Properties and Application of Cell-Free DNA as a Clinical Biomarker
Abstract
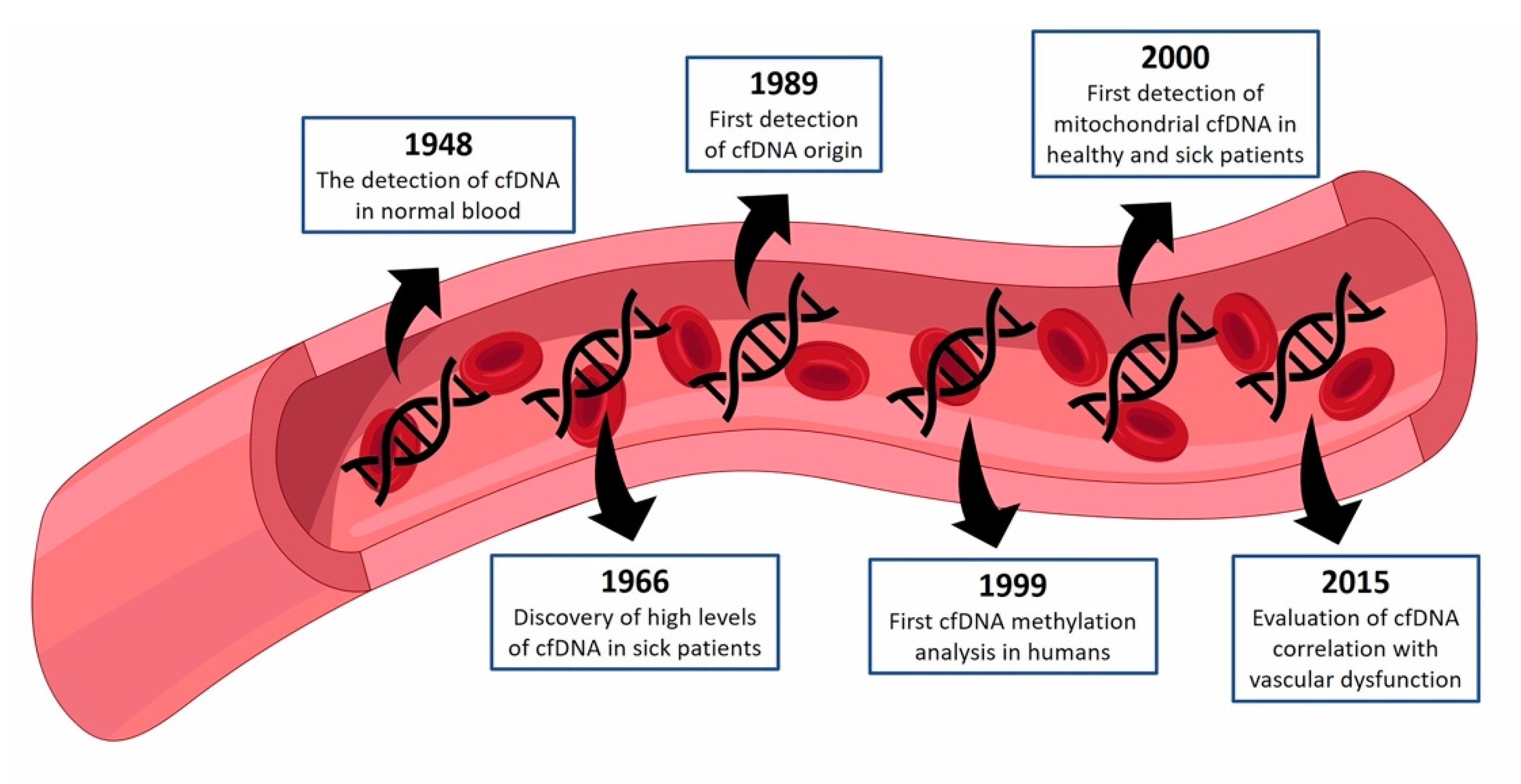
:1. cfDNA—Historical Perspective
2. cfDNA—Source and Mechanism of Release
3. cfDNA—Molecular Features
3.1. cfDNA Integrity
3.2. Genetic and Epigenetic Profile
3.3. Copy-Number Variations (CNVs)
3.4. Mutations
3.5. cfDNA Composition
3.6. Epigenetic
3.7. cfDNA Concentration
3.8. mt-cfDNA
4. Clinical Findings
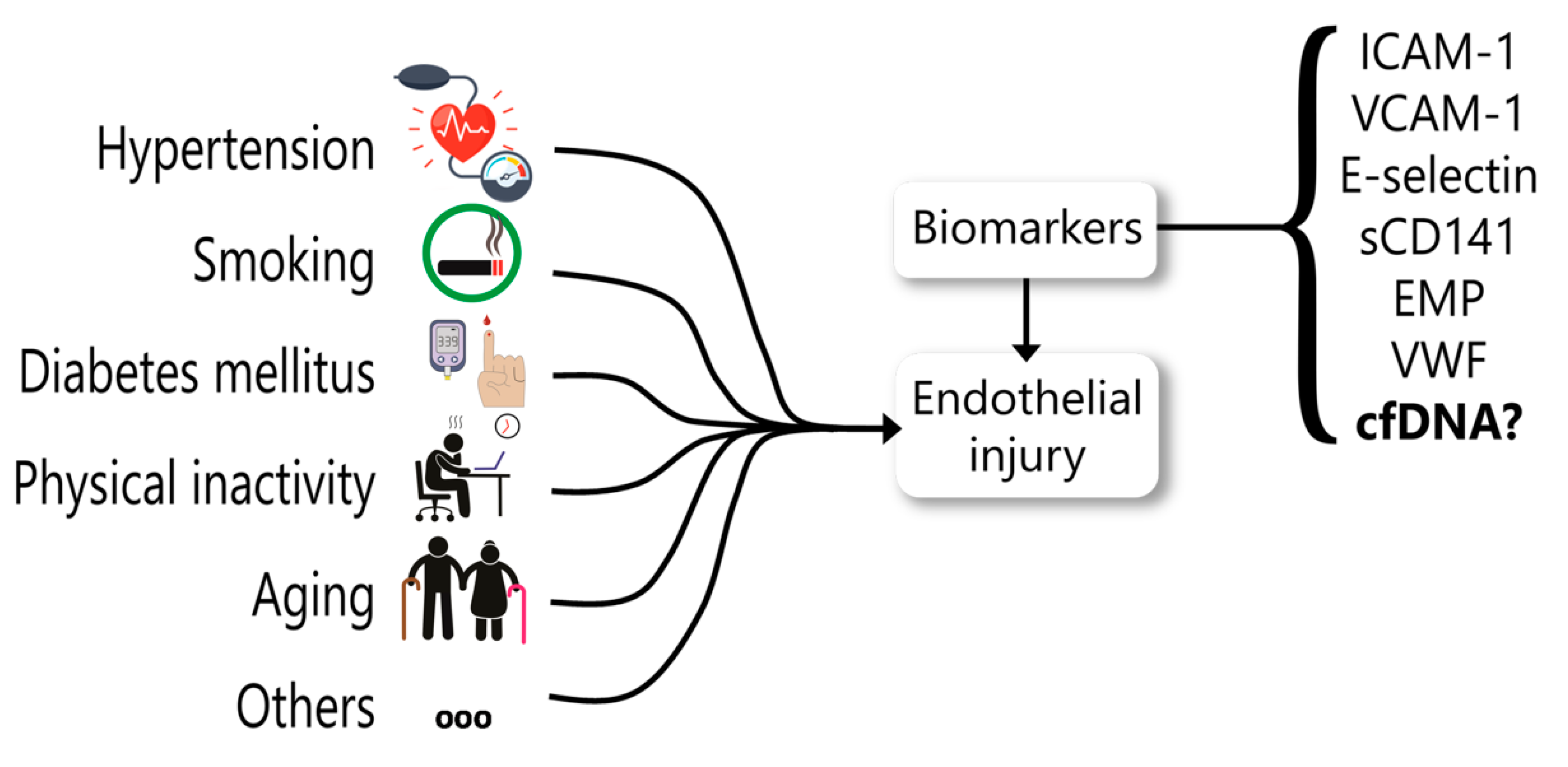
5. cfDNA as a Biomarker for Endothelial Dysfunction
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.S.; Pretorius, P.J. The Diverse Origins of Circulating Cell-Free DNA in the Human Body: A Critical Re-Evaluation of the Literature. Biol. Rev. 2018, 93, 1649–1683. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Les Acides Nucléiques Du Plasma Sanguin Chez l’homme. C R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Bendich, A.; Wilczok, T.; Borenfreund, E. Circulating DNA as a Possible Factor in Oncogenesis. Science 1965, 148, 374–376. [Google Scholar] [CrossRef]
- Tan, E.M.; Schur, P.H.; Carr, R.I.; Kunkel, H.G. Deoxybonucleic Acid (DNA) and Antibodies to DNA in the Serum of Patients with Systemic Lupus Erythematosus. J. Clin. Investig. 1966, 45, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Vasioukhin, V.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Point Mutations of the N-Ras Gene in the Blood Plasma DNA of Patients with Myelodysplastic Syndrome or Acute Myelogenous Leukaemia. Br. J. Haematol. 1994, 86, 774–779. [Google Scholar] [CrossRef]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble Normal and Mutated DNA Sequences from Single-Copy Genes in Human Blood. Cancer Epidemiol. Biomark. Prev. 1994, 3, 67–71. [Google Scholar]
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.G.; Wainscoat, J.S.; Dennis Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; et al. Presence of Fetal DNA in Maternal Plasma and Serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef]
- Esteller, M. Erratum: Detection of Aberrant Promoter Hypermethylation of Tumor Suppressor Genes in Serum Dna from Non-Small Cell Lung Cancer Patients. Cancer Res. 1999, 59, 3853. [Google Scholar]
- Wong, I.H.N.; Lo, Y.M.D.; Zhang, J.; Liew, C.T.; Ng, M.H.L.; Wong, N.; Lai, P.B.S.; Lau, W.Y.; Hjelm, N.M.; Johnson, P.J. Detection of Aberrant P16 Methylation in the Plasma and Serum of Liver Cancer Patients. Cancer Res. 1999, 59, 71–73. [Google Scholar]
- Zhong, S.; Ng, M.C.Y.; Lo, Y.M.D.; Chan, J.C.N.; Johnson, P.J. Presence of Mitochondrial TRNA(Leu(UUR)) A to G 3243 Mutation in DNA Extracted from Serum and Plasma of Patients with Type 2 Diabetes Mellitus. J. Clin. Pathol. 2000, 53, 466–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bronkhorst, A.J.; Ungerer, V.; Diehl, F.; Anker, P.; Dor, Y.; Fleischhacker, M.; Gahan, P.B.; Hui, L.; Holdenrieder, S.; Thierry, A.R. Towards Systematic Nomenclature for Cell-Free DNA. Hum. Genet. 2020. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Tein, M.S.C.; Pang, C.C.P.; Yeung, C.K.; Tong, K.L.; Magnus Hjelm, N. Presence of Donor-Specific DNA in Plasma of Kidney and Liver-Transplant Recipients. Lancet 1998, 351, 1329–1330. [Google Scholar] [CrossRef]
- Chang, C.P.Y.; Chia, R.H.; Wu, T.L.; Tsao, K.C.; Sun, C.F.; Wu, J.T. Elevated Cell-Free Serum DNA Detected in Patients with Myocardial Infarction. Clin. Chim. Acta 2003, 327, 95–101. [Google Scholar] [CrossRef]
- Rainer, T.H.; Wong, L.K.S.; Lam, W.; Yuen, E.; Lam, N.Y.L.; Metreweli, C.; Lo, Y.M.D. Prognostic Use of Circulating Plasma Nucleic Acid Concentrations in Patients with Acute Stroke. Clin. Chem. 2003, 49, 562–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, C.G.; Wenceslau, C.F.; Goulopoulou, S.; Ogbi, S.; Baban, B.; Sullivan, J.C.; Matsumoto, T.; Webb, R.C. Circulating Mitochondrial DNA and Toll-like Receptor 9 Are Associated with Vascular Dysfunction in Spontaneously Hypertensive Rats. Cardiovasc. Res. 2015, 107, 119–130. [Google Scholar] [CrossRef]
- MARTINS, G.A.; KAWAMURA, M.T.; CARVALHO, M.D.G.D.C. Detection of DNA in the Plasma of Septic Patients. Ann. N. Y. Acad. Sci. 2006, 906, 134–140. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Rainer, T.H.; Chan, L.Y.S.; Hjelm, N.M.; Cocks, R.A. Plasma DNA as a Prognostic Marker in Trauma Patients. Clin. Chem. 2000, 46, 319–323. [Google Scholar] [CrossRef] [Green Version]
- Golstein, P.; Kroemer, G. Cell Death by Necrosis: Towards a Molecular Definition. Trends Biochem. Sci. 2007, 32, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Adigun, R.; Basit, H.; Murray, J. Necrosis. Cell Liquefactive, Coagulative, Caseous, Fat, Fibrinoid and Gangrenous; StatPearls Publishing: Treasure Island, CA, USA, 2019; pp. 1–7. [Google Scholar]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin from Apoptotic and Necrotic Cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [CrossRef]
- Viorritto, I.C.B.; Nikolov, N.P.; Siegel, R.M. Autoimmunity versus Tolerance: Can Dying Cells Tip the Balance? Clin. Immunol. 2007, 122, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Kamataki, A.; Yamaki, J.; Homma, Y. Characterization of Circulating DNA in Healthy Human Plasma. Clin. Chim. Acta 2008, 387, 55–58. [Google Scholar] [CrossRef]
- Van Der Vaart, M.; Pretorius, P.J. The Origin of Circulating Free DNA [1]. Clin. Chem. 2007, 53, 2215. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.-J.; Reich, C.F.; Pisetsky, D.S. The Role of Macrophages in the in Vitro Generation of Extracellular DNA from Apoptotic and Necrotic Cells. Immunology 2005, 115, 55–62. [Google Scholar] [CrossRef]
- Proskuryakov, S.Y.; Konoplyannikov, A.G.; Gabai, V.L. Necrosis: A Specific Form of Programmed Cell Death? Exp. Cell Res. 2003, 283, 1–16. [Google Scholar] [CrossRef]
- Kroemer, G.; El-Deiry, W.S.; Golstein, P.; Peter, M.E.; Vaux, D.; Vandenabeele, P.; Zhivotovsky, B.; Blagosklonny, M.V.; Malorni, W.; Knight, R.A.; et al. Classification of Cell Death: Recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2005, 12 (Suppl. S2), 1463–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, P.; Chan, C.W.M.; Chan, K.C.A.; Cheng, S.H.; Wong, J.; Wong, V.W.S.; Wong, G.L.H.; Chan, S.L.; Mok, T.S.K.; Chan, H.L.Y.; et al. Lengthening and Shortening of Plasma DNA in Hepatocellular Carcinoma Patients. Proc. Natl. Acad. Sci. USA 2015, 112, E1317–E1325. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zhu, L.; Wu, X.; Bao, H.; Wang, X.; Chang, Z.; Shao, Y.W.; Wang, Z. Cell-Free DNA Provides a Good Representation of the Tumor Genome despite Its Biased Fragmentation Patterns. PLoS ONE 2017, 12, e0169231. [Google Scholar] [CrossRef]
