Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs
Abstract
:1. Introduction
2. Results
2.1. Motif Profile of RNA Aptamers
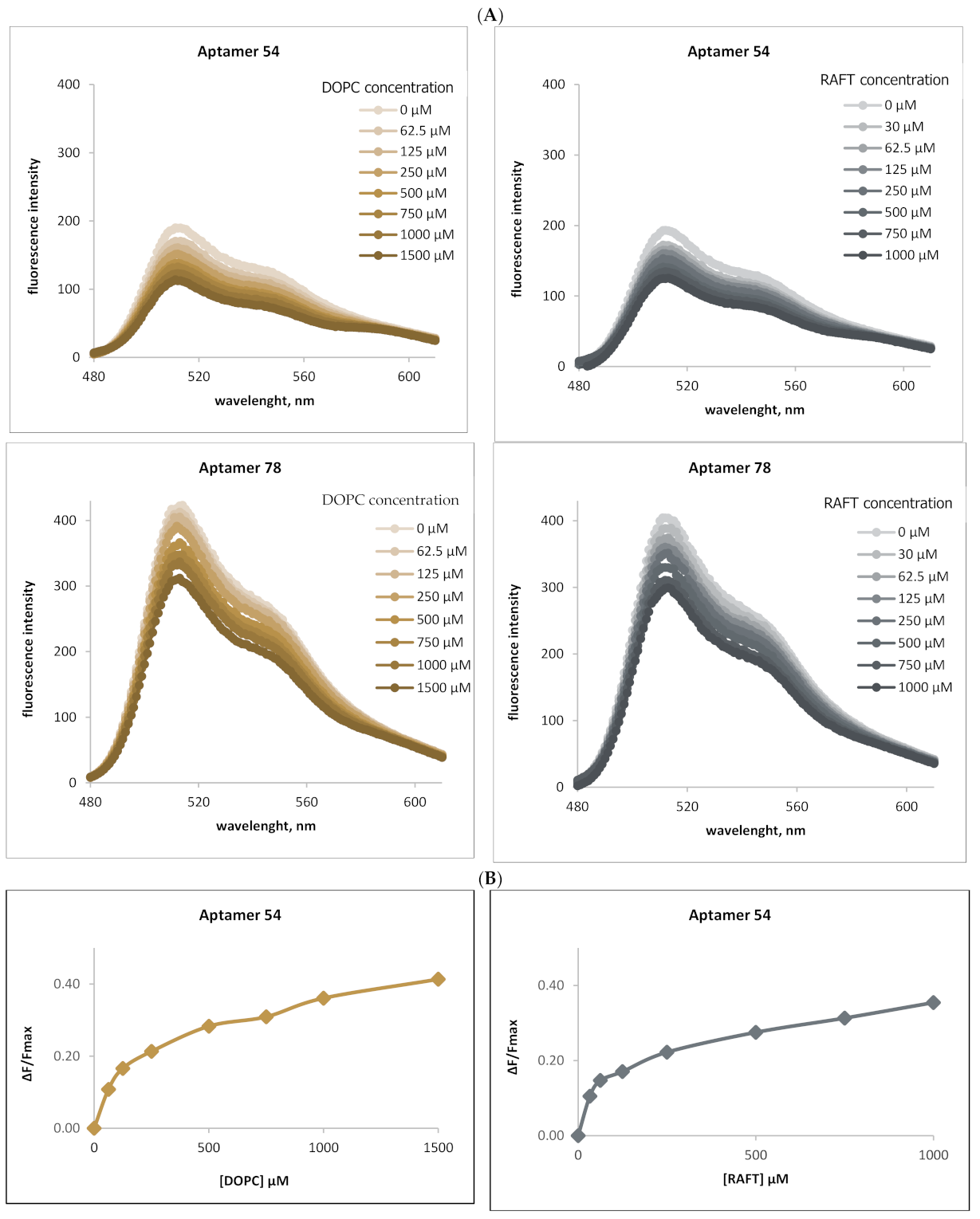
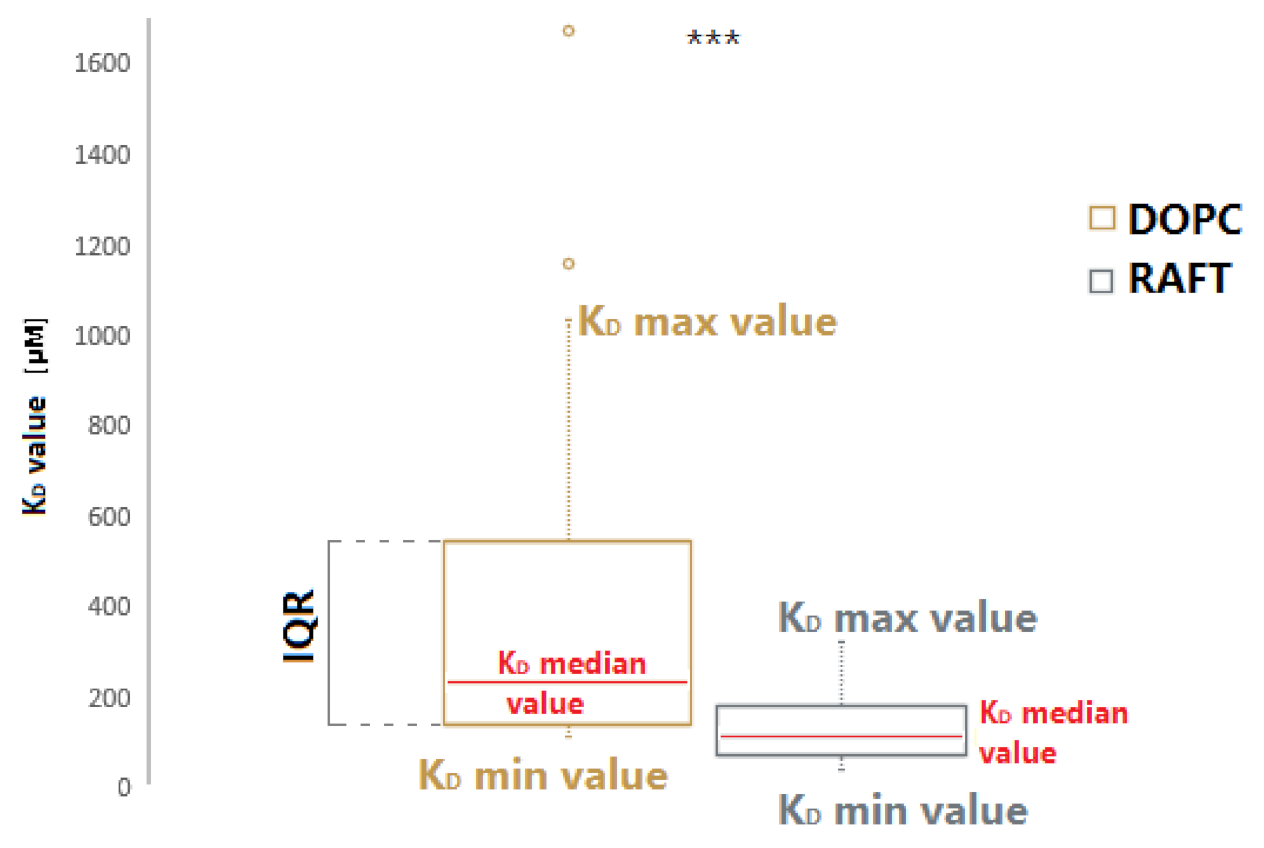
2.2. Binding of RNA Aptamers to the Ordered and Disordered Lipid Membranes
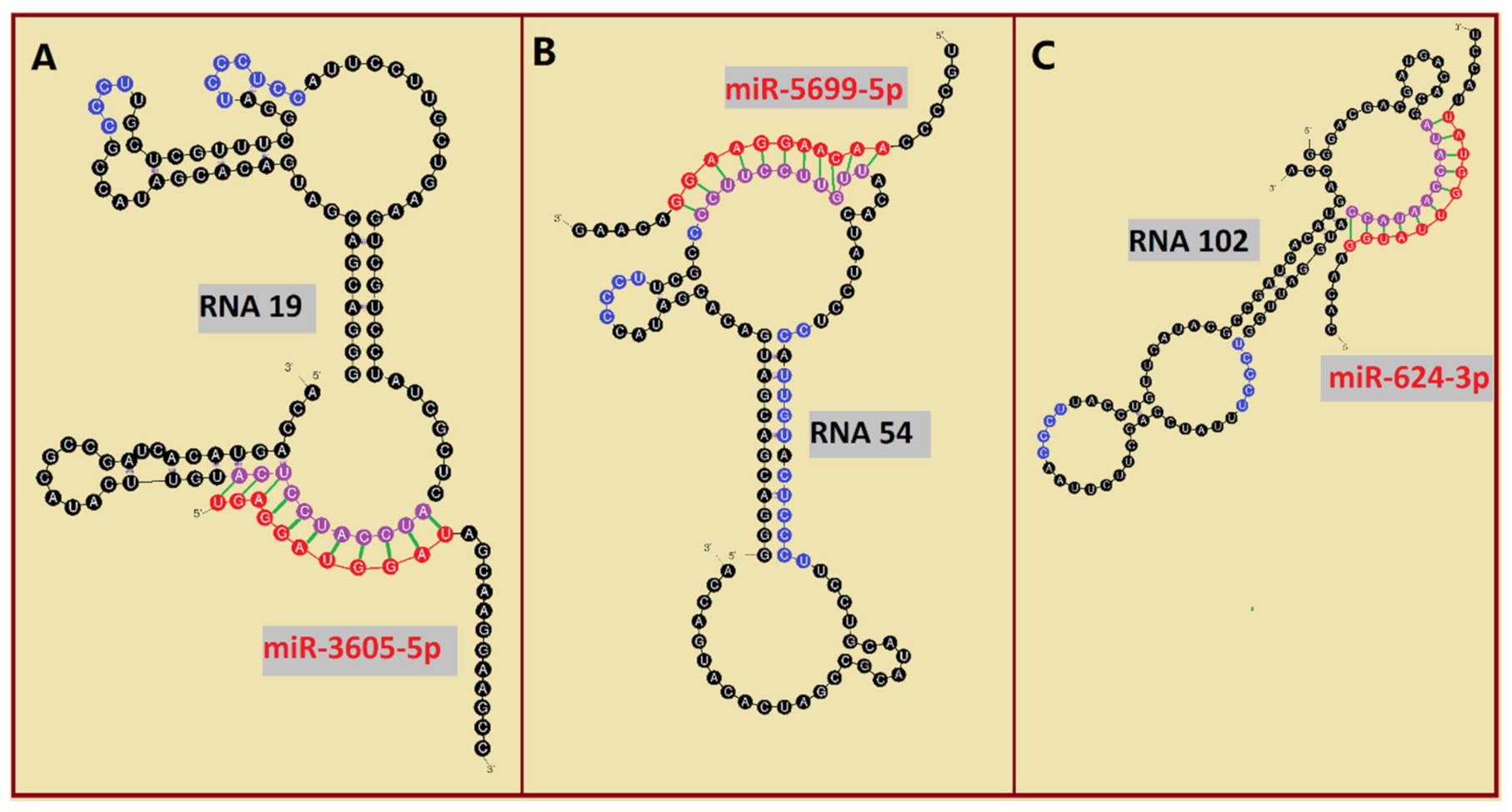
2.3. Analysis of EXO- and RAFT-Motifs within RNA Aptamers
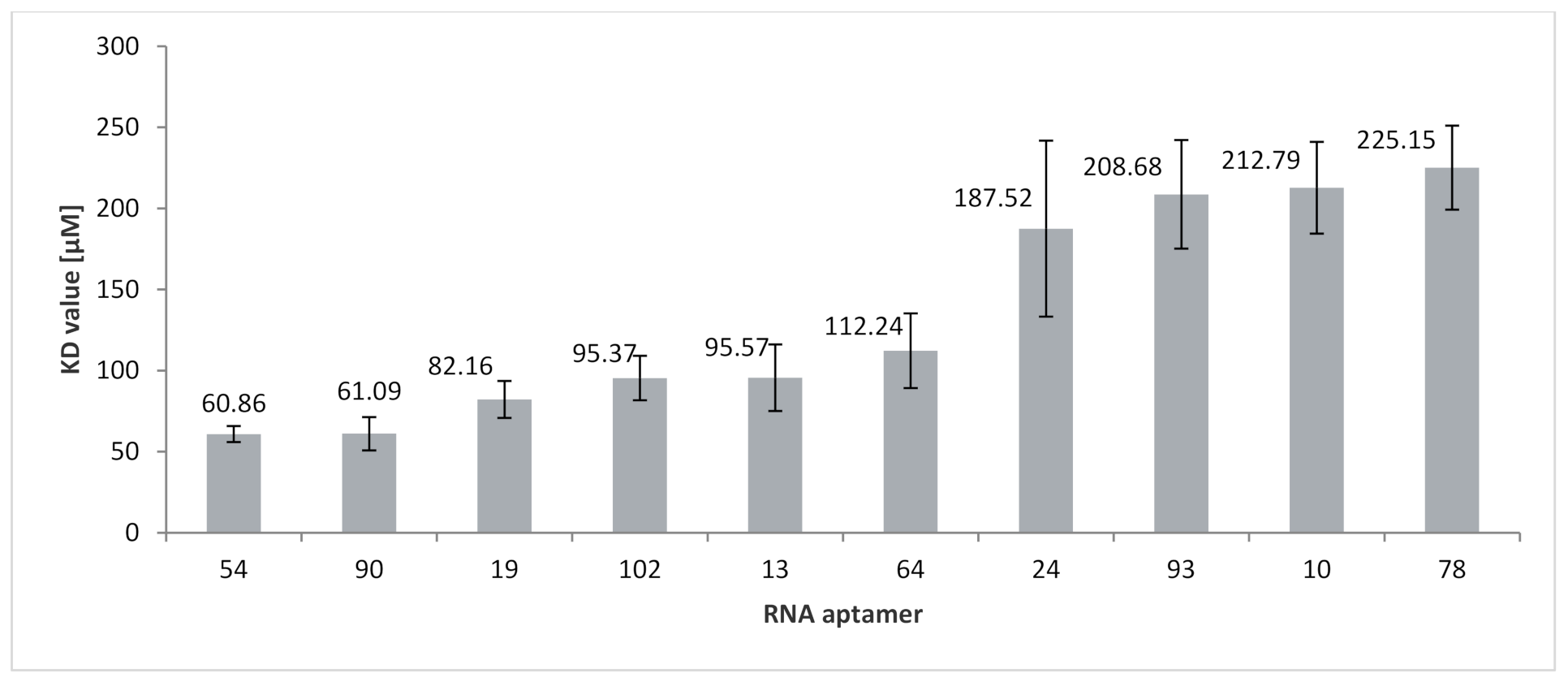
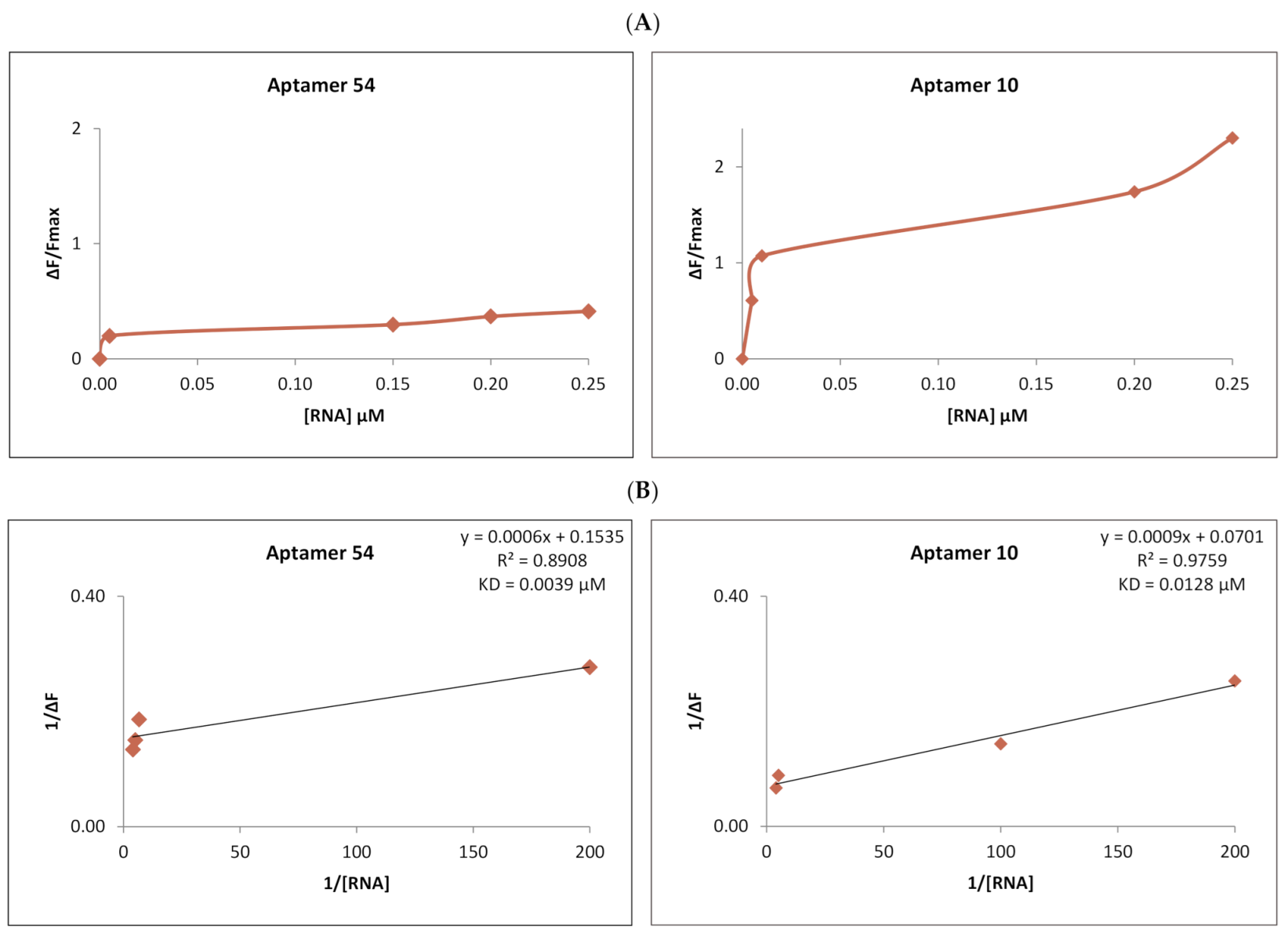
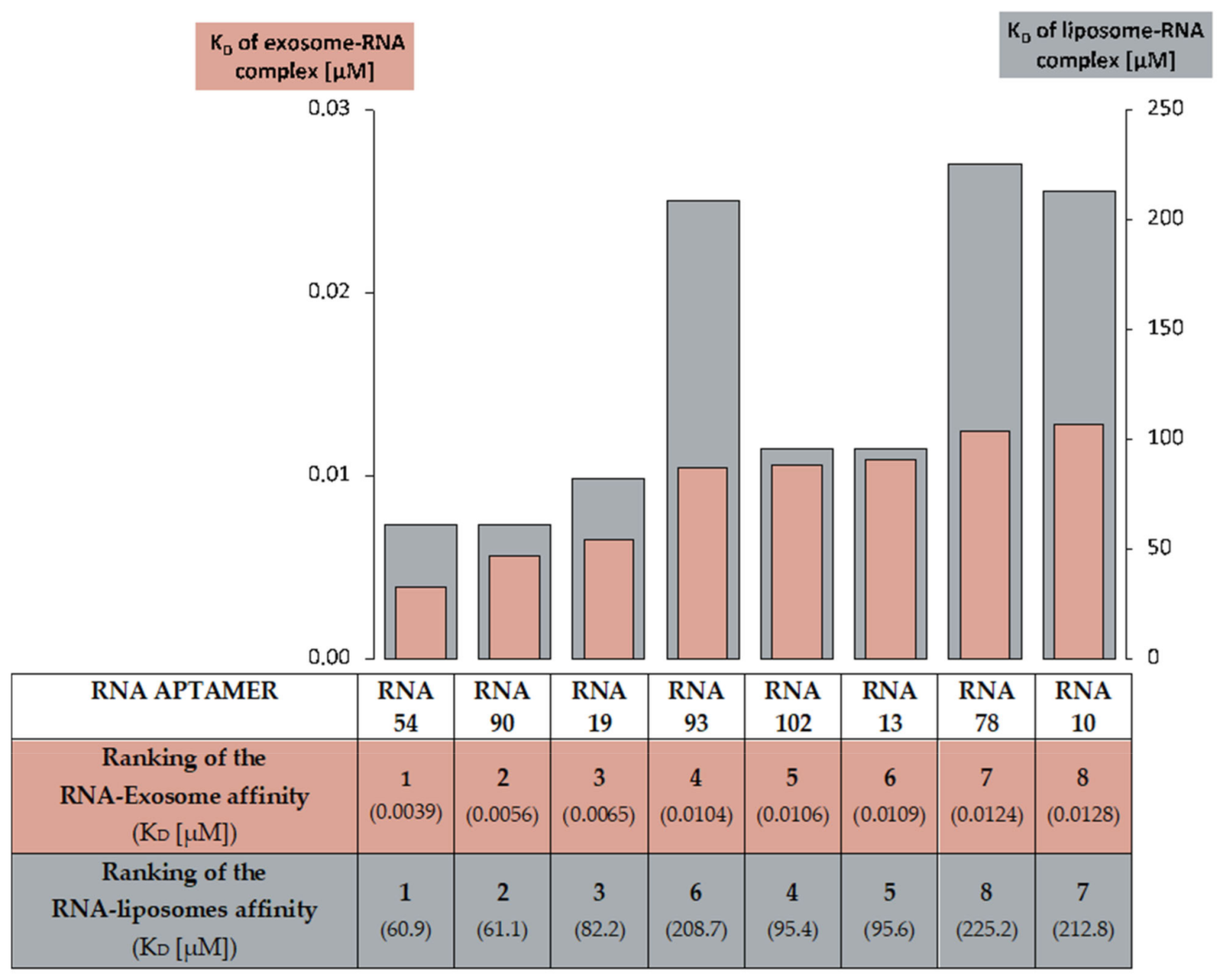
2.4. Analysis of RNA Aptamers Affinity to Exosomal Membranes
3. Discussion
3.1. The Importance of Lipid Rafts for Binding RNA Aptamers to Membranes
3.2. The Importance of EXO- and RAFT-Motifs in Binding of RNA Molecules to Membranes
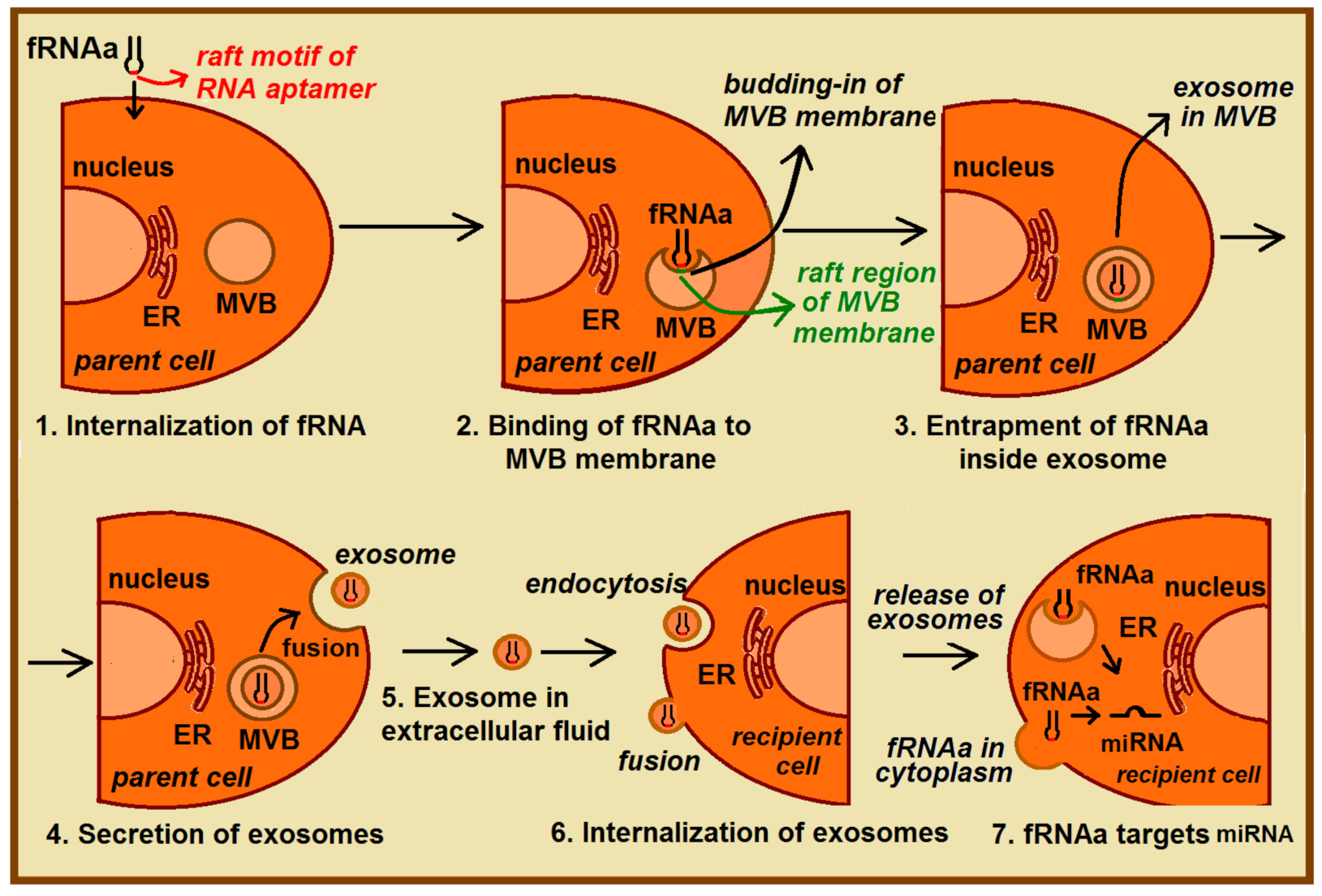
3.3. Potential Medical Applications of Functional RNA Aptamers (fRNAa)
4. Materials and Methods
4.1. Materials
4.2. Preparation of Large Unilamellar Vesicles (LUVs)
4.3. Preparation of RNA Aptamers
4.4. Fluorescent Measurements
4.5. Determinaton of the Dissociation Constant KD for RNA-Liposome Complex
4.6. Isolation and Labelling of Exosomes
4.7. Fluorescence Measurements and Determinaton of the Dissociation Constant KD for RNA-Exosome Complex
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.M.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Blanchard, N.; Lankar, D.; Faure, F.; Regnault, A.; Dumont, C.; Raposo, G.; Hivroz, C. TCR Activation of human T cells induces the production of exosomes bearing the TCR/CD3/ζ Complex. J. Immunol. 2002, 168, 3235–3241. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular characterization of dendritic cell-derived exosomes: Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef] [Green Version]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Théry, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom1, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitt, J.M.; Charrier, M.; Viaud, S.; Andre, F.; Besse, B.; Chaput, N.; Zitvogel, L. Dendritic cell—Derived exosomes as immunotherapies in the fight against cancer. J. Immunol. 2014, 193, 1006–1011. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigenpresenting cells. Nat. Comm. 2011, 2, 282. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, S.; Losordo, D.W. Exosomes and cardiac repair after myocardial infarction. Circ. Res. 2014, 114, 315–324. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhovena, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janas, A.M.; Sapoń, K.; Janas, T.; Stowell, M.H.; Janas, T. Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. Biochim. Biophys. Acta 2016, 1858, 1139–1151. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, H.; Tachikawa, M.; Yagi, Y.; Umetsu, M.; Nurdin, A.; Miyauchi, E.; Watanabe, M.; Uchida, Y.; Terasaki, T. Cluster of differentiation 46 Is the major receptor in human blood–brain barrier endothelial cells for uptake of exosomes derived from brain-metastatic melanoma cells (SK-Mel-28). Mol. Pharm. 2018, 16, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Ma, R.; Zhang, L.; Li, H.; Tang, Y.; Du, G.; Niu, D.; Yin, D. The role of exosomes in thyroid cancer and their potential clinical application. Front. Oncol. 2020, 10, 2675. [Google Scholar] [CrossRef]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007, 89, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta. 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Osteikoetxea, X.; Balogh, A.; Szabó-Taylor, K.; Németh, A.; Szabó, T.G.; Pálóczi, K.; Sódar, B.; Kittel, A.; György, B.; Pállinger, E.; et al. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PLoS ONE 2015, 10, e0121184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pike, L.J. Rafts defined: A report on the keystone symposium on lipid rafts and cell function. J. Lipid Res. 2006, 47, 1597–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [Green Version]
- De Gassart, A.; Géminard, C.; Février, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Rohrer, B.; Levental, K.R.; Levental, I. Rafting through traffic: Membrane domains in cellular logistics. Biochim. Biophys. Acta Biomembr. 2014, 1838, 3003–3013. [Google Scholar] [CrossRef] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and-dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leidal, A.M.; Huang, H.H.; Marsh, T.; Solvik, T.; Zhang, D.; Ye, J.; Kai, F.; Goldsmith, J.; Liu, J.Y.; Huang, Y.; et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020, 22, 187–199. [Google Scholar] [CrossRef]
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177. [Google Scholar] [CrossRef]
- Pigati, L.; Yaddanapudi, S.C.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef] [Green Version]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; ’t Hoen, P.A. Deep sequencing of RNA from immune cellderived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.P.; Li, L.; Schorey, J.S. Exosomal RNA from Mycobacterium tuberculosis-infected cells is functional in recipient macrophages. Traffic 2015, 16, 555–571. [Google Scholar] [CrossRef] [Green Version]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collén, A.; Sunnerhagen, P.; et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; de Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Li, C.; Zhang, Y.; Zhang, D.; Otterbein, L.E.; Jin, Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 2019, 216, 2202–2220. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 2016, 5, e19276. [Google Scholar] [CrossRef]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef]
- Janas, T.; Janas, P.; Sapoń, K.; Janas, T. Binding of RNA aptamers to membrane lipid rafts: Implications for exosomal miRNAs transfer from cancer to immune cells. Int. J. Mol. Sci. 2020, 21, 8503. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hoshina, S.; Sekizuka, T.; Kataoka, M.; Hasegawa, H.; Hamada, H.; Kuroda, M.; Katano, H. Profile of exosomal and intracellular microRNA in gamma-herpesvirus-infected lymphoma cell lines. PLoS ONE 2016, 11, e0162574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahadi, A.; Brennan, S.; Kennedy, P.J.; Hutvagner, G.; Tran, N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci. Rep. 2016, 6, 24922. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Sapoń, K.; Janas, T.; Yarus, M. Specific binding of VegT mRNA localization signal to membranes in Xenopus oocytes. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118952. [Google Scholar] [CrossRef]
- Janas, T.; Janas, T.; Yarus, M. Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res. 2006, 34, 2128–2136. [Google Scholar] [CrossRef] [Green Version]
- Czerniak, T.; Saenz, J.P. Lipid membranes modulate the activity of RNA through sequence-specific interactions. Biorxiv 2021. [Google Scholar] [CrossRef]
- Wei, H.; Malcor, J.D.M.; Harper, M.T. Lipid rafts are essential for release of phosphatidylserine-exposing extracellular vesicles from platelets. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sapoń, K.; Maziarz, D.; Janas, T.; Sikorski, A.F.; Janas, T. Cholera toxin subunit B for sensitive and rapid determination of exosomes by gel filtration. Membranes 2020, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.; Ajito, S.; Sato, S.; Ohta, N.; Igarashi, N.; Shimizu, N. Preferential Intercalation of human amyloid-β peptide into interbilayer region of lipid-raft membrane in macromolecular crowding environment. J. Phys. Chem. B 2018, 122, 9482–9489. [Google Scholar] [CrossRef]
