M.; Ma, X.; Zhou, C.; Schultz, R.M.; Ross, P.J. Chromatin remodeling in bovine embryos indicates species-specific regulation of genome activation. Nat. Commun. 2020, 11, 4654. [Google Scholar] [CrossRef]
- Hug, C.B.; Vaquerizas, J.M. The Birth of the 3D Genome during Early Embryonic Development. Trends Genet. 2018, 34, 903–914. [Google Scholar] [CrossRef]
- Aoki, F.; Worrad, D.M.; Schultz, R.M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997, 181, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Latham, K.E.; Garrels, J.I.; Chang, C.; Solter, D. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development 1991, 112, 921–932. [Google Scholar] [CrossRef]
- Hamatani, T.; Carter, M.G.; Sharov, A.A.; Ko, M.S.H. Dynamics of Global Gene Expression Changes during Mouse Preimplantation Development. Dev. Cell 2004, 6, 117–131. [Google Scholar] [CrossRef]
- Vassena, R.; Boué, S.; González-Roca, E.; Aran, B.; Auer, H.; Veiga, A.; Izpisua Belmonte, J.C. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 2011, 138, 3699–3709. [Google Scholar] [CrossRef]
- Memili, E.; First, N.L. Zygotic and embryonic gene expression in cow: A review of timing and mechanisms of early gene expression as compared with other species. Zygote 2000, 8, 87–96. [Google Scholar] [CrossRef]
- Misirlioglu, M.; Page, G.P.; Sagirkaya, H.; Kaya, A.; Parrish, J.J.; First, N.L.; Memili, E. Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc. Natl. Acad. Sci. USA 2006, 103, 18905–18910. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.; Krebs, S.; Zakhartchenko, V.; Schwalb, B.; Blum, H.; Wolf, E. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 4139–4144. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Yamamoto, R.; Franke, V.; Cao, M.; Suzuki, Y.; Suzuki, M.G.; Vlahovicek, K.; Svoboda, P.; Schultz, R.M.; Aoki, F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3’ processing. EMBO J. 2015, 34, 1523–1537. [Google Scholar] [CrossRef] [PubMed]
- Hochegger, H.; Takeda, S.; Hunt, T. Cyclin-dependent kinases and cell-cycle transitions: Does one fit all. Nat. Rev. Mol. Cell Biol. 2008, 9, 910–916. [Google Scholar] [CrossRef] [PubMed]
- Koliopoulos, M.G.; Alfieri, C. Cell cycle regulation by complex nanomachines. FEBS J. 2021. [Google Scholar] [CrossRef]
- Gelens, L.; Qian, J.; Bollen, M.; Saurin, A.T. The Importance of Kinase-Phosphatase Integration: Lessons from Mitosis. Trends Cell Biol. 2018, 28, 6–21. [Google Scholar] [CrossRef]
- Holder, J.; Poser, E.; Barr, F.A. Getting out of mitosis: Spatial and temporal control of mitotic exit and cytokinesis by PP1 and PP2A. FEBS Lett. 2019, 593, 2908–2924. [Google Scholar] [CrossRef]
- Peters, J.M. The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell 2002, 9, 931–943. [Google Scholar] [CrossRef]
- Acquaviva, C.; Pines, J. The anaphase-promoting complex/cyclosome: APC/C. J. Cell Sci. 2006, 119, 2401–2404. [Google Scholar] [CrossRef]
- Cogswell, J.P.; Godlevski, M.M.; Bonham, M.; Bisi, J.; Babiss, L. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol. Cell Biol. 1995, 15, 2782–2790. [Google Scholar] [CrossRef][Green Version]
- King, R.W.; Peters, J.M.; Tugendreich, S.; Rolfe, M.; Hieter, P.; Kirschner, M.W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 1995, 81, 279–288. [Google Scholar] [CrossRef]
- Brandeis, M.; Hunt, T. The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J. 1996, 15, 5280–5289. [Google Scholar] [CrossRef] [PubMed]
