Changes in Sphingolipid Profile of Benzo[a]pyrene-Transformed Human Bronchial Epithelial Cells Are Reflected in the Altered Composition of Sphingolipids in Their Exosomes
Abstract
:1. Introduction
2. Results
2.1. Characterization of Isolated Exosomes
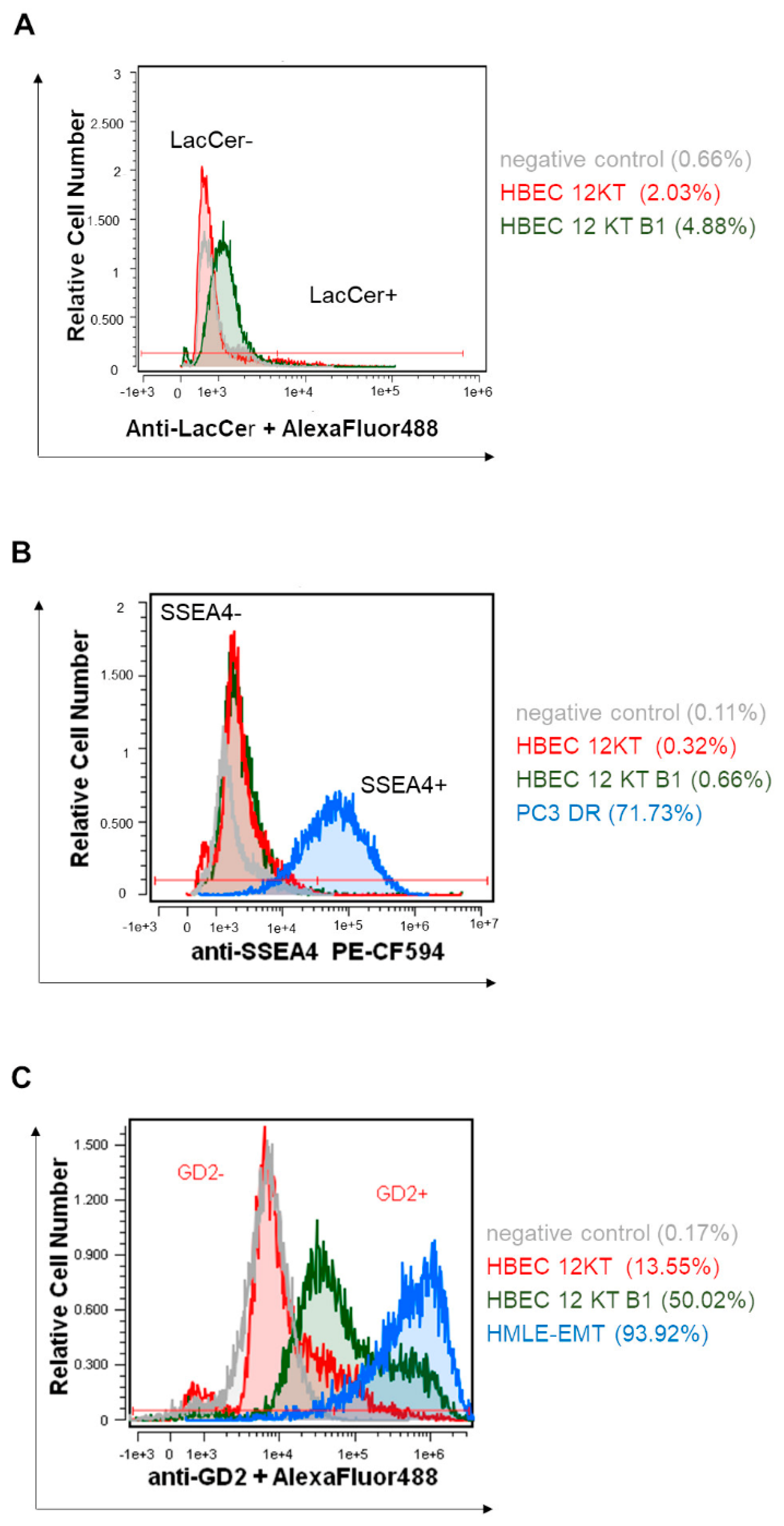
2.2. HPLC-MS/MS and Flow Cytometric Analyses of SL and GSL Profiles in Transformed and Parental HBEC-12KT Cells, and in Their Exosomes
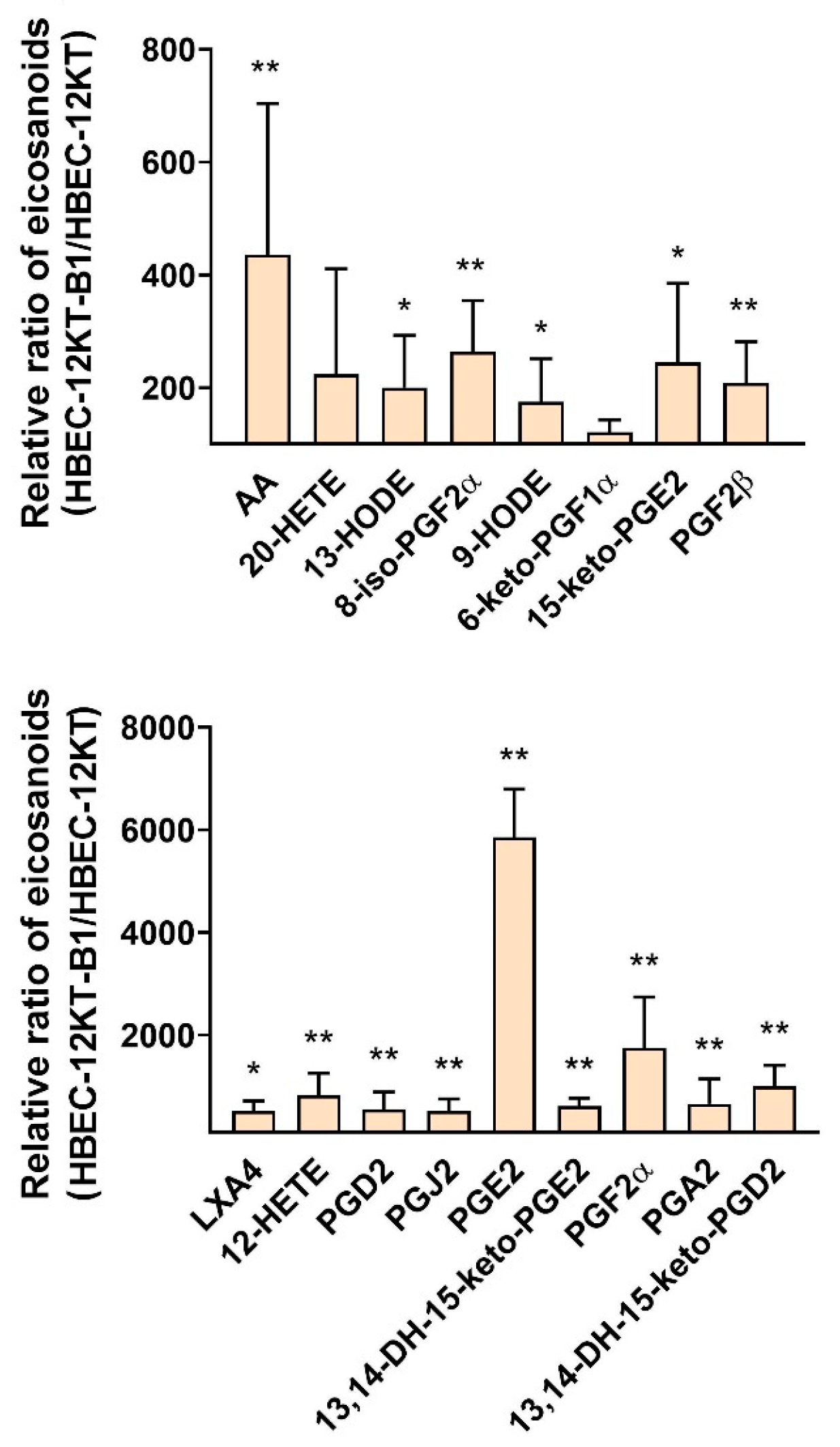
2.3. HPLC-MS/MS Analysis of Eicosanoids Released from Transformed and Parental HBEC-12KT Cells
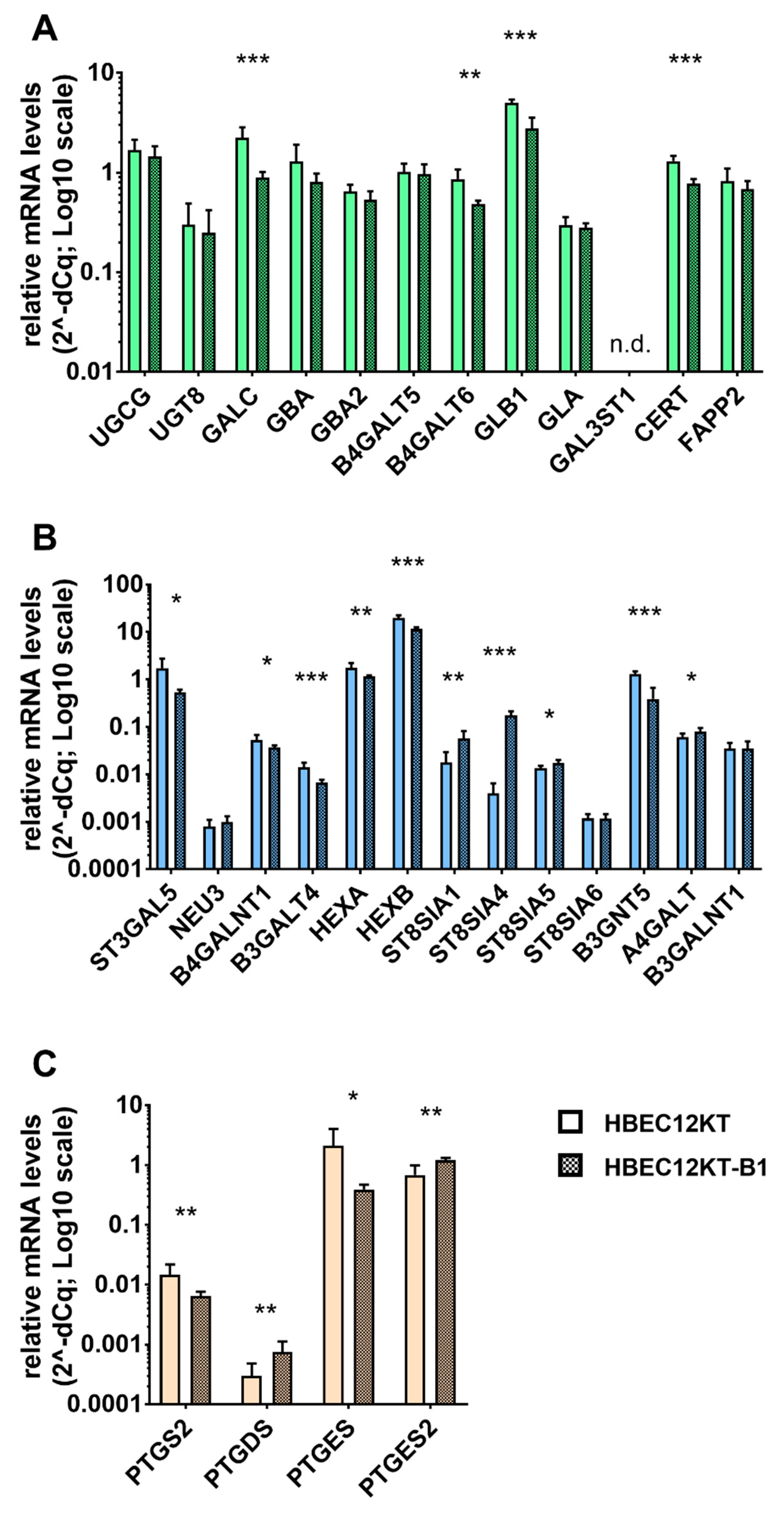
2.4. Expression Analysis of Genes Participating in the GSL and Eicosanoid Metabolism
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Isolation of Exosomes from Cell Conditioned Media
4.3. Size and Concentration Characterization of Isolated Exosomes
4.4. Transmission Electron Microscopy and Immunogold Labeling of Isolated Exosomes
4.5. Flow Cytometry
4.6. Sphingolipid Extraction and Analysis
4.7. Liquid Chromatography Separation and Tandem Mass Spectrometry Conditions for Eicosanoid Analysis
4.8. RNA Isolation and RT-qPCR Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [Green Version]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Qasim, M.; Khan, K.; Kim, J.-H. Biogenesis, membrane trafficking, functions, and next generation nanotherapeutics medicine of extracellular vesicles. Int. J. Nanomed. 2021, 16, 3357–3383. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; König, A.K.; Marmé, F.; Runz, S.; Wolterink, S.; Koensgen, D.; Mustea, A.; Sehouli, J.; Altevogt, P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009, 278, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Di Vizio, D.; Morello, M.; Dudley, A.C.; Schow, P.W.; Adam, R.M.; Morley, S.; Mulholland, D.; Rotinen, M.; Hager, M.H.; Insabato, L.; et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am. J. Pathol. 2012, 181, 1573–1584. [Google Scholar] [CrossRef]