- Betz, J.; Bielaszewska, M.; Thies, A.; Humpf, H.U.; Dreisewerd, K.; Karch, H.; Kim, K.S.; Friedrich, A.W.; Müthing, J. Shiga toxin glycosphingolipid receptors in microvascular and macrovascular endothelial cells: Differential association with membrane lipid raft microdomains. J. Lipid Res. 2011, 52, 618–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shishina, A.K.; Kovrigina, E.A.; Galiakhmetov, A.R.; Rathore, R.; Kovrigin, E.L. Study of forster resonance energy transfer to lipid domain markers ascertains partitioning of semisynthetic lipidated N-ras in lipid raft nanodomains. Biochemistry 2018, 57, 872–881. [Google Scholar] [CrossRef]
- Takeda, M.; Leser, G.P.; Russell, C.J.; Lamb, R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 2003, 100, 14610–14617. [Google Scholar] [CrossRef] [Green Version]
- Vlassov, A.; Khvorova, A.; Yarus, M. Binding and disruption of phospholipid bilayers by supramolecular RNA complexes. Proc. Natl. Acad. Sci. USA 2001, 98, 7706–7711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iida, M.; Mashima, T.; Yamaoki, Y.; So, M.; Nagata, T.; Katahira, M. The anti-prion RNA aptamer R12 disrupts the Alzheimer’s disease-related complex between prion and amyloid β. FEBS J. 2019, 286, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Cha, D.J.; Franklin, J.L.; Dou, Y.; Liu, Q.; Higginbotham, J.N.; Beckler, M.D.; Weaver, A.M.; Vickers, K.; Prasad, N.; Levy, S.; et al. KRAS-dependent sorting of miRNA to exosomes. Elife 2015, 4, e07197. [Google Scholar] [CrossRef]
- Corradi, E.; Dalla Costa, I.; Gavoci, A.; Iyer, A.; Roccuzzo, M.; Otto, T.A.; Oliani, E.; Bridi, S.; Strohbuecker, S.; Santos-Rodriguez, G.; et al. Axonal precursor miRNAs hitchhike on endosomes and locally regulate the development of neural circuits. EMBO J. 2020, 39, e102513. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 30, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on aptamer research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Li, L.; Wang, Z.; Irfan, M.; Qu, F. Recent advances of aptasensors for exosomes detection. Biosens. Bioelectron. 2020, 160, 112213. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Sapon, K.; Stowell, M.H.B.; Janas, T. Selection of membrane RNA aptamers to amyloid beta peptide: Implications for exosome-based antioxidant strategies. Int. J. Mol. Sci. 2019, 20, 299. [Google Scholar] [CrossRef] [Green Version]