- Wasner, M.; Tschöp, K.; Spiesbach, K.; Haugwitz, U.; Johne, C.; Mössner, J.; Mantovani, R.; Engeland, K. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003, 536, 66–70. [Google Scholar] [CrossRef]
- Palozola, K.C.; Lerner, J.; Zaret, K.S. A changing paradigm of transcriptional memory propagation through mitosis. Nat. Rev. Mol. Cell Biol. 2019, 20, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Palozola, K.C.; Donahue, G.; Liu, H.; Grant, G.R.; Becker, J.S.; Cote, A.; Yu, H.; Raj, A.; Zaret, K.S. Mitotic transcription and waves of gene reactivation during mitotic exit. Science 2017, 358, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Susor, A.; Jansova, D.; Anger, M.; Kubelka, M. Translation in the mammalian oocyte in space and time. Cell Tissue Res. 2016, 363, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Sirard, M.A.; Florman, H.M.; Leibfried-Rutledge, M.L.; Barnes, F.L.; Sims, M.L.; First, N.L. Timing of nuclear progression and protein synthesis necessary for meiotic maturation of bovine oocytes. Biol. Reprod. 1989, 40, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Balakier, H.; MacLusky, N.J.; Casper, R.F. Characterization of the first cell cycle in human zygotes: Implications for cryopreservation. Fertil. Steril. 1993, 59, 359–365. [Google Scholar] [CrossRef]
- Holm, P.; Shukri, N.N.; Vajta, G.; Booth, P.; Bendixen, C.; Callesen, H. Developmental kinetics of the first cell cycles of bovine in vitro produced embryos in relation to their in vitro viability and sex. Theriogenology 1998, 50, 1285–1299. [Google Scholar] [CrossRef]
- Fancsovits, P.; Toth, L.; Takacs, Z.F.; Murber, A.; Papp, Z.; Urbancsek, J. Early pronuclear breakdown is a good indicator of embryo quality and viability. Fertil. Steril. 2005, 84, 881–887. [Google Scholar] [CrossRef]
- Jones, K.T. Mammalian egg activation: From Ca2+ spiking to cell cycle progression. Reproduction 2005, 130, 813–823. [Google Scholar] [CrossRef]
- Radonova, L.; Svobodova, T.; Anger, M. Regulation of the cell cycle in early mammalian embryos and its clinical implications. Int. J. Dev. Biol. 2019, 63, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.X.; Fakhreddin, R.I.; Shimerov, H.K.; Kedziora, K.M.; Kumar, R.J.; Perez, J.; Limas, J.C.; Grant, G.D.; Cook, J.G.; Gupta, G.P.; et al. Evidence that the human cell cycle is a series of uncoupled, memoryless phases. Mol. Syst. Biol. 2019, 15, e8604. [Google Scholar] [CrossRef] [PubMed]
- Ciemerych, M.A.; Sicinski, P. Cell cycle in mouse development. Oncogene 2005, 24, 2877–2898. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, A.; Kaldis, P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009, 28, 2925–2939. [Google Scholar] [CrossRef] [PubMed]
- Palmer, N.; Kaldis, P. Regulation of the Embryonic Cell Cycle During Mammalian Preimplantation Development. Curr. Top. Dev. Biol. 2016, 120, 1–53. [Google Scholar]
- Chotiner, J.Y.; Wolgemuth, D.J.; Wang, P.J. Functions of cyclins and CDKs in mammalian gametogenesis†. Biol. Reprod. 2019, 101, 591–601. [Google Scholar] [CrossRef]
- Campbell, G.J.; Hands, E.L.; Van de Pette, M. The Role of CDKs and CDKIs in Murine Development. Int. J. Mol. Sci. 2020, 21, 6343. [Google Scholar] [CrossRef]
- Kozar, K.; Ciemerych, M.A.; Rebel, V.I.; Shigematsu, H.; Zagozdzon, A.; Sicinska, E.; Geng, Y.; Yu, Q.; Bhattacharya, S.; Bronson, R.T.; et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell 2004, 118, 477–491. [Google Scholar] [CrossRef]