- Kim, J.; Kim, T.Y.; Lee, M.S.; Mun, J.Y.; Ihm, C.; Kim, S.A. Exosome cargo reflects TGF-β1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2016, 478, 643–648. [Google Scholar] [CrossRef]
- El-Sayed, I.Y.; Daher, A.; Destouches, D.; Firlej, V.; Kostallari, E.; Maillé, P.; Huet, E.; Haidar-Ahmad, N.; Jenster, G.; de la Taille, A.; et al. Extracellular vesicles released by mesenchymal-like prostate carcinoma cells modulate EMT state of recipient epithelial-like carcinoma cells through regulation of AR signaling. Cancer Lett. 2017, 410, 100–111. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Latifkar, A.; Cerione, R.A.; Antonyak, M.A. Probing the mechanisms of extracellular vesicle biogenesis and function in cancer. Biochem. Soc. Trans. 2018, 46, 1137–1146. [Google Scholar] [CrossRef]
- Jabalee, J.; Towle, R.; Garnis, C. The role of extracellular vesicles in cancer: Cargo, function, and therapeutic implications. Cells 2018, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef]
- Record, M.; Silvente-Poirot, S.; Poirot, M.; Wakelam, M.J.O. Extracellular vesicles: Lipids as key components of their biogenesis and functions. J. Lipid Res. 2018, 59, 1316–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Lydic, T.A.; Townsend, S.; Adda, C.G.; Collins, C.; Mathivanan, S.; Reid, G.E. Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods 2015, 87, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Haraszti, R.A.; Didiot, M.C.; Sapp, E.; Leszyk, J.; Shaffer, S.A.; Rockwell, H.E.; Gao, F.; Narain, N.R.; DiFiglia, M.; Kiebish, M.A.; et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 2016, 5, 32570. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell. Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- García-González, V.; Díaz-Villanueva, J.F.; Galindo-Hernández, O.; Martínez-Navarro, I.; Hurtado-Ureta, G.; Pérez-Arias, A.A. Ceramide metabolism balance, a multifaceted factor in critical steps of breast cancer development. Int. J. Mol. Sci. 2018, 19, 2527. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, K.; Ohmi, Y.; Ohkawa, Y.; Bhuiyan, R.H.; Zhang, P.; Tajima, O.; Hashimoto, N.; Hamamura, K.; Furukawa, K. New era of research on cancer-associated glycosphingolipids. Cancer Sci. 2019, 10, 1544–1551. [Google Scholar] [CrossRef] [Green Version]
- Cumin, C.; Huang, Y.L.; Everest-Dass, A.; Jacob, F. Deciphering the importance of glycosphingolipids on cellular and molecular mechanisms associated with epithelial-to-mesenchymal transition in cancer. Biomolecules 2021, 11, 62. [Google Scholar] [CrossRef]
- Furukawa, K.; Ohkawa, Y.; Yamauchi, Y.; Hamamura, K.; Ohmi, Y.; Furukawa, K. Fine tuning of cell signals by glycosylation. J. Biochem. 2012, 151, 573–578. [Google Scholar] [CrossRef] [Green Version]
- Machala, M.; Procházková, J.; Hofmanová, J.; Králiková, L.; Slavík, J.; Tylichová, Z.; Ovesná, P.; Kozubík, A.; Vondráček, J. Colon cancer and perturbations of the sphingolipid metabolism. Int. J. Mol. Sci. 2019, 20, 6051. [Google Scholar] [CrossRef] [Green Version]
- Russo, D.; Capolupo, L.; Loomba, J.S.; Sticco, L.; D’Angelo, G. Glycosphingolipid metabolism in cell fate specification. J. Cell Sci. 2018, 131, jcs219204. [Google Scholar] [CrossRef] [Green Version]