- Patel, K.A.; Chaudhary, R.K.; Roy, I. RNA aptamers rescue mitochondrial dysfunction in a yeast model of Huntington’s disease. Mol. Ther. Nucleic Acids 2018, 12, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Esposito, C.L.; Nuzzo, S.; Catuogno, S.; Romano, S.; de Nigris, F.; de Franciscis, V. STAT3 gene silencing by aptamer-siRNA chimera as selective therapeutic for glioblastoma. Mol. Ther. Nucleic Acids 2018, 10, 398–411. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.N.; Tsai, Y.C.; Hsu, C.C.; Liang, Y.L.; Wu, Y.Y.; Kang, C.Y.; Lin, C.-H.; Hsu, P.-H.; Lee, G.-B.; Hsu, K.-F. An aptamer interacting with heat shock protein 70 shows therapeutic effects and prognostic ability in serous ovarian cancer. Mol. Ther. Nucleic Acids 2021, 23, 757–768. [Google Scholar] [CrossRef]
- Didiot, M.C.; Hall, L.M.; Coles, A.H.; Haraszti, R.A.; Godinho, B.M.; Chase, K.; Sapp, E.; Ly, S.; Alterman, J.F.; Hassler, M.R.; et al. Exosome-mediated delivery of hydrophobically modified siRNA for huntingtin mRNA silencing. Mol. Ther. 2016, 24, 1836–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, Y.; Bai, M.; Wang, J.; Zhu, K.; Liu, R.; Ge, S.; Li, J.; Ning, T.; Deng, T.; et al. Exosomes serve as nanoparticles to suppress tumor growth and angiogenesis in gastric cancer by delivering hepatocyte growth factor si RNA. Cancer Sci. 2018, 109, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setten, R.L.; Rossi, J.J.; Han, S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Phillips, A.J. The challenge of gene therapy and DNA delivery. J. Pharm. Pharmacol. 2001, 53, 1169–1174. [Google Scholar] [CrossRef]
- Sekhon, S.S.; Ahn, G.; Park, G.Y.; Park, D.Y.; Lee, S.H.; Ahn, J.Y.; Kim, Y.H. The Role of Aptamer Loaded Exosome Complexes in the Neurodegenerative Diseases. Toxicol. Environ. Health. Sci. 2019, 11, 85–93. [Google Scholar] [CrossRef]
- Susa, F.; Limongi, T.; Dumontel, B.; Vighetto, V.; Cauda, V. Engineered extracellular vesicles as a reliable tool in cancer nanomedicine. Cancers 2019, 11, 1979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, A.T.; Rahman, S.; Badierah, R.; Lee, E.J.; Mattar, E.H.; Redwan, E.M.; Choi, I. Expedition into exosome biology: A perspective of progress from discovery to therapeutic development. Cancers 2021, 13, 1157. [Google Scholar] [CrossRef] [PubMed]
- Gobbo, J.; Marcion, G.; Cordonnier, M.; Dias, A.M.; Pernet, N.; Hammann, A.; Richaud, S.; Mjahed, H.; Isambert, N.; Clausse, V.; et al. Restoring anticancer immune response by targeting tumor-derived exosomes with a HSP70 peptide aptamer. J. Natl. Cancer Inst. 2016, 108, djv330. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zhao, Y.; Xue, F.; Zheng, Y.; Huang, H.; Wang, W.; Chang, Y.; Yang, H.; Zhang, J. Exosomal DNA aptamer targeting α-synuclein aggregates reduced neuropathological deficits in a mouse Parkinson’s disease model. Mol. Ther. Nucleic Acids 2019, 17, 726–740. [Google Scholar] [CrossRef] [Green Version]