- Geng, Y.; Yu, Q.; Sicinska, E.; Das, M.; Schneider, J.E.; Bhattacharya, S.; Rideout, W.M.; Bronson, R.T.; Gardner, H.; Sicinski, P. Cyclin E Ablation in the Mouse. Cell 2003, 114, 431–443. [Google Scholar] [CrossRef]
- Parisi, T.; Beck, A.R.; Rougier, N.; McNeil, T.; Lucian, L.; Werb, Z.; Amati, B. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 2003, 22, 4794–4803. [Google Scholar] [CrossRef] [PubMed]
- Brandeis, M.; Rosewell, I.; Carrington, M.; Crompton, T.; Jacobs, M.A.; Kirk, J.; Gannon, J.; Hunt, T. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc. Natl. Acad. Sci. USA 1998, 95, 4344–4349. [Google Scholar] [CrossRef] [PubMed]
- Strauss, B.; Harrison, A.; Coelho, P.A.; Yata, K.; Zernicka-Goetz, M.; Pines, J. Cyclin B1 is essential for mitosis in mouse embryos, and its nuclear export sets the time for mitosis. J. Cell Biol. 2018, 217, 179–193. [Google Scholar] [CrossRef]
- Zhang, Q.H.; Yuen, W.S.; Adhikari, D.; Flegg, J.A.; FitzHarris, G.; Conti, M.; Sicinski, P.; Nabti, I.; Marangos, P.; Carroll, J. Cyclin A2 modulates kinetochore-microtubule attachment in meiosis II. J. Cell Biol. 2017, 216, 3133–3143. [Google Scholar] [CrossRef]
- Murphy, M. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat. Genet. 1999, 23, 481. [Google Scholar] [CrossRef]
- Hara, K.T.; Oda, S.; Naito, K.; Nagata, M.; Schultz, R.M.; Aoki, F. Cyclin A2-CDK2 regulates embryonic gene activation in 1-cell mouse embryos. Dev. Biol. 2005, 286, 102–113. [Google Scholar] [CrossRef]
- Pagliuca, F.W.; Collins, M.O.; Lichawska, A.; Zegerman, P.; Choudhary, J.S.; Pines, J. Quantitative proteomics reveals the basis for the biochemical specificity of the cell-cycle machinery. Mol. Cell 2011, 43, 406–417. [Google Scholar] [CrossRef]
- Hégarat, N.; Crncec, A.; Suarez Peredo Rodriguez, M.F.; Echegaray Iturra, F.; Gu, Y.; Busby, O.; Lang, P.F.; Barr, A.R.; Bakal, C.; Kanemaki, M.T.; et al. Cyclin A triggers Mitosis either via the Greatwall kinase pathway or Cyclin B. EMBO J. 2020, 39, e104419. [Google Scholar] [CrossRef]
- Kanakkanthara, A.; Jeganathan, K.B.; Limzerwala, J.F.; Baker, D.J.; Hamada, M.; Nam, H.J.; van Deursen, W.H.; Hamada, N.; Naylor, R.M.; Becker, N.A.; et al. Cyclin A2 is an RNA binding protein that controls Mre11 mRNA translation. Science 2016, 353, 1549–1552. [Google Scholar] [CrossRef] [PubMed]
- Kalaszczynska, I.; Geng, Y.; Iino, T.; Mizuno, S.; Choi, Y.; Kondratiuk, I.; Silver, D.P.; Wolgemuth, D.J.; Akashi, K.; Sicinski, P. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell 2009, 138, 352–365. [Google Scholar] [CrossRef]
- Santamaría, D.; Barrière, C.; Cerqueira, A.; Hunt, S.; Tardy, C.; Newton, K.; Cáceres, J.F.; Dubus, P.; Malumbres, M.; Barbacid, M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007, 448, 811–815. [Google Scholar] [CrossRef]
- Diril, M.K.; Ratnacaram, C.K.; Padmakumar, V.C.; Du, T.; Wasser, M.; Coppola, V.; Tessarollo, L.; Kaldis, P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc. Natl. Acad. Sci. USA 2012, 109, 3826–3831. [Google Scholar] [CrossRef] [PubMed]
- Pagano, M.; Jackson, P.K. Wagging the dogma; tissue-specific cell cycle control in the mouse embryo. Cell 2004, 118, 535–538. [Google Scholar] [CrossRef]
- Susor, A.; Kubelka, M. Translational Regulation in the Mammalian Oocyte. Results Probl. Cell Differ. 2017, 63, 257–295. [Google Scholar] [PubMed]