- Finetti, F.; Travelli, C.; Ercoli, J.; Colombo, G.; Buoso, E.; Trabalzini, L. Prostaglandin E2 and cancer: Insight into tumor progression and immunity. Biology 2020, 9, 434. [Google Scholar] [CrossRef]
- Pettus, B.J.; Chalfant, C.E.; Hannun, Y.A. Sphingolipids in inflammation: Roles and implications. Curr. Mol. Med. 2004, 4, 405–418. [Google Scholar] [CrossRef]
- Kawamori, T.; Kaneshiro, T.; Okumura, M.; Maalouf, S.; Uflacker, A.; Bielawski, J.; Hannun, Y.A.; Obeid, L.M. Role for sphingosine kinase 1 in colon carcinogenesis. FASEB J. 2009, 23, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Bersaas, A.; Arnoldussen, Y.J.; Sjøberg, M.; Haugen, A.; Mollerup, S. Epithelial-mesenchymal transition and FOXA genes during tobacco smoke carcinogen induced transformation of human bronchial epithelial cells. Toxicol. In Vitro 2016, 35, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Menter, D.G.; Dubois, R.N. Prostaglandins in cancer cell adhesion, migration and invasion. Int. J. Cell Biol. 2012, 2012, 723419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biringer, R.G. A review of prostanoid receptors: Expression, characterization, regulation and mechanism of action. J. Cell Commun. Signal. 2021, 15, 155–184. [Google Scholar] [CrossRef] [PubMed]
- Vangaveti, V.; Baune, B.T.; Kennedy, R.L. Hydroxyoctadecadienoic acids: Novel regulators of macrophage differentiation and atherogenesis. Ther. Adv. Endocrinol. Metab. 2010, 1, 51–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van’t Erve, T.J.; Lih, F.B.; Jelsema, C.; Deterding, L.J.; Eling, T.E.; Mason, R.P.; Kadiiska, M.B. Reinterpreting the best biomarker of oxidative stress: The 8-iso-prostaglandin F2α/prostaglandin F2α ratio shows complex origins of lipid peroxidation biomarkers in animal models. Free Radic. Biol. Med. 2016, 95, 65–73. [Google Scholar] [CrossRef]