- Pan, Q.; Ramakrishnaiah, V.; Henry, S.; Fouraschen, S.; de Ruiter, P.E.; Kwekkeboom, J.; Tilanus, H.W.; Janssen, H.L.A.; van der Laan, L.J.W. Hepatic cell-to-cell transmission of small silencing RNA can extend the therapeutic reach of RNA interference (RNAi). Gut 2012, 61, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Askenase, P.W. COVID-19 therapy with mesenchymal stromal cells (MSC) and convalescent plasma must consider exosome involvement: Do the exosomes in convalescent plasma antagonize the weak immune antibodies? J. Extracell. Vesicles 2020, 10, e12004. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Shi, M.; Liu, X.; Jin, C.; Xing, X.; Qiu, L.; Tan, W. Aptamer-functionalized exosomes: Elucidating the cellular uptake mechanism and the potential for cancer-targeted chemotherapy. Anal. Chem. 2019, 91, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Danquah, M.; Chaudhary, A.K.; Mahato, R.I. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J. Control. Release 2015, 219, 237–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghoshal, B.; Bertrand, E.; Bhattacharyya, S.N. Non-canonical argonaute loading of extracellular vesicle-derived exogenous single-stranded miRNA in recipient cells. J. Cell Sci. 2021, 134, jcs253914. [Google Scholar] [CrossRef]
- Janas, T.; Janas, T.; Yarus, M. A membrane transporter for tryptophan composed of RNA. RNA 2004, 10, 1541–1549. [Google Scholar] [CrossRef] [Green Version]
- Sapoń, K.; Janas, T.; Sikorski, A.F.; Janas, T. Polysialic acid chains exhibit enhanced affinity for ordered regions of membranes. Biochim. Biophys. Acta Biomembr. 2019, 1861, 245–255. [Google Scholar] [CrossRef]
- Sapoń, K.; Gawrońska, I.; Janas, T.; Sikorski, A.F.; Janas, T. Exosome-associated polysialic acid modulates membrane potentials, membrane thermotropic properties, and raft-dependent interactions between vesicles. FEBS Lett. 2020, 594, 1685–1697. [Google Scholar] [CrossRef]

| Searched RNA Motifs | % of RNA Motif In Original RNA Pool (148 RNAs) | % of RNA Motif in Analysed RNAs (10 RNAs) | |
|---|---|---|---|
| both EXO- and RAFT-MOTIF | CCCU | 20% | 29% |
| EXO-MOTIFS | GGAG | 2% | 4.5% |
| UGAG | 6% | 4.5% | |
| UCCU | 19% | 18% | |