- Sha, Q.Q.; Zhang, J.; Fan, H.Y. A story of birth and death: mRNA translation and clearance at the onset of Maternal-to-Zygotic transition in mammals. Biol. Reprod. 2019, 101, 579–590. [Google Scholar] [CrossRef]
- Esencan, E.; Kallen, A.; Zhang, M.; Seli, E. Translational activation of maternally derived mRNAs in oocytes and early embryos and the role of embryonic poly(A) binding protein (EPAB). Biol. Reprod. 2019, 100, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.; Nebreda, A.R. RINGO/Speedy proteins, a family of non-canonical activators of CDK1 and CDK2. Semin. Cell Dev. Biol. 2020, 107, 21–27. [Google Scholar] [CrossRef]
- Vassalli, J.D.; Huarte, J.; Belin, D.; Gubler, P.; Vassalli, A.; O’Connell, M.L.; Parton, L.A.; Rickles, R.J.; Strickland, S. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev. 1989, 3, 2163–2171. [Google Scholar] [CrossRef]
- McGrew, L.L.; Dworkin-Rastl, E.; Dworkin, M.B.; Richter, J.D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989, 3, 803–815. [Google Scholar] [CrossRef]
- Groisman, I.; Jung, M.Y.; Sarkissian, M.; Cao, Q.; Richter, J.D. Translational control of the embryonic cell cycle. Cell 2002, 109, 473–483. [Google Scholar] [CrossRef]
- Richter, J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007, 32, 279–285. [Google Scholar] [CrossRef]
- Seli, E.; Lalioti, M.D.; Flaherty, S.M.; Sakkas, D.; Terzi, N.; Steitz, J.A. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc. Natl. Acad. Sci. USA 2005, 102, 367–372. [Google Scholar] [CrossRef]
- Kronja, I.; Orr-Weaver, T.L. Translational regulation of the cell cycle: When, where, how and why. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3638–3652. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, C.R.; Han, S.J.; Daldello, E.M.; Cho, A.; Martins, J.P.S.; Xia, G.; Conti, M. Maternal mRNAs with distinct 3’ UTRs define the temporal pattern of Ccnb1 synthesis during mouse oocyte meiotic maturation. Genes Dev. 2017, 31, 1302–1307. [Google Scholar] [CrossRef]
- Yatskevich, S.; Rhodes, J.; Nasmyth, K. Organization of Chromosomal DNA by SMC Complexes. Annu. Rev. Genet. 2019, 53, 445–482. [Google Scholar] [CrossRef]
- Waizenegger, I.C.; Hauf, S.; Meinke, A.; Peters, J.M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 2000, 103, 399–410. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Bury, L.; Cheeseman, I.M.; Blower, M.D. Cohesin Removal Reprograms Gene Expression upon Mitotic Entry. Mol. Cell 2020, 78, 127–140.e7. [Google Scholar] [CrossRef]
- Ciosk, R.; Zachariae, W.; Michaelis, C.; Shevchenko, A.; Mann, M.; Nasmyth, K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 1998, 93, 1067–1076. [Google Scholar] [CrossRef]
- Kamenz, J.; Hauf, S. Time To Split Up: Dynamics of Chromosome Separation. Trends Cell Biol. 2017, 27, 42–54. [Google Scholar] [CrossRef]
- Kudo, N.R.; Wassmann, K.; Anger, M.; Schuh, M.; Wirth, K.G.; Xu, H.; Helmhart, W.; Kudo, H.; McKay, M.; Maro, B.; et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell 2006, 126, 135–146. [Google Scholar] [CrossRef]
- Tsou, M.F.; Stearns, T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 2006, 442, 947–951. [Google Scholar] [CrossRef]
- Kumar, R. Separase: Function Beyond Cohesion Cleavage and an Emerging Oncogene. J. Cell Biochem. 2017, 118, 1283–1299. [Google Scholar] [CrossRef]