- Patwardhan, G.A.; Liu, Y.Y. Sphingolipids and expression regulation of genes in cancer. Prog. Lipid Res. 2011, 50, 104–114. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Muñoz, A.; Presa, N.; Gomez-Larrauri, A.; Rivera, I.G.; Trueba, M.; Ordoñez, M. Control of inflammatory responses by ceramide, sphingosine 1-phosphate and ceramide 1-phosphate. Prog. Lipid Res. 2016, 61, 51–62. [Google Scholar] [CrossRef]
- Basu, S.; Rossary, A.; Vasson, M. Role of inflammation and eicosanoids in breast cancer. Lipid Technol. 2016, 28, 60–64. [Google Scholar] [CrossRef]
- Moreno, J.J. New aspects of the role of hydroxyeicosatetraenoic acids in cell growth and cancer development. Biochem. Pharmacol. 2009, 77, 1–10. [Google Scholar] [CrossRef]
- Nakamura, H.; Murayama, T. The role of sphingolipids in arachidonic acid metabolism. J. Pharmacol. Sci. 2014, 124, 307–312. [Google Scholar] [CrossRef] [Green Version]
- Sampaio, J.L.; Gerl, M.J.; Klose, C.; Ejsing, C.S.; Beug, H.; Simons, K.; Shevchenko, A. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA 2011, 108, 1903–1907. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Chen, B.; Shen, Z.; Deng, H.; Liu, D.; Xie, X.; Gan, X.; Xu, X.; Huang, Z.; Chen, J. DNA promoter hypermethylation contributes to down-regulation of galactocerebrosidase gene in lung and head and neck cancers. Int. J. Clin. Exp. Pathol. 2015, 8, 11042–11050. [Google Scholar]
- Kovbasnjuk, O.; Mourtazina, R.; Baibakov, B.; Wang, T.; Elowsky, C.; Choti, M.A.; Kane, A.; Donowitz, M. The glycosphingolipid globotriaosylceramide in the metastatic transformation of colon cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 19087–19092. [Google Scholar] [CrossRef] [Green Version]