| RAFT-MOTIFS | UCCC | 19% | 22% |
| CUCC | 16% | 13% | |
| UUGU | 18% | 9% | |
| RNA Name | RNA Length [nts] | RNA Nucleotides Composition | RNA 2° Structure | RNA Motifs (Location) both EXO- and RAFT-MOTIF EXO-MOTIFS RAFT-MOTIFS | KD Value (±SE) [µM] |
|---|---|---|---|---|---|
| 54 | 91 | 19 A’s 35 C’s 14 G’s 23 U’s | 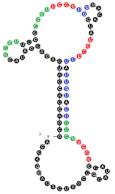 | CCCU x3 (4L; 4L; 3S/1L) UCCU x3 (4L) UCCC x1 (3S/1L) CUCC x2 (3L/1S; 1L/3S) UUGU x2 (2L/1S/1L; 4S) overlaps: UCCU & UUGU UCCU & CUCC CUCC, UCCC & CCCU | 60.9 (±5.0) |
| 90 | 91 | 24 A’s 22 C’s 23 G’s 22 U’s |  | GGAG (3L/1S) UUGU (1S/1L/2S) | 61.1 (±10.3) |
| 19 | 111 | 22 A’s 39 C’s 21 G’s 29 U’s | 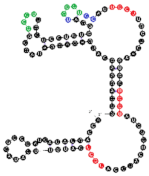 | CCCU x2 (1S/3L; 3L/1S) UCCU x3 (4L; 4S; 3L/1S) UCCC (4L) CUCC (1L/3S) overlaps UCCC, CCCU & CUCC | 82.2 (±11.4) |
| 102 | 91 | 24 A’s 28 C’s 16 G’s 23 U’s | 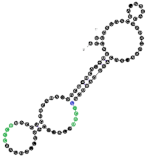 | CCCU x2 (4L; 4L) UCCC (1S/3L) overlaps UCCC & CCCU | 95.4 (±13.7) |
| 13 | 91 | 20 A’s 23 C’s 23 G’s 25 U’s |  | CCCU (2S/2L) UCCC (3S/1L) overlap | 95.6 (±20.5) |
| 64 | 90 | 21 A’s 25 C’s 20 G’s 24 U’s | 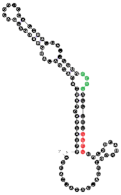 | CCCU (3L/1S) UCCU (4S) | 112.2 (±23.1) |
| 24 | 111 | 23 A’s 33 C’s 28 G’s 27 U’s | 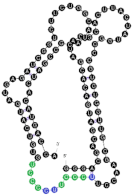 | CCCU x2 (3S/1L; 4L) UGAG (3L/1S) UCCC x2 (4S; 4L) overlaps UCCC & CCCU 2x | 187.5 (±54.3) |
| 93 | 111 | 26 A’s 34 C’s 24 G’s 27 U’s | 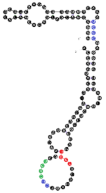 | CCCU (4L) UCCU (2S/2L) UCCC x2 (4S; 4L) CUCC (4L) overlaps CUCC, UCCC & CCCU | 208.7 (±35.5) |
| 10 | 111 | 19 A’s 33 C’s 25 G’s 34 U’s |  | CCCU (4L) GGAG (1S/3L) UCCC x2 (4S; 4L) CUCC x2 (1L/3S; 1S/3L) UUGU (2L/2S) overlaps CUCC, UCCC & CCCU CUCC & UCCC | 212.8 (±28.3) |
| 78 | 91 | 21 A’s 26 C’s 23 G’s 21 U’s | 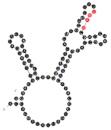 | UGAG (1L/3S) No RAFT-motif | 225.2 (±25.9) |
| Raft RNA Aptamer with miRNA- and Raft-Binding Motifs | Human miRNA with Reverse/Complement Motif | |||
|---|---|---|---|---|
| Name | Human miRNA-Binding Motif | Raft-Binding Motifs | Name | Sequence, Reversed/Complement Motif in Red Color |
RNA 60 RNA 103 RNA 113 | 12-nts long AUGCUCUCUGUA UGUCCAUUGUAU AGCCUUCUCCAU | CUCC UUGU, CUCC CUCC, UCCC, CCCU | miR-6818-5p miR-2355-5p miR-4531 | UUGUGUGAGUACAGAGAGCAUC AUCCCCAGAUACAAUGGACAA AUGGAGAAGGCUUCUGA |