- Maier, N.K.; Ma, J.; Lampson, M.A.; Cheeseman, I.M. Separase cleaves the kinetochore protein Meikin at the meiosis I/II transition. Dev. Cell 2021, 56, 2192–2206.e8. [Google Scholar] [CrossRef]
- Vijayakumari, D.; Müller, J.; Hauf, S. Cdc48 influence on separase levels is independent of mitosis and suggests translational sensitivity of separase. bioRxiv 2021. [Google Scholar] [CrossRef]
- Meadows, J.C.; Millar, J.B. Sharpening the anaphase switch. Biochem. Soc. Trans. 2015, 43, 19–22. [Google Scholar] [CrossRef]
- Hellmuth, S.; Gómez-H, L.; Pendás, A.M.; Stemmann, O. Securin-independent regulation of separase by checkpoint-induced shugoshin-MAD2. Nature 2020, 580, 536–541. [Google Scholar] [CrossRef]
- Huang, X.; Andreu-Vieyra, C.V.; Wang, M.; Cooney, A.J.; Matzuk, M.M.; Zhang, P. Preimplantation mouse embryos depend on inhibitory phosphorylation of separase to prevent chromosome missegregation. Mol. Cell Biol. 2009, 29, 1498–1505. [Google Scholar] [CrossRef]
- Li, M.; York, J.P.; Zhang, P. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell Biol. 2007, 27, 3481–3488. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef]
- Pesenti, M.E.; Weir, J.R.; Musacchio, A. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 2016, 37, 152–163. [Google Scholar] [CrossRef]
- Nagaoka, S.I.; Hodges, C.A.; Albertini, D.F.; Hunt, P.A. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr. Biol. 2011, 21, 651–657. [Google Scholar] [CrossRef]
- Sebestova, J.; Danylevska, A.; Novakova, L.; Kubelka, M.; Anger, M. Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle 2012, 11, 3011–3018. [Google Scholar] [CrossRef]
- Lane, S.I.; Yun, Y.; Jones, K.T. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development 2012, 139, 1947–1955. [Google Scholar] [CrossRef]
- Kyogoku, H.; Kitajima, T.S. Large Cytoplasm Is Linked to the Error-Prone Nature of Oocytes. Dev. Cell 2017, 41, 287–298.e4. [Google Scholar] [CrossRef]
- Lane, S.I.R.; Jones, K.T. Chromosome biorientation and APC activity remain uncoupled in oocytes with reduced volume. J. Cell Biol. 2017, 216, 3949–3957. [Google Scholar] [CrossRef]
- Vázquez-Diez, C.; Paim, L.M.G.; FitzHarris, G. Cell-Size-Independent Spindle Checkpoint Failure Underlies Chromosome Segregation Error in Mouse Embryos. Curr. Biol. 2019, 29, 865–873. [Google Scholar] [CrossRef]
- Bolton, H.; Graham, S.J.; Van der Aa, N.; Kumar, P.; Theunis, K.; Fernandez Gallardo, E.; Voet, T.; Zernicka-Goetz, M. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat. Commun. 2016, 7, 11165. [Google Scholar] [CrossRef]
- Pauerova, T.; Radonova, L.; Kovacovicova, K.; Novakova, L.; Skultety, M.; Anger, M. Aneuploidy during the onset of mouse embryo development. Reproduction 2020, 160, 773–782. [Google Scholar] [CrossRef]
- Tsurumi, C.; Hoffmann, S.; Geley, S.; Graeser, R.; Polanski, Z. The spindle assembly checkpoint is not essential for CSF arrest of mouse oocytes. J. Cell Biol. 2004, 167, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Kouznetsova, A.; Kitajima, T.S.; Brismar, H.; Höög, C. Post-metaphase correction of aberrant kinetochore-microtubule attachments in mammalian eggs. EMBO Rep. 2019, 20, e47905. [Google Scholar] [CrossRef]
- Yun, Y.; Lane, S.I.; Jones, K.T. Premature dyad separation in meiosis II is the major segregation error with maternal age in mouse oocytes. Development 2014, 141, 199–208. [Google Scholar] [CrossRef]