- Tyler, A.; Johansson, A.; Karlsson, T.; Gudey, S.K.; Brännström, T.; Grankvist, K.; Behnam-Motlagh, P. Targeting glucosylceramide synthase induction of cell surface globotriaosylceramide (Gb3) in acquired cisplatin-resistance of lung cancer and malignant pleural mesothelioma cells. Exp. Cell Res. 2015, 336, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, R.D.; Sheridan, S.; Girard, L.; Sato, M.; Kim, Y.; Pollack, J.; Peyton, M.; Zou, Y.; Kurie, J.M.; Dimaio, J.M.; et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004, 64, 9027–9034. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Pospichalova, V.; Svoboda, J.; Dave, Z.; Kotrbova, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E.; et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef]
- Prochazkova, J.; Slavik, J.; Bouchal, J.; Levkova, M.; Huskova, Z.; Ehrmann, J.; Ovesna, P.; Kolar, Z.; Skalicky, P.; Strakova, N.; et al. Specific alterations of sphingolipid metabolism identified in EpCAM-positive cells isolated from human colon tumors. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158742. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
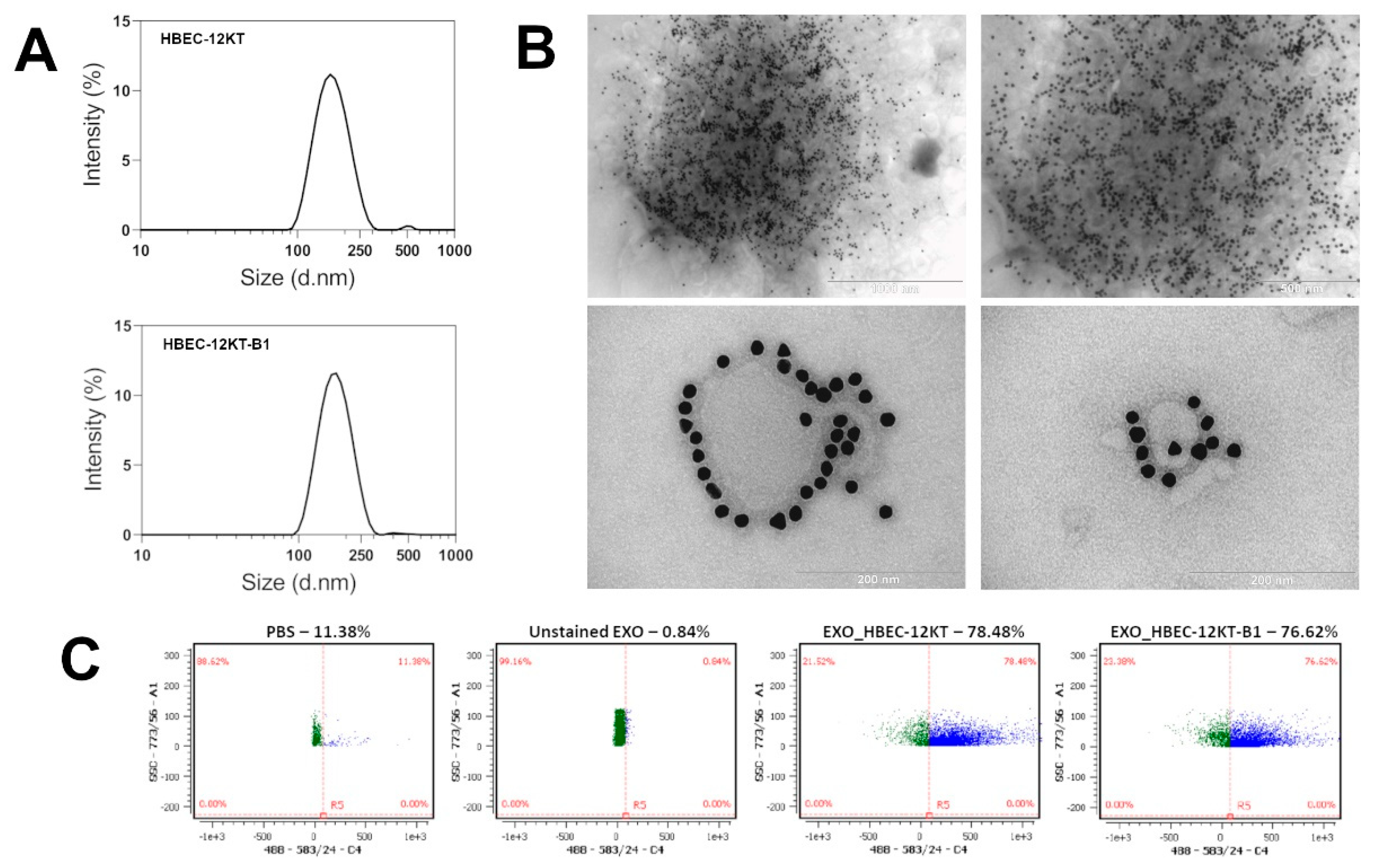


| Cell Line | Z-Average (d.nm) | PdI | Number of Particles per mL | Number of Particles per Sample | Number of Cells per Sample | Number of Particles per Million Cells |
|---|---|---|---|---|---|---|
| HBEC-12KT | 158 ± 7 | 0.130 ± 0.009 | 5.58 ± 2.46 × 1011 | 2.51 ± 1.11 × 1010 | 1.01 ± 0.31 × 108 | 2.53 ± 1.23 × 108 |