- Wang, W.; Kong, P.; Ma, G.; Li, L.; Zhu, J.; Xia, T.; Xie, H.; Zhou, W.; Wang, S. Characterization of the Release and Biological Significance of Cell-Free DNA from Breast Cancer Cell Lines. Oncotarget 2017, 8, 43180–43191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aucamp, J.; Bronkhorst, A.J.; Peters, D.L.; Van Dyk, H.C.; Van der Westhuizen, F.H.; Pretorius, P.J. Kinetic Analysis, Size Profiling, and Bioenergetic Association of DNA Released by Selected Cell Lines in Vitro. Cell. Mol. Life Sci. 2017, 74, 2689–2707. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fadiel, A.; Naftolin, F.; Eichenbaum, K.D.; Xia, Y. Circulation DNA: Biological Implications for Cancer Metastasis and Immunology. Med. Hypotheses 2005, 65, 956–961. [Google Scholar] [CrossRef]
- Diamond, J.; Vanpouille-Box, C.; Spada, S.; Rudqvist, N.; Chapman, J.; Ueberheide, B.; Pilones, K.; Sarfraz, Y.; Formenti, S.; Demaria, S. Exosomes Shuttle TREX1-Sensitive IFN-Stimulatory DsDNA from Irradiated Cancer Cells to DCs. Cancer Immunol. Res. 2019, 6, 910–920. [Google Scholar] [CrossRef] [Green Version]
- Vanpouille-Box, C.; Alard, A.; Aryankalayil, M.J.; Sarfraz, Y.; Diamond, J.M.; Schneider, R.J.; Inghirami, G.; Coleman, C.N.; Formenti, S.C.; Demaria, S. DNA Exonuclease Trex1 Regulates Radiotherapy-Induced Tumour Immunogenicity. Nat. Commun. 2017, 8, 15618. [Google Scholar] [CrossRef] [PubMed]
- McArthur, K.; Whitehead, L.W.; Heddleston, J.M.; Li, L.; Padman, B.S.; Oorschot, V.; Geoghegan, N.D.; Chappaz, S.; Davidson, S.; Chin, H.S.; et al. BAK/BAX Macropores Facilitate Mitochondrial Herniation and MtDNA Efflux during Apoptosis. Science 2018, 359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal Instability Drives Metastasis through a Cytosolic DNA Response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Sun, L.; Chen, Z.J. Regulation and Function of the CGAS-STING Pathway of Cytosolic DNA Sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Vanpouille-Box, C.; Demaria, S.; Formenti, S.C.; Galluzzi, L. Cytosolic DNA Sensing in Organismal Tumor Control. Cancer Cell 2018, 34, 361–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil Elastase and Myeloperoxidase Regulate the Formation of Neutrophil Extracellular Traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef] [Green Version]
- Branzk, N.; Papayannopoulos, V. Molecular Mechanisms Regulating NETosis in Infection and Disease. Semin. Immunopathol. 2013, 35, 513–530. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, E.; Yalavarthi, S.; Berthier, C.C.; Hodgin, J.B.; Khandpur, R.; Lin, A.M.; Rubin, C.J.; Zhao, W.; Olsen, S.H.; Klinker, M.; et al. Netting Neutrophils Induce Endothelial Damage, Infiltrate Tissues, and Expose Immunostimulatory Molecules in Systemic Lupus Erythematosus. J. Immunol. 2011, 187, 538–552. [Google Scholar] [CrossRef] [Green Version]
- Rangé, H.; Labreuche, J.; Louedec, L.; Rondeau, P.; Planesse, C.; Sebbag, U.; Bourdon, E.; Michel, J.B.; Bouchard, P.; Meilhac, O. Periodontal Bacteria in Human Carotid Atherothrombosis as a Potential Trigger for Neutrophil Activation. Atherosclerosis 2014, 236, 448–455. [Google Scholar] [CrossRef]
- Warnatsch, A.; Ioannou, M.; Wang, Q.; Papayannopoulos, V. Inflammation. Neutrophil Extracellular Traps License Macrophages for Cytokine Production in Atherosclerosis. Science 2015, 349, 316–320. [Google Scholar] [CrossRef] [Green Version]
- Saffarzadeh, M.; Juenemann, C.; Queisser, M.A.; Lochnit, G.; Barreto, G.; Galuska, S.P.; Lohmeyer, J.; Preissner, K.T. Neutrophil Extracellular Traps Directly Induce Epithelial and Endothelial Cell Death: A Predominant Role of Histones. PLoS ONE 2012, 7, e32366. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Krumbholz, M.; Schönermarck, U.; Back, W.; Gross, W.L.; Werb, Z.; Gröne, H.J.; Brinkmann, V.; Jenne, D.E. Netting Neutrophils in Autoimmune Small-Vessel Vasculitis. Nat. Med. 2009, 15, 623–625. [Google Scholar] [CrossRef]
- Relja, B.; Land, W.G. Damage-Associated Molecular Patterns in Trauma. Eur. J. Trauma Emerg. Surg. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, T.A.; Brill, A.; Duerschmied, D.; Schatzberg, D.; Monestier, M.; Myers, D.D.; Wrobleski, S.K.; Wakefield, T.W.; Hartwig, J.H.; Wagner, D.D. Extracellular DNA Traps Promote Thrombosis. Proc. Natl. Acad. Sci. USA 2010, 107, 15880–15885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil Extracellular Traps Sequester Circulating Tumor Cells and Promote Metastasis. J. Clin. Investig. 2013, 123, 3446–3458. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Pelayo, R.; Monestier, M.; Ammollo, C.T.; Semeraro, F.; Taylor, F.B.; Esmon, N.L.; Lupu, F.; Esmon, C.T. Extracellular Histones Are Major Mediators of Death in Sepsis. Nat. Med. 2009, 15, 1318–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Zhang, S.; Wang, Y.; Rahman, M.; Syk, I.; Zhang, E.; Thorlacius, H. Proinflammatory Role of Neutrophil Extracellular Traps in Abdominal Sepsis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 307, L586–L596. [Google Scholar] [CrossRef] [PubMed]
- Demers, M.; Wong, S.L.; Martinod, K.; Gallant, M.; Cabral, J.E.; Wang, Y.; Wagner, D.D. Priming of Neutrophils toward NETosis Promotes Tumor Growth. Oncoimmunology 2016, 5, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinegin, B.; Vorobjeva, N.; Pinegin, V. Neutrophil Extracellular Traps and Their Role in the Development of Chronic Inflammation and Autoimmunity. Autoimmun. Rev. 2015, 14, 633–640. [Google Scholar] [CrossRef]
- Beiter, T.; Fragasso, A.; Hudemann, J.; Schild, M.; Steinacker, J.; Mooren, F.C.; Niess, A.M. Neutrophils Release Extracellular DNA Traps in Response to Exercise. J. Appl. Physiol. 2014, 117, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Breitbach, S.; Tug, S.; Simon, P. Circulating Cell-Free DNA: An up-Coming Molecular Marker in Exercise Physiology. Sport. Med. 2012, 42, 565–586. [Google Scholar] [CrossRef]
- Yipp, B.G.; Petri, B.; Salina, D.; Jenne, C.N.; Scott, B.N.V.; Zbytnuik, L.D.; Pittman, K.; Asaduzzaman, M.; Wu, K.; Meijndert, H.C.; et al. Infection-Induced NETosis Is a Dynamic Process Involving Neutrophil Multitasking in Vivo. Nat. Med. 2012, 18, 1386–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yipp, B.G.; Kubes, P. NETosis: How Vital Is It? Blood 2013, 122, 2784–2794. [Google Scholar] [CrossRef] [PubMed]
- Pilsczek, F.H.; Salina, D.; Poon, K.K.H.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.Y.; Surette, M.G.; Sugai, M.; et al. A Novel Mechanism of Rapid Nuclear Neutrophil Extracellular Trap Formation in Response to Staphylococcus Aureus. J. Immunol. 2010, 185, 7413–7425. [Google Scholar] [CrossRef] [Green Version]