- Dobles, M.; Liberal, V.; Scott, M.L.; Benezra, R.; Sorger, P.K. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 2000, 101, 635–645. [Google Scholar] [CrossRef]
- Tian, Q.; Hanlon Newell, A.E.; Wang, Y.; Olson, S.B.; Fedorov, L.M. Complex cytogenetic analysis of early lethality mouse embryos. Chromosome Res 2011, 19, 567–574. [Google Scholar] [CrossRef]
- Kalitsis, P.; Earle, E.; Fowler, K.J.; Choo, K.H. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000, 14, 2277–2282. [Google Scholar] [CrossRef]
- Iwanaga, Y.; Chi, Y.H.; Miyazato, A.; Sheleg, S.; Haller, K.; Peloponese, J.M.; Li, Y.; Ward, J.M.; Benezra, R.; Jeang, K.T. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007, 67, 160–166. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, T.; Fang, Y.; Xie, S.; Huang, X.; Mahmood, R.; Ramaswamy, G.; Sakamoto, K.M.; Darzynkiewicz, Z.; Xu, M.; et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood 2004, 103, 1278–1285. [Google Scholar] [CrossRef]
- Jeganathan, K.; Malureanu, L.; Baker, D.J.; Abraham, S.C.; van Deursen, J.M. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 2007, 179, 255–267. [Google Scholar] [CrossRef]
- Wells, D.; Bermudez, M.G.; Steuerwald, N.; Thornhill, A.R.; Walker, D.L.; Malter, H.; Delhanty, J.D.; Cohen, J. Expression of genes regulating chromosome segregation, the cell cycle and apoptosis during human preimplantation development. Hum. Reprod. 2005, 20, 1339–1348. [Google Scholar] [CrossRef]
- Eliscovich, C.; Peset, I.; Vernos, I.; Méndez, R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat. Cell Biol. 2008, 10, 858–865. [Google Scholar] [CrossRef]
- Hassold, T.; Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001, 2, 280–291. [Google Scholar] [CrossRef]
- Nagaoka, S.I.; Hassold, T.J.; Hunt, P.A. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012, 13, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Van Echten-Arends, J.; Mastenbroek, S.; Sikkema-Raddatz, B.; Korevaar, J.C.; Heineman, M.J.; van der Veen, F.; Repping, S. Chromosomal mosaicism in human preimplantation embryos: A systematic review. Hum. Reprod. Update 2011, 17, 620–627. [Google Scholar] [CrossRef]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2020, 21, 44–62. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anger, M.; Radonova, L.; Horakova, A.; Sekach, D.; Charousova, M. Impact of Global Transcriptional Silencing on Cell Cycle Regulation and Chromosome Segregation in Early Mammalian Embryos. Int. J. Mol. Sci. 2021, 22, 9073. https://doi.org/10.3390/ijms22169073
Anger M, Radonova L, Horakova A, Sekach D, Charousova M. Impact of Global Transcriptional Silencing on Cell Cycle Regulation and Chromosome Segregation in Early Mammalian Embryos. International Journal of Molecular Sciences. 2021; 22(16):9073. https://doi.org/10.3390/ijms22169073
Chicago/Turabian StyleAnger, Martin, Lenka Radonova, Adela Horakova, Diana Sekach, and Marketa Charousova. 2021. "Impact of Global Transcriptional Silencing on Cell Cycle Regulation and Chromosome Segregation in Early Mammalian Embryos" International Journal of Molecular Sciences 22, no. 16: 9073. https://doi.org/10.3390/ijms22169073
APA StyleAnger, M., Radonova, L., Horakova, A., Sekach, D., & Charousova, M. (2021). Impact of Global Transcriptional Silencing on Cell Cycle Regulation and Chromosome Segregation in Early Mammalian Embryos. International Journal of Molecular Sciences, 22(16), 9073. https://doi.org/10.3390/ijms22169073