| HBEC-12KT-B1 | 162 ± 8 | 0.123 ± 0.030 | 8.55 ± 2.08 × 1011 | 3.85 ± 0.94 × 1010 | 1.85 ± 0.38 × 108 | 2.26 ± 0.70 × 108 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machala, M.; Slavík, J.; Kováč, O.; Procházková, J.; Pěnčíková, K.; Pařenicová, M.; Straková, N.; Kotouček, J.; Kulich, P.; Mollerup, S.; et al. Changes in Sphingolipid Profile of Benzo[a]pyrene-Transformed Human Bronchial Epithelial Cells Are Reflected in the Altered Composition of Sphingolipids in Their Exosomes. Int. J. Mol. Sci. 2021, 22, 9195. https://doi.org/10.3390/ijms22179195
Machala M, Slavík J, Kováč O, Procházková J, Pěnčíková K, Pařenicová M, Straková N, Kotouček J, Kulich P, Mollerup S, et al. Changes in Sphingolipid Profile of Benzo[a]pyrene-Transformed Human Bronchial Epithelial Cells Are Reflected in the Altered Composition of Sphingolipids in Their Exosomes. International Journal of Molecular Sciences. 2021; 22(17):9195. https://doi.org/10.3390/ijms22179195
Chicago/Turabian StyleMachala, Miroslav, Josef Slavík, Ondrej Kováč, Jiřina Procházková, Kateřina Pěnčíková, Martina Pařenicová, Nicol Straková, Jan Kotouček, Pavel Kulich, Steen Mollerup, and et al. 2021. "Changes in Sphingolipid Profile of Benzo[a]pyrene-Transformed Human Bronchial Epithelial Cells Are Reflected in the Altered Composition of Sphingolipids in Their Exosomes" International Journal of Molecular Sciences 22, no. 17: 9195. https://doi.org/10.3390/ijms22179195
APA StyleMachala, M., Slavík, J., Kováč, O., Procházková, J., Pěnčíková, K., Pařenicová, M., Straková, N., Kotouček, J., Kulich, P., Mollerup, S., Vondráček, J., & Hýžďalová, M. (2021). Changes in Sphingolipid Profile of Benzo[a]pyrene-Transformed Human Bronchial Epithelial Cells Are Reflected in the Altered Composition of Sphingolipids in Their Exosomes. International Journal of Molecular Sciences, 22(17), 9195. https://doi.org/10.3390/ijms22179195