- Hristov, M.; Erl, W.; Linder, S.; Weber, P.C. Apoptotic Bodies from Endothelial Cells Enhance the Number and Initiate the Differentiation of Human Endothelial Progenitor Cells in Vitro. Blood 2004, 104, 2761–2766. [Google Scholar] [CrossRef]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding Microvesicles: Artefacts No More. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane Vesicles, Current State-of-the-Art: Emerging Role of Extracellular Vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, J.R. Q & A: What Are Exosomes, Exactly? BMC Biol. 2016, 14, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Torralba, D.; Baixauli, F.; Villarroya-Beltri, C.; Fernández-Delgado, I.; Latorre-Pellicer, A.; Acín-Pérez, R.; Martín-Cófreces, N.B.; Jaso-Tamame, Á.L.; Iborra, S.; Jorge, I.; et al. Priming of Dendritic Cells by DNA-Containing Extracellular Vesicles from Activated T Cells through Antigen-Driven Contacts. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New Evidence That a Large Proportion of Human Blood Plasma Cell-Free DNA Is Localized in Exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zocco, D.; Bernardi, S.; Novelli, M.; Astrua, C.; Fava, P.; Zarovni, N.; Carpi, F.M.; Bianciardi, L.; Malavenda, O.; Quaglino, P.; et al. Isolation of Extracellular Vesicles Improves the Detection of Mutant DNA from Plasma of Metastatic Melanoma Patients. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Lázaro-Ibáñez, E.; Lässer, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; García-Rodríguez, A.; Lötvall, J. DNA Analysis of Low- and High-Density Fractions Defines Heterogeneous Subpopulations of Small Extracellular Vesicles Based on Their DNA Cargo and Topology. J. Extracell. Vesicles 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- McGough, I.J.; Vincent, J.P. Exosomes in Developmental Signalling. Development. 2016, 143, 2482–2493. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes Maintain Cellular Homeostasis by Excreting Harmful DNA from Cells. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Palis, J. Primitive and Definitive Erythropoiesis in Mammals. Front. Physiol. 2014, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Notta, F.; Zandi, S.; Takayama, N.; Dobson, S.; Gan, O.I.; Wilson, G.; Kaufmann, K.B.; McLeod, J.; Laurenti, E.; Dunant, C.F.; et al. Distinct Routes of Lineage Development Reshape the Human Blood Hierarchy across Ontogeny. Science 2016, 351. [Google Scholar] [CrossRef] [Green Version]
- Gnanapragasam, M.N.; Bieker, J.J. Orchestration of Late Events in Erythropoiesis by KLF1/EKLF. Curr. Opin. Hematol. 2017, 24, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Gnanapragasam, M.N.; Mcgrath, K.E.; Catherman, S.; Xue, L.; Palis, J.; Bieker, J.J. Regular Article Red Cells, Iron, and Erythropoiesis Eklf/Klf1-Regulated Cell Cycle Exit Is Essential for Erythroblast Enucleation. Blood 2016, 128, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Keerthivasan, G.; Wickrema, A.; Crispino, J.D. Erythroblast Enucleation. Stem Cells Int. 2011, 2011, 139851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.C.M.; Gimm, J.A.; Lo, A.J.; Koury, M.J.; Krauss, S.W.; Mohandas, N.; Chasis, J.A. Mechanism of Protein Sorting during Erythroblast Enucleation: Role of Cytoskeletal Connectivity. Blood 2004, 103, 1912–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soni, S.; Bala, S.; Gwynn, B.; Sahr, K.E.; Peters, L.L.; Hanspal, M. Absence of Erythroblast Macrophage Protein (Emp) Leads to Failure of Erythroblast Nuclear Extrusion. J. Biol. Chem. 2006, 281, 20181–20189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawane, K.; Fukuyama, H.; Kondoh, G.; Takeda, J.; Ohsawa, Y.; Uchiyama, Y.; Nagata, S. Requirement of DNase II for Definitive Erythropoiesis in the Mouse Fetal Liver. Science 2001, 292, 1546–1549. [Google Scholar] [CrossRef]
- Lam, W.K.J.; Gai, W.; Sun, K.; Wong, R.S.M.; Chan, R.W.Y.; Jiang, P.; Chan, N.P.H.; Hui, W.W.I.; Chan, A.W.H.; Szeto, C.C.; et al. DNA of Erythroid Origin Is Present in Human Plasma and Informs the Types of Anemia. Clin. Chem. 2017, 63, 1614–1623. [Google Scholar] [CrossRef]
- Lui, Y.Y.N.; Chik, K.W.; Chiu, R.W.K.; Ho, C.Y.; Lam, C.W.K.; Lo, Y.M.D. Predominant Hematopoietic Origin of Cell-Free Dna in Plasma and Serum after Sex-Mismatched Bone Marrow Transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Sandquist, M.; Wong, H.R. Biomarkers of Sepsis and Their Potential Value in Diagnosis, Prognosis and Treatment. Expert Rev. Clin. Immunol. 2014, 10, 1349–1356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spisák, S.; Solymosi, N.; Ittzés, P.; Bodor, A.; Kondor, D.; Vattay, G.; Barták, B.K.; Sipos, F.; Galamb, O.; Tulassay, Z.; et al. Complete Genes May Pass from Food to Human Blood. PLoS ONE 2013, 8, e69805. [Google Scholar] [CrossRef] [Green Version]
- Wichmann, D.; Panning, M.; Quack, T.; Kramme, S.; Burchard, G.D.; Grevelding, C.; Drosten, C. Diagnosing Schistosomiasis by Detection of Cell-Free Parasite DNA in Human Plasma. PLoS Negl. Trop. Dis. 2009, 3, e422. [Google Scholar] [CrossRef]
- De Vlaminck, I.; Martin, L.; Kertesz, M.; Patel, K.; Kowarsky, M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; et al. Noninvasive Monitoring of Infection and Rejection after Lung Transplantation. Proc. Natl. Acad. Sci. USA 2015, 112, 13336–13341. [Google Scholar] [CrossRef] [Green Version]
- Burnham, P.; Kim, M.S.; Agbor-Enoh, S.; Luikart, H.; Valantine, H.A.; Khush, K.K.; De Vlaminck, I. Single-Stranded DNA Library Preparation Uncovers the Origin and Diversity of Ultrashort Cell-Free DNA in Plasma. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Blauwkamp, T.A.; Thair, S.; Rosen, M.J.; Blair, L.; Lindner, M.S.; Vilfan, I.D.; Kawli, T.; Christians, F.C.; Venkatasubrahmanyam, S.; Wall, G.D.; et al. Analytical and Clinical Validation of a Microbial Cell-Free DNA Sequencing Test for Infectious Disease. Nat. Microbiol. 2019, 4, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Kowarsky, M.; Camunas-Soler, J.; Kertesz, M.; De Vlaminck, I.; Koh, W.; Pan, W.; Martin, L.; Neff, N.F.; Okamoto, J.; Wong, R.J.; et al. Numerous Uncharacterized and Highly Divergent Microbes Which Colonize Humans Are Revealed by Circulating Cell-Free DNA. Proc. Natl. Acad. Sci. USA 2017, 114, 9623–9628. [Google Scholar] [CrossRef] [Green Version]
- Grumaz, S.; Stevens, P.; Grumaz, C.; Decker, S.O.; Weigand, M.A.; Hofer, S.; Brenner, T.; von Haeseler, A.; Sohn, K. Next-Generation Sequencing Diagnostics of Bacteremia in Septic Patients. Genome Med. 2016, 8, 1–13. [Google Scholar] [CrossRef]
- Ngan, R.K.C.; Yip, T.T.C.; Cheng, W.W.; Chan, J.K.C.; Cho, W.C.S.; Ma, V.W.S.; Wan, K.K.; Au, S.K.; Law, C.K.; Lau, W.H. Circulating Epstein-Barr Virus DNA in Serum of Patients with Lymphoepithelioma-like Carcinoma of the Lung: A Potential Surrogate Marker for Monitoring Disease. Clin. Cancer Res. 2002, 8, 986–994. [Google Scholar] [PubMed]