RNA 9 RNA 10 RNA 10 RNA 14 RNA 14 RNA 19 RNA 32 RNA 41 RNA 43 RNA 48 RNA 49 RNA 53 RNA 54 RNA 67 RNA 68 RNA 102 RNA 108 RNA 119 RNA 140 RNA 142 RNA 153 RNA 157 RNA 167 RNA 167 | 11-nts long CCUUCUCUCCA GUGAAAGGAGU UACUUUCUCUU GUAGUUACUUC CCCAGACCAGA AUCCAUCCUCA UCCCUCACUAG UCUGAAGGUUC AGAAUCUGCAU GCUCUUUUCAU GUUCGUCUUAU CCCCAGGACUC CCUUCCUUGUU CCUUUUGUUCG CUCCUCUGGCC AUACCAAUACC UUGUCCACCGC UCCCCUGGUCA ACUGAUCCAGC UCAUGCCCUCU CUUUUAGCCCU AUAUCAGCUCA UUCAGUUCUCA UUCAGUUCUCA | CUCC CUCC, UCCC, UUGU, CUCC, UCCC, CCCU CUCC, UCCC, UUGU, CUCC, UCCC, CCCU UCCC UCCC CCCU, UCCC, CCCU, CUCC UUGU, UCCC, CCCU UCCC, CCCU UUGU, CUCC, CCCU, CUCC, UCCC UUGU, UUGU, CUCC, UUGU CUCC, UCCC UCCC, CUCC, UCCC, CCCU CCCU, CCCU, UUGU, CUCC, UUGU, CUCC, UCCC, CCCU CCCU, UUGU CUCC, CUCC UCCC, CCCU, CCCU UUGU UCCC, CCCU, UUGU, UCCC, CCCU CUCC, CUCC, UCCC, UCCC CCCU, CUCC CCCU, UUGU, UUGU, CUCC CCCU, CUCC, CUCC, UCCC, CCCU CUCC, CCCU CUCC, CCCU | miR-7846-3p miR-4764-3p miR-3686 miR-548ao-5p miR-4786-3p miR-3605-5p miR-921 miR-3142 miR-5683 miR-3618 miR-208b-3p miR-3198 miR-5699-5p miR-208b-3p miR-4726-5p miR-624-3p miR-4638-5p miR-4465 miR-1287-5p miR-8052 miR-135b-3p miR-24-1-5p miR-146b-5p miR-146a-5p | CAGCGGAGCCUGGAGAGAAGG UUAACUCCUUUCACACCCAUGG AUCUGUAAGAGAAAGUAAAUGA AGAAGUAACUACGGUUUUUGCA UGAAGCCAGCUCUGGUCUGGGC UGAGGAUGGAUAGCAAGGAAGCC CUAGUGAGGGACAGAACCAGGAUUC AAGGCCUUUCUGAACCUUCAGA UACAGAUGCAGAUUCUCUGACUUC UGUCUACAUUAAUGAAAAGAGC AUAAGACGAACAAAAGGUUUGU GUGGAGUCCUGGGGAAUGGAGA UGCCCCAACAAGGAAGGACAAG AUAAGACGAACAAAAGGUUUGU AGGGCCAGAGGAGCCUGGAGUGG CACAAGGUAUUGGUAUUACCU ACUCGGCUGCGGUGGACAAGU CUCAAGUAGUCUGACCAGGGGA UGCUGGAUCAGUGGUUCGAGUC CGGGACUGUAGAGGGCAUGAGC AUGUAGGGCUAAAAGCCAUGGG UGCCUACUGAGCUGAUAUCAGU UGAGAACUGAAUUCCAUAGGCUG UGAGAACUGAAUUCCAUGGGU |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mańka, R.; Janas, P.; Sapoń, K.; Janas, T.; Janas, T. Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs. Int. J. Mol. Sci. 2021, 22, 9416. https://doi.org/10.3390/ijms22179416
Mańka R, Janas P, Sapoń K, Janas T, Janas T. Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs. International Journal of Molecular Sciences. 2021; 22(17):9416. https://doi.org/10.3390/ijms22179416
Chicago/Turabian StyleMańka, Rafał, Pawel Janas, Karolina Sapoń, Teresa Janas, and Tadeusz Janas. 2021. "Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs" International Journal of Molecular Sciences 22, no. 17: 9416. https://doi.org/10.3390/ijms22179416
APA StyleMańka, R., Janas, P., Sapoń, K., Janas, T., & Janas, T. (2021). Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs. International Journal of Molecular Sciences, 22(17), 9416. https://doi.org/10.3390/ijms22179416