- Lo, Y.M.D.; Tein, M.S.C.; Lau, T.K.; Haines, C.J.; Leung, T.N.; Poon, P.M.K.; Wainscoat, J.S.; Johnson, P.J.; Chang, A.M.Z.; Hjelm, N.M. Quantitative Analysis of Fetal DNA in Maternal Plasma and Serum: Implications for Noninvasive Prenatal Diagnosis. Am. J. Hum. Genet. 1998, 62, 768–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gielis, E.M.; Ledeganck, K.J.; De Winter, B.Y.; Del Favero, J.; Bosmans, J.L.; Claas, F.H.J.; Abramowicz, D.; Eikmans, M. Cell-Free DNA: An Upcoming Biomarker in Transplantation. Am. J. Transplant. 2015, 15, 2541–2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehra, N.; Penning, M.; Maas, J.; Van Daal, N.; Giles, R.H.; Voest, E.E. Circulating Mitochondrial Nucleic Acids Have Prognostic Value for Survival in Patients with Advanced Prostate Cancer. Clin. Cancer Res. 2007, 13, 421–426. [Google Scholar] [CrossRef] [Green Version]
- Sergeeva, V.A.; Kostyuk, S.V.; Ershova, E.S.; Malinovskaya, E.M.; Smirnova, T.D.; Kameneva, L.V. Circulating Nucleic Acids in Serum and Plasma. CNAPS IX 2016, 924, 2–5. [Google Scholar] [CrossRef]
- Pinti, M.; Cevenini, E.; Nasi, M.; De Biasi, S.; Salvioli, S.; Monti, D.; Benatti, S.; Gibellini, L.; Cotichini, R.; Stazi, M.A.; et al. Circulating Mitochondrial DNA Increases with Age and Is a Familiar Trait: Implications for “Inflamm-Aging”. Eur. J. Immunol. 2014, 44, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Umetani, N.; Kim, J.; Hiramatsu, S.; Reber, H.A.; Hines, O.J.; Bilchik, A.J.; Hoon, D.S.B. Increased Integrity of Free Circulating DNA in Sera of Patients with Colorectal or Periampullary Cancer: Direct Quantitative PCR for ALU Repeats. Clin. Chem. 2006, 52, 1062–1069. [Google Scholar] [CrossRef] [Green Version]
- Tangkijvanich, P.; Hourpai, N.; Rattanatanyong, P.; Wisedopas, N.; Mahachai, V.; Mutirangura, A. Serum LINE-1 Hypomethylation as a Potential Prognostic Marker for Hepatocellular Carcinoma. Clin. Chim. Acta 2007, 379, 127–133. [Google Scholar] [CrossRef]
- Umetani, N.; Giuliano, A.E.; Hiramatsu, S.H.; Amersi, F.; Nakagawa, T.; Martino, S.; Hoon, D.S.B. Prediction of Breast Tumor Progression by Integrity of Free Circulating DNA in Serum. J. Clin. Oncol. 2006, 24, 4270–4276. [Google Scholar] [CrossRef]
- Ivanov, M.; Chernenko, P.; Breder, V.; Laktionov, K.; Rozhavskaya, E.; Musienko, S.; Baranova, A.; Mileyko, V. Utility of CfDNA Fragmentation Patterns in Designing the Liquid Biopsy Profiling Panels to Improve Their Sensitivity. Front. Genet. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Wentzel, J.F.; Aucamp, J.; van Dyk, E.; du Plessis, L.; Pretorius, P.J. Characterization of the Cell-Free DNA Released by Cultured Cancer Cells. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Guibert, J.; Benachi, A.; Grebille, A.-G.; Ernault, P.; Zorn, J.-R.; Costa, J.-M. Kinetics of SRY Gene Appearance in Maternal Serum: Detection by Real Time PCR in Early Pregnancy after Assisted Reproductive Technique. Hum. Reprod. 2003, 18, 1733–1736. [Google Scholar] [CrossRef] [Green Version]
- Wataganara, T.; LeShane, E.S.; Chen, A.Y.; Borgatta, L.; Peter, I.; Johnson, K.L.; Bianchi, D.W. Plasma Gamma-Globin Gene Expression Suggests That Fetal Hematopoietic Cells Contribute to the Pool of Circulating Cell-Free Fetal Nucleic Acids during Pregnancy. Clin. Chem. 2004, 50, 689–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finning, K.; Martin, P.; Summers, J.; Massey, E.; Poole, G.; Daniels, G. Effect of High Throughput RHD Typing of Fetal DNA in Maternal Plasma on Use of Anti-RhD Immunoglobulin in RhD Negative Pregnant Women: Prospective Feasibility Study. BMJ 2008, 336, 816–818. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early-and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couraud, S.; Vaca-Paniagua, F.; Villar, S.; Oliver, J.; Schuster, T.; Blanché, H.; Girard, N.; Trédaniel, J.; Guilleminault, L.; Gervais, R.; et al. Noninvasive Diagnosis of Actionable Mutations by Deep Sequencing of Circulating Free DNA in Lung Cancer from Never-Smokers: A Proof-of-Concept Study from BioCAST/IFCT-1002. Clin. Cancer Res. 2014, 20, 4613–4624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Olmo, D.C.; Domínguez, C.; García-Arranz, M.; Anker, P.; Stroun, M.; García-Verdugo, J.M.; García-Olmo, D. Cell-Free Nucleic Acids Circulating in the Plasma of Colorectal Cancer Patients Induce the Oncogenic Transformation of Susceptible Cultured Cells. Cancer Res. 2010, 70, 560–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Tong, K.L.; Li, P.K.; Chan, A.Y.; Yeung, C.K.; Pang, C.C.; Wong, T.Y.; Lee, K.C.; Lo, Y.M. Presence of Donor- and Recipient-Derived DNA in Cell-Free Urine Samples of Renal Transplantation Recipients: Urinary DNA Chimerism. Clin. Chem. 1999, 45, 1741–1746. [Google Scholar] [CrossRef]
- García Moreira, V.; Prieto García, B.; Baltar Martín, J.M.; Ortega Suárez, F.; Alvarez, F.V. Cell-Free DNA as a Noninvasive Acute Rejection Marker in Renal Transplantation. Clin. Chem. 2009, 55, 1958–1966. [Google Scholar] [CrossRef] [Green Version]
- Macher, H.C.; Suárez-Artacho, G.; Guerrero, J.M.; Gómez-Bravo, M.A.; Álvarez-Gómez, S.; Bernal-Bellido, C.; Dominguez-Pascual, I.; Rubio, A. Monitoring of Transplanted Liver Health by Quantification of Organ-Specific Genomic Marker in Circulating DNA from Receptor. PLoS ONE 2014, 9, e113987. [Google Scholar] [CrossRef] [Green Version]
- Tsui, N.B.Y.; Kadir, R.A.; Chan, K.C.A.; Chi, C.; Mellars, G.; Tuddenham, E.G.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.K.; Lo, Y.M.D. Noninvasive Prenatal Diagnosis of Hemophilia by Microfluidics Digital PCR Analysis of Maternal Plasma DNA. Blood 2011, 117, 3684–3691. [Google Scholar] [CrossRef] [Green Version]
- Beck, J.; Bierau, S.; Balzer, S.; Andag, R.; Kanzow, P.; Schmitz, J.; Gaedcke, J.; Moerer, O.; Slotta, J.E.; Walson, P.; et al. Digital Droplet PCR for Rapid Quantification of Donor DNA in the Circulation of Transplant Recipients as a Potential Universal Biomarker of Graft Injury. Clin. Chem. 2013, 59, 1732–1741. [Google Scholar] [CrossRef] [Green Version]
- Snyder, T.M.; Khush, K.K.; Valantine, H.A.; Quake, S.R. Universal Noninvasive Detection of Solid Organ Transplant Rejection. Proc. Natl. Acad. Sci. USA 2011, 108, 6229–6234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurkmans, D.P.; Verhoeven, J.G.H.P.; de Leur, K.; Boer, K.; Joosse, A.; Baan, C.C.; von der Thüsen, J.H.; van Schaik, R.H.N.; Mathijssen, R.H.J.; van der Veldt, A.A.M.; et al. Donor-Derived Cell-Free DNA Detects Kidney Transplant Rejection during Nivolumab Treatment. J. Immunother. Cancer 2019, 7, 182. [Google Scholar] [CrossRef]
- Schütz, E.; Fischer, A.; Beck, J.; Harden, M.; Koch, M.; Wuensch, T.; Stockmann, M.; Nashan, B.; Kollmar, O.; Matthaei, J.; et al. Graft-Derived Cell-Free DNA, a Noninvasive Early Rejection and Graft Damage Marker in Liver Transplantation: A Prospective, Observational, Multicenter Cohort Study. PLoS Med. 2017, 14, e1002286. [Google Scholar] [CrossRef]
- Oellerich, M.; Schütz, E.; Kanzow, P.; Schmitz, J.; Beck, J.; Kollmar, O.; Streit, F.; Walson, P.D. Use of Graft-Derived Cell-Free DNA as an Organ Integrity Biomarker to Reexamine Effective Tacrolimus Trough Concentrations after Liver Transplantation. Ther. Drug Monit. 2014, 36, 136–140. [Google Scholar] [CrossRef] [Green Version]
- Liao, G.J.W.; Lun, F.M.F.; Zheng, Y.W.L.; Chan, K.C.A.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.K.; Lo, Y.M.D. Targeted Massively Parallel Sequencing of Maternal Plasma DNA Permits Efficient and Unbiased Detection of Fetal Alleles. Clin. Chem. 2011, 57, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.W.K.; Chan, K.C.A.; Gao, Y.; Lau, V.Y.M.; Zheng, W.; Leung, T.Y.; Foo, C.H.F.; Xie, B.; Tsui, N.B.Y.; Lun, F.M.F.; et al. Noninvasive Pre-Natal Diagnosis of Fetal Chromosomal Aneuploidy by Massively Parallel Genomic Sequencing of DNA in Maternal Plasma. Proc. Natl. Acad. Sci. USA 2008, 105, 20458–20463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchi, D.W.; Parker, R.L.; Wentworth, J.; Madankumar, R.; Saffer, C.; Das, A.F.; Craig, J.A.; Chudova, D.I.; Devers, P.L.; Jones, K.W.; et al. DNA Sequencing versus Standard Prenatal Aneuploidy Screening. N. Engl. J. Med. 2014, 370, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Jeon, Y.J.; Zhou, Y.; Li, Y.; Guo, Q.; Chen, J.; Quan, S.; Zhang, A.; Zheng, H.; Zhu, X.; Lin, J.; et al. The Feasibility Study of Non-Invasive Fetal Trisomy 18 and 21 Detection with Semiconductor Sequencing Platform. PLoS ONE 2014, 9, e110240. [Google Scholar] [CrossRef] [Green Version]
- Niba, E.T.E.; Tran, V.K.; Tuan-Pham, L.A.; Vu, D.C.; Nguyen, N.K.; Ta, V.T.; Tran, T.H.; Lee, T.; Takeshima, Y.; Matsuo, M. Validation of Ambiguous MLPA Results by Targeted Next-Generation Sequencing Discloses a Nonsense Mutation in the DMD Gene. Clin. Chim. Acta. 2014, 436, 155–159. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Noninvasive Diagnosis of Fetal Aneuploidy by Shotgun Sequencing DNA from Maternal Blood. Proc. Natl. Acad. Sci. USA 2008, 105, 16266–16271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.; Batey, A.; Struble, C.; Musci, T.; Song, K.; Oliphant, A. Gestational Age and Maternal Weight Effects on Fetal Cell-Free DNA in Maternal Plasma. Prenat. Diagn. 2013, 33, 662–666. [Google Scholar] [CrossRef]
- Yeo, W.; Wong, N.; Wong, W.-L.; Lai, P.B.S.; Zhong, S.; Johnson, P.J. High Frequency of Promoter Hypermethylation of RASSF1A in Tumor and Plasma of Patients with Hepatocellular Carcinoma. Liver Int. 2005, 25, 266–272. [Google Scholar] [CrossRef] [PubMed]
- De Vlaminck, I.; Valantine, H.A.; Snyder, T.M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; Bernstein, D.; Weisshaar, D.; et al. Circulating Cell-Free DNA Enables Noninvasive Diagnosis of Heart Transplant Rejection. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef] [Green Version]
- Sharon, E.; Shi, H.; Kharbanda, S.; Koh, W.; Martin, L.R.; Khush, K.K.; Valantine, H.; Pritchard, J.K.; De Vlaminck, I. Quantification of Transplant-Derived Circulating Cell-Free DNA in Absence of a Donor Genotype. PLoS Comput. Biol. 2017, 13, e1005629. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Archila, F.A.; Constantin, T.; Prins, S.A.; Liberto, J.; Damm, I.; Towfighi, P.; Navarro, S.; Kirkizlar, E.; Demko, Z.P.; et al. Optimizing Detection of Kidney Transplant Injury by Assessment of Donor-Derived Cell-Free DNA via Massively Multiplex PCR. J. Clin. Med. 2018, 8, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurdyukov, S.; Bullock, M. DNA Methylation Analysis: Choosing the Right Method. Biology 2016, 5, 3. [Google Scholar] [CrossRef]
- Oellerich, M.; Schütz, E.; Beck, J.; Walson, P.D. Circulating Cell-Free DNA-Diagnostic and Prognostic Applications in Personalized Cancer Therapy. Ther. Drug Monit. 2019, 41, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Z.; Jia, E.; Ouyang, T.; Pan, M.; Lu, J.; Ge, Q.; Bai, Y. Analysis of Genome-Wide in Cell Free DNA Methylation: Progress and Prospect. Analyst 2019, 144, 5912–5922. [Google Scholar] [CrossRef]
- Sant, K.E.; Nahar, M.S.; Dolinoy, D.C. DNA Methylation Screening and Analysis. Methods Mol. Biol. 2012, 889, 385–406. [Google Scholar] [CrossRef] [Green Version]
- Galardi, F.; De Luca, F.; Romagnoli, D.; Biagioni, C.; Moretti, E.; Biganzoli, L.; Di Leo, A.; Migliaccio, I.; Malorni, L.; Benelli, M. Cell-Free Dna-Methylation-Based Methods and Applications in Oncology. Biomolecules 2020, 10, 1677. [Google Scholar] [CrossRef]
- Meddeb, R.; Pisareva, E.; Thierry, A.R. Guidelines for the Preanalytical Conditions for Analyzing Circulating Cell-Free DNA. Clin. Chem. 2019, 65, 623–633. [Google Scholar] [CrossRef]
- El Messaoudi, S.; Rolet, F.; Mouliere, F.; Thierry, A.R. Circulating Cell Free DNA: Preanalytical Considerations. Clin. Chim. Acta 2013, 424, 222–230. [Google Scholar] [CrossRef]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-Free DNA in Human Blood Plasma. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, Structures, and Functions of Circulating DNA in Oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; Pantel, K. Circulating DNA as Biomarker in Breast Cancer. Breast Cancer Res. 2015, 17, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, Y.M.D.; Chan, K.C.A.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.F.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.W.L.; Chan, K.C.A.; Sun, H.; Jiang, P.; Su, X.; Chen, E.Z.; Lun, F.M.F.; Hung, E.C.W.; Lee, V.; Wong, J.; et al. Nonhematopoietically Derived DNA Is Shorter than Hematopoietically Derived DNA in Plasma: A Transplantation Model. Clin. Chem. 2012, 58, 549–558. [Google Scholar] [CrossRef] [Green Version]
- Zovico, P.V.C.; Gasparini, V.H.; Venâncio, F.A.; Miguel, G.P.S.; Pedrosa, R.G.; Haraguchi, F.K.; Barauna, V.G.V.G.; Neto, V.H.G.; Venâncio, F.A.; Miguel, G.P.S.; et al. Cell-Free DNA as an Obesity Biomarker. Physiol. Res. 2020, in press, 515–520. [Google Scholar] [CrossRef]
- Grunt, M.; Hillebrand, T.; Schwarzenbach, H. Clinical Relevance of Size Selection of Circulating DNA. Transl. Cancer Res. 2018, 7 (Suppl. S2), S171–S184. [Google Scholar] [CrossRef]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and Death of Circulating Cell-Free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Chan, K.C.A.; Zhang, J.; Hui, A.B.Y.; Wong, N.; Lau, T.K.; Leung, T.N.; Lo, K.W.; Huang, D.W.S.; Lo, Y.M.D. Size Distributions of Maternal and Fetal DNA in Maternal Plasma. Clin. Chem. 2004, 50, 88–92. [Google Scholar] [CrossRef] [Green Version]
- Mouliere, F.; Messaoudi, S.E.; Gongora, C.; Guedj, A.S.; Robert, B.; del Rio, M.; Molina, F.; Lamy, P.J.; Lopez-Crapez, E.; Mathonnet, M.; et al. Circulating Cell-Free DNA from Colorectal Cancer Patients May Reveal High KRAS or BRAF Mutation Load. Transl. Oncol. 2013, 6, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.C.A.; Jiang, P.; Zheng, Y.W.L.; Liao, G.J.W.; Sun, H.; Wong, J.; Siu, S.S.N.; Chan, W.C.; Chan, S.L.; Chan, A.T.C.; et al. Cancer Genome Scanning in Plasma: Detection of Tumor-Associated Copy Number Aberrations, Single-Nucleotide Variants, and Tumoral Heterogeneity by Massively Parallel Sequencing. Clin. Chem. 2013, 59, 211–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heitzer, E.; Ulz, P.; Belic, J.; Gutschi, S.; Quehenberger, F.; Fischereder, K.; Benezeder, T.; Auer, M.; Pischler, C.; Mannweiler, S.; et al. Tumor-Associated Copy Number Changes in the Circulation of Patients with Prostate Cancer Identified through Whole-Genome Sequencing. Genome Med. 2013, 5, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yung, T.K.F.; Chan, K.C.A.; Mok, T.S.K.; Tong, J.; To, K.F.; Lo, Y.M.D. Single-Molecule Detection of Epidermal Growth Factor Receptor Mutations in Plasma by Microfluidics Digital PCR in Non-Small Cell Lung Cancer Patients. Clin. Cancer Res. 2009, 15, 2076–2084. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.C.A.; Lai, P.B.S.; Mok, T.S.K.; Chan, H.L.Y.; Ding, C.; Yeung, S.W.; Lo, Y.M.D. Quantitative Analysis of Circulating Methylated DNA as a Biomarker for Hepatocellular Carcinoma. Clin. Chem. 2008, 54, 1528–1536. [Google Scholar] [CrossRef]
- Andor, N.; Maley, C.C.; Ji, H.P. Genomic Instability in Cancer: Teetering on the Limit of Tolerance. Cancer Res. 2017, 77, 2179–2185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Molparia, B.; Nichani, E.; Torkamani, A. Assessment of Circulating Copy Number Variant Detection for Cancer Screening. PLoS ONE 2017, 12, e0180647. [Google Scholar] [CrossRef] [Green Version]
- Heitzer, E.; Auer, M.; Hoffmann, E.M.; Pichler, M.; Gasch, C.; Ulz, P.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Mohan, S.; et al. Establishment of Tumor-Specific Copy Number Alterations from Plasma DNA of Patients with Cancer. Int. J. Cancer 2013, 133, 346–356. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Dittmar, R.L.; Xia, S.; Zhang, H.; Du, M.; Huang, C.C.; Druliner, B.R.; Boardman, L.; Wang, L. Cell-Free DNA Copy Number Variations in Plasma from Colorectal Cancer Patients. Mol. Oncol. 2017, 11, 1099–1111. [Google Scholar] [CrossRef]
- Xia, Y.; Huang, C.C.; Dittmar, R.; Du, M.; Wang, Y.; Liu, H.; Shenoy, N.; Wang, L.; Kohli, M. Copy Number Variations in Urine Cell Free DNA as Biomarkers in Advanced Prostate Cancer. Oncotarget 2016, 7, 35818–35831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutilin, D.S.; Airapetova, T.G.; Anistratov, P.A.; Pyltsin, S.P.; Leiman, I.A.; Karnaukhov, N.S.; Kit, O.I. Copy Number Variation in Tumor Cells and Extracellular DNA in Patients with Lung Adenocarcinoma. Bull. Exp. Biol. Med. 2019, 167, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Galanopoulos, M.; Papanikolaou, I.S.; Zografos, E.; Viazis, N.; Papatheodoridis, G.; Karamanolis, D.; Marinos, E.; Mantzaris, G.J.; Gazouli, M. Comparative Study of Mutations in Single Nucleotide Polymorphism Loci of KRAS and BRAF Genes in Patients Who Underwent Screening Colonoscopy, with and without Premalignant Intestinal Polyps. Anticancer Res. 2017, 37, 651–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatami, F.; Larijani, B.; Heshmat, R.; Nasiri, S.; Haddadi-Aghdam, M.; Teimoori-Toolabi, L.; Tavangar, S.M. Hypermethylated RASSF1 and SLC5A8 Promoters alongside BRAFV600E Mutation as Biomarkers for Papillary Thyroid Carcinoma. J. Cell. Physiol. 2020. [Google Scholar] [CrossRef]
- Tzanikou, E.; Haselmann, V.; Markou, A.; Duda, A.; Utikal, J.; Neumaier, M.; Lianidou, E.S. Direct Comparison Study between Droplet Digital PCR and a Combination of Allele-Specific PCR, Asymmetric Rapid PCR and Melting Curve Analysis for the Detection of BRAF V600E Mutation in Plasma from Melanoma Patients. Clin. Chem. Lab. Med. 2020. [Google Scholar] [CrossRef]
- Fratte, C.D.; Guardascione, M.; De Mattia, E.; Borsatti, E.; Boschetto, R.; Farruggio, A.; Canzonieri, V.; Romanato, L.; Borsatti, R.; Gagno, S.; et al. Clonal Selection of a Novel Deleterious TP53 Somatic Mutation Discovered in CtDNA of a KIT/PDGFRA Wild-Type Gastrointestinal Stromal Tumor Resistant to Imatinib. Front. Pharmacol. 2020, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Boldrin, E.; Nardo, G.; Zulato, E.; Bonanno, L.; Polo, V.; Frega, S.; Pavan, A.; Indraccolo, S.; Saggioro, D. Detection of Loss of Heterozygosity in CfDNA of Advanced EGFR-or KRAS-Mutated Non-Small-Cell Lung Cancer Patients. Int. J. Mol. Sci. 2020, 21, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karachaliou, N.; Mayo-De Las Casas, C.; Queralt, C.; De Aguirre, I.; Melloni, B.; Cardenal, F.; Garcia-Gomez, R.; Massuti, B.; Sánchez, J.M.; Porta, R.; et al. Association of EGFR L858R Mutation in Circulating Free DNA with Survival in the EURTAC Trial. JAMA Oncol. 2015, 1, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Hamakawa, T.; Kukita, Y.; Kurokawa, Y.; Miyazaki, Y.; Takahashi, T.; Yamasaki, M.; Miyata, H.; Nakajima, K.; Taniguchi, K.; Takiguchi, S.; et al. Monitoring Gastric Cancer Progression with Circulating Tumour DNA. Br. J. Cancer 2015, 112, 352–356. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Weigelt, B.; Cortes, J.; Won, H.H.; Ng, C.K.Y.; Nuciforo, P.; Bidard, F.-C.; Aura, C.; Saura, C.; Peg, V.; et al. Capturing Intra-Tumor Genetic Heterogeneity by de Novo Mutation Profiling of Circulating Cell-Free Tumor DNA: A Proof-of-Principle. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 1729–1735. [Google Scholar] [CrossRef]
- Min, S.; Shin, S.; Chung, Y.J. Detection of KRAS Mutations in Plasma Cell-Free DNA of Colorectal Cancer Patients and Comparison with Cancer Panel Data for Tissue Samples of the Same Cancers. Genom. Inform. 2019, 17, 1–6. [Google Scholar] [CrossRef]
- Ermakov, A.V.; Konkova, M.S.; Kostyuk, S.V.; Izevskaya, V.L.; Baranova, A.; Veiko, N.N. Oxidized Extracellular DNA as a Stress Signal in Human Cells. Oxid. Med. Cell. Longev. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Loseva, P.; Kostyuk, S.; Malinovskaya, E.; Clement, N.; Dechesne, C.A.; Dani, C.; Smirnova, T.; Glebova, K.; Baidakova, G.; Baranova, A.; et al. Extracellular DNA Oxidation Stimulates Activation of NRF2 and Reduces the Production of ROS in Human Mesenchymal Stem Cells. Expert Opin. Biol. Ther. 2012, 12 (Suppl. S1), 85–97. [Google Scholar] [CrossRef]
- Bulicheva, N.; Fidelina, O.; Mkrtumova, N.; Neverova, M.; Bogush, A.; Bogush, M.; Roginko, O.; Veiko, N. Effect of Cell-Free DNA of Patients with Cardiomyopathy and RDNA on the Frequency of Contraction of Electrically Paced Neonatal Rat Ventricular Myocytes in Culture. Ann. Acad. Sci. 2008, 1137, 273–277. [Google Scholar] [CrossRef]
- Veiko, N.N.; Bulycheva, N.A.; Roginko, O.A.; Veiko, R.V.; Ershova, E.S.; Kozdoba, O.A.; Kuzmin, V.A.; Vinogradov, A.M.; Yudin, A.A.; Speranskyi, A.I. Ribosomal Repeat in Cell Free DNA as a Marker for Cell Death. Biochem. Suppl. Ser. B Biomed. Chem. 2008, 2, 198–207. [Google Scholar] [CrossRef]
- Veiko, N.N.; Shubaeva, N.O.; Ivanova, S.M.; Speranskii, A.I.; Lyapunova, N.A.; Spitkovskii, D.M. Blood Serum DNA in Patients with Rheumatoid Arthritis Is Considerably Enriched with Fragments of Ribosomal Repeats Containing Immunostimulatory CpG-Motifs. Bull. Exp. Biol. Med. 2006, 142, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Guz, J.; Foksinski, M.; Siomek, A.; Gackowski, D.; Rozalski, R.; Dziaman, T.; Szpila, A.; Olinski, R. The Relationship between 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine Level and Extent of Cytosine Methylation in Leukocytes DNA of Healthy Subjects and in Patients with Colon Adenomas and Carcinomas. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2008, 640, 170–173. [Google Scholar] [CrossRef]
- Mangal, D.; Vudathala, D.; Park, J.H.; Seon, H.L.; Penning, T.M.; Blair, I.A. Analysis of 7,8-Dihydro-8-Oxo-2′-Deoxyguanosine in Cellular DNA during Oxidative Stress. Chem. Res. Toxicol. 2009, 22, 788–797. [Google Scholar] [CrossRef]
- Ermakov, A.V.; Konkova, M.S.; Kostyuk, S.V.; Egolina, N.A.; Efremova, L.V.; Veiko, N.N. Oxidative Stress as a Significant Factor for Development of an Adaptive Response in Irradiated and Nonirradiated Human Lymphocytes after Inducing the Bystander Effect by Low-Dose X-Radiation. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2009, 669, 155–161. [Google Scholar] [CrossRef]
- Kostyuk, S.V.; Malinovskaya, E.M.; Ermakov, A.V.; Smirnova, T.D.; Kameneva, L.V.; Chvartatskaya, O.V.; Loseva, P.A.; Ershova, E.S.; Lyubchenko, L.N.; Veiko, N.N. Fragments of Cell-Free DNA Increase Transcription in Human Mesenchymal Stem Cells, Activate TLR-Dependent Signal Pathway, and Suppress Apoptosis. Biochem. Suppl. Ser. B Biomed. Chem. 2012, 6, 68–74. [Google Scholar] [CrossRef]
- Callinan, P.A.; Feinberg, A.P. The Emerging Science of Epigenomics. Hum. Mol. Genet. 2006, 15, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Laird, P.W. Cancer Epigenetics Comes of Age. Nat. Genet. 1999, 21, 163–167. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive Human Cell-Type Methylation Atlas Reveals Origins of Circulating Cell-Free DNA in Health and Disease. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Andreatta, M.V.; Curty, V.M.; Coutinho, J.V.S.; Santos, M.Â.A.; Vassallo, P.F.; de Sousa, N.F.; Barauna, V.G. CfDNA as an Earlier Predictor of Exercise-Induced Performance Decrement Related to Muscle Damage. Int. J. Sports Physiol. Perform. 2018, 13, 953–956. [Google Scholar] [CrossRef] [PubMed]
- Breitbach, S.; Tug, S.; Helmig, S.; Zahn, D.; Kubiak, T.; Michal, M.; Gori, T.; Ehlert, T.; Beiter, T.; Simon, P. Direct Quantification of Cell-Free, Circulating DNA from Unpurified Plasma. PLoS ONE 2014, 9, e87838. [Google Scholar] [CrossRef] [Green Version]
- Tug, S.; Helmig, S.; Deichmann, E.R.; Schmeier-Jürchott, A.; Wagner, E.; Zimmermann, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-Induced Increases in Cell Free DNA in Human Plasma Originate Predominantly from Cells of the Haematopoietic Lineage. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar] [PubMed]
- Zhong, X.Y.; Laivuori, H.; Livingston, J.C.; Ylikorkala, O.; Sibai, B.M.; Holzgreve, W.; Hahn, S. Elevation of Both Maternal and Fetal Extracellular Circulating Deoxyribonucleic Acid Concentrations in the Plasma of Pregnant Women with Preeclampsia. Am. J. Obstet. Gynecol. 2001, 184, 414–419. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, G.C.; Petrone, A.B.; Tennant, C.S.; Lucke-Wold, N.; Kabbani, Y.; Tarabishy, A.R.; Chantler, P.D.; Barr, T.L. Circulating Extracellular DNA Levels Are Acutely Elevated in Ischaemic Stroke and Associated with Innate Immune System Activation. Brain Inj. 2017, 31, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.Y.; Von Mühlenen, I.; Li, Y.; Kang, A.; Gupta, A.K.; Tyndall, A.; Holzgreve, W.; Hahn, S.; Hasler, P. Increased Concentrations of Antibody-Bound Circulatory Cell-Free DNA in Rheumatoid Arthritis. Clin. Chem. 2007, 53, 1609–1614. [Google Scholar] [CrossRef] [PubMed]
- Swarup, V.; Rajeswari, M.R. Circulating (Cell-Free) Nucleic Acids-A Promising, Non-Invasive Tool for Early Detection of Several Human Diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saukkonen, K.; Lakkisto, P.; Pettilä, V.; Varpula, M.; Karlsson, S.; Ruokonen, E.; Pulkki, K. Cell-Free Plasma DNA as a Predictor of Outcome in Severe Sepsis and Septic Shock. Clin. Chem. 2008, 54, 1000–1007. [Google Scholar] [CrossRef] [Green Version]
- Lam, N.Y.L.; Rainer, T.H.; Chan, L.Y.S.; Joynt, G.M.; Lo, Y.M.D. Time Course of Early and Late Changes in Plasma DNA in Trauma Patients. Clin. Chem. 2003, 49, 1286–1291. [Google Scholar] [CrossRef]
- Leng, S.; Zheng, J.; Jin, Y.; Zhang, H.; Zhu, Y.; Wu, J.; Xu, Y.; Zhang, P. Plasma Cell-Free DNA Level and Its Integrity as Biomarkers to Distinguish Non-Small Cell Lung Cancer from Tuberculosis. Clin. Chim. Acta 2018, 477, 160–165. [Google Scholar] [CrossRef]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, K.; Aggarwal, S.; Bhardwaj, S.; Ramakrishna, G.; Pandey, C.K. Serial Perioperative Cell-Free DNA Levels in Donors and Recipients Undergoing Living Donor Liver Transplantation. Acta Anaesthesiol. Scand. 2017, 61, 1084–1094. [Google Scholar] [CrossRef]
- Kamat, A.A.; Baldwin, M.; Urbauer, D.; Dang, D.; Han, L.Y.; Godwin, A.; Karlan, B.Y.; Simpson, J.L.; Gershenson, D.M.; Coleman, R.L.; et al. Plasma Cell-Free DNA in Ovarian Cancer: An Independent Prognostic Biomarker. Cancer 2010, 116, 1918–1925. [Google Scholar] [CrossRef] [Green Version]
- Tissot, C.; Toffart, A.C.; Villar, S.; Souquet, P.J.; Merle, P.; Moro-Sibilot, D.; Pérol, M.; Zavadil, J.; Brambilla, C.; Olivier, M.; et al. Circulating Free DNA Concentration Is an Independent Prognostic Biomarker in Lung Cancer. Eur. Respir. J. 2015, 46, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Radpour, R.; Barekati, Z.; Asadollahi, R.; Bitzer, J.; Wight, E.; Bürki, N.; Diesch, C.; Holzgreve, W.; Zhong, X.Y. Levels of Plasma Circulating Cell Free Nuclear and Mitochondrial DNA as Potential Biomarkers for Breast Tumors. Mol. Cancer 2009, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Tsai, N.W.; Lin, T.K.; Chen, S.D.; Chang, W.N.; Wang, H.C.; Yang, T.M.; Lin, Y.J.; Jan, C.R.; Huang, C.R.; Liou, C.W.; et al. The Value of Serial Plasma Nuclear and Mitochondrial DNA Levels in Patients with Acute Ischemic Stroke. Clin. Chim. Acta 2011, 412, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Nakahira, K.; Guo, X.; Choi, A.M.K.; Gu, Z. Very Short Mitochondrial DNA Fragments and Heteroplasmy in Human Plasma. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Mair, R.; Mouliere, F.; Smith, C.G.; Chandrananda, D.; Gale, D.; Marass, F.; Tsui, D.W.Y.; Massie, C.E.; Wright, A.J.; Watts, C.; et al. Measurement of Plasma Cell-Free Mitochondrial Tumor DNA Improves Detection of Glioblastoma in Patient-Derived Orthotopic Xenograft Models. Cancer Res. 2019, 79, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Yu, M. Circulating Cell-Free Mitochondrial DNA as a Novel Cancer Biomarker: Opportunities and Challenges. Mitochondrial DNA 2012, 23, 329–332. [Google Scholar] [CrossRef]
- Ingelsson, B.; Söderberg, D.; Strid, T.; Söderberg, A.; Bergh, A.C.; Loitto, V.; Lotfi, K.; Segelmark, M.; Spyrou, G.; Rosén, A. Lymphocytes Eject Interferogenic Mitochondrial DNA Webs in Response to CpG and Non-CpG Oligodeoxynucleotides of Class C. Proc. Natl. Acad. Sci. USA 2018, 115, E478–E487. [Google Scholar] [CrossRef] [Green Version]
- Itagaki, K.; Kaczmarek, E.; Lee, Y.T.; Tang, I.T.; Isal, B.; Adibnia, Y.; Sandler, N.; Grimm, M.J.; Segal, B.H.; Otterbein, L.E.; et al. Mitochondrial DNA Released by Trauma Induces Neutrophil Extracellular Traps. PLoS ONE 2015, 10, e0120549. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.; Hill, M.; Chitty, L.S. Offering Non-Invasive Prenatal Testing as Part of Routine Clinical Service. Can High Levels of Informed Choice Be Maintained? Prenat. Diagn. 2017, 37, 1130–1137. [Google Scholar] [CrossRef]
- Allyse, M.; Minear, M.A.; Berson, E.; Sridhar, S.; Rote, M.; Hung, A.; Chandrasekharan, S. Non-Invasive Prenatal Testing: A Review of International Implementation and Challenges. Int. J. Womens. Health 2015, 7, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Kwapisz, D. The First Liquid Biopsy Test Approved. Is It a New Era of Mutation Testing for Non-Small Cell Lung Cancer? Ann. Transl. Med. 2017, 5, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FDA. Nucleic Acid Based Tests. Available online: https://www.fda.gov/medical-devices/in-vitro-diagnostics/nucleic-acid-based-tests (accessed on 17 April 2021).
- Ossandon, M.R.; Agrawal, L.; Bernhard, E.J.; Conley, B.A.; Dey, S.M.; Divi, R.L.; Guan, P.; Lively, T.G.; McKee, T.C.; Sorg, B.S.; et al. Circulating Tumor DNA Assays in Clinical Cancer Research. J. Natl. Cancer Inst. 2018, 110, 929–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volckmar, A.-L.; Sültmann, H.; Riediger, A.; Fioretos, T.; Schirmacher, P.; Endris, V.; Stenzinger, A.; Dietz, S. A Field Guide for Cancer Diagnostics Using Cell-Free DNA: From Principles to Practice and Clinical Applications. Genes Chromosomes Cancer 2018, 57, 123–139. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, Y.; Li, S.; Zeng, F.; Meng, Y.; Chen, Z.; Liu, S.; Tao, Y.; Yu, F. The Interplay of Circulating Tumor DNA and Chromatin Modification, Therapeutic Resistance, and Metastasis. Mol. Cancer 2019, 18, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Weng, J.-L.; Atyah, M.; Zhou, C.-H.; Ren, N. Progress in Quantitative Technique of Circulating Cell Free DNA and Its Role in Cancer Diagnosis and Prognosis. Cancer Genet. 2019, 239, 75–84. [Google Scholar] [CrossRef]
- Cervena, K.; Vodicka, P.; Vymetalkova, V. Diagnostic and Prognostic Impact of Cell-Free DNA in Human Cancers: Systematic Review. Mutat. Res. 2019, 781, 100–129. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.; Taus, A.; Montagut, C. Dynamic Treatment Stratification Using CtDNA. Recent Results Cancer Res. 2020, 215, 263–273. [Google Scholar] [CrossRef]
- Shao, X.; He, Y.; Ji, M.; Chen, X.; Qi, J.; Shi, W.; Hao, T.; Ju, S. Quantitative Analysis of Cell-Free DNA in Ovarian Cancer. Oncol. Lett. 2015, 10, 3478–3482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanzow, P.; Kollmar, O.; Schütz, E.; Oellerich, M.; Schmitz, J.; Beck, J.; Walson, P.D.; Slotta, J.E. Graft-Derived Cell-Free DNA as an Early Organ Integrity Biomarker after Transplantation of a Marginal HELLP Syndrome Donor Liver. Transplantation 2014, 98, e43–e45. [Google Scholar] [CrossRef]
- Krüger-Genge, A.; Blocki, A.; Franke, R.P.; Jung, F. Vascular Endothelial Cell Biology: An Update. Int. J. Mol. Sci. 2019, 20, 4411. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.; Buchanan, M.R.; Anderson, T.J. Endothelial Function Testing as a Biomarker of Vascular Disease. Circulation 2003, 108, 2054–2059. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.A. Coronary Risk Factors, Endothelial Function, and Atherosclerosis: A Review. Clin. Cardiol. 1997, 20, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Leite, A.R.; Borges-Canha, M.; Cardoso, R.; Neves, J.S.; Castro-Ferreira, R.; Leite-Moreira, A. Novel Biomarkers for Evaluation of Endothelial Dysfunction. Angiology 2020, 3319720903586. [Google Scholar] [CrossRef] [PubMed]
- Constans, J.; Conri, C. Circulating Markers of Endothelial Function in Cardiovascular Disease. Clin. Chim. Acta 2006, 368, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Coscas, R.; Bensussan, M.; Jacob, M.P.; Louedec, L.; Massy, Z.; Sadoine, J.; Daudon, M.; Chaussain, C.; Bazin, D.; Michel, J.B. Free DNA Precipitates Calcium Phosphate Apatite Crystals in the Arterial Wall in Vivo. Atherosclerosis 2017, 259, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Kapustin, A.N.; Chatrou, M.L.L.; Drozdov, I.; Zheng, Y.; Davidson, S.M.; Soong, D.; Furmanik, M.; Sanchis, P.; De Rosales, R.T.M.; Alvarez-Hernandez, D.; et al. Vascular Smooth Muscle Cell Calcification Is Mediated by Regulated Exosome Secretion. Circ. Res. 2015, 116, 1312–1323. [Google Scholar] [CrossRef] [Green Version]
- Paunel-Görgülü, A.; Wacker, M.; El Aita, M.; Hassan, S.; Schlachtenberger, G.; Deppe, A.; Choi, Y.H.; Kuhn, E.; Mehler, T.O.; Wahlers, T. CfDNA Correlates with Endothelial Damage after Cardiac Surgery with Prolonged Cardiopulmonary Bypass and Amplifies NETosis in an Intracellular TLR9-Independent Manner. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Yang, Y.; Zeng, C.; Lu, X.; Song, Y.; Nie, J.; Ran, R.; Zhang, Z.; He, C.; Zhang, W.; Liu, S.-M. 5-Hydroxymethylcytosines in Circulating Cell-Free DNA Reveal Vascular Complications of Type 2 Diabetes. Clin. Chem. 2019, 65, 1414–1425. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Miranda, F.S.; Barauna, V.G.; dos Santos, L.; Costa, G.; Vassallo, P.F.; Campos, L.C.G. Properties and Application of Cell-Free DNA as a Clinical Biomarker. Int. J. Mol. Sci. 2021, 22, 9110. https://doi.org/10.3390/ijms22179110
de Miranda FS, Barauna VG, dos Santos L, Costa G, Vassallo PF, Campos LCG. Properties and Application of Cell-Free DNA as a Clinical Biomarker. International Journal of Molecular Sciences. 2021; 22(17):9110. https://doi.org/10.3390/ijms22179110
Chicago/Turabian Stylede Miranda, Felipe Silva, Valério Garrone Barauna, Leandro dos Santos, Gustavo Costa, Paula Frizera Vassallo, and Luciene Cristina Gastalho Campos. 2021. "Properties and Application of Cell-Free DNA as a Clinical Biomarker" International Journal of Molecular Sciences 22, no. 17: 9110. https://doi.org/10.3390/ijms22179110
APA Stylede Miranda, F. S., Barauna, V. G., dos Santos, L., Costa, G., Vassallo, P. F., & Campos, L. C. G. (2021). Properties and Application of Cell-Free DNA as a Clinical Biomarker. International Journal of Molecular Sciences, 22(17), 9110. https://doi.org/10.3390/ijms22179110





