Non-Coding RNAs in Normal B-Cell Development and in Mantle Cell Lymphoma: From Molecular Mechanism to Biomarker and Therapeutic Agent Potential
Abstract
1. Introduction
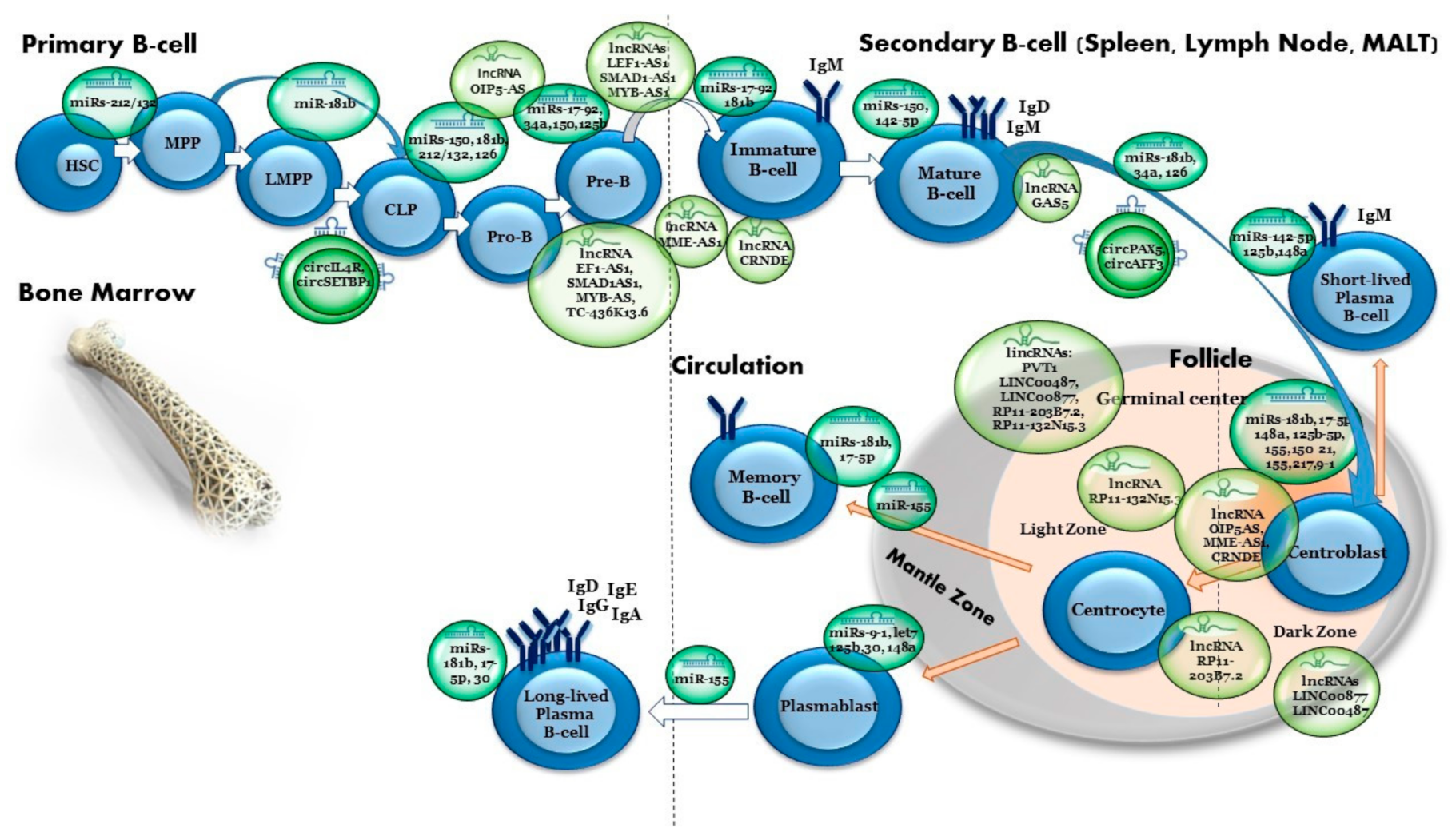
2. Normal B-Cell Development
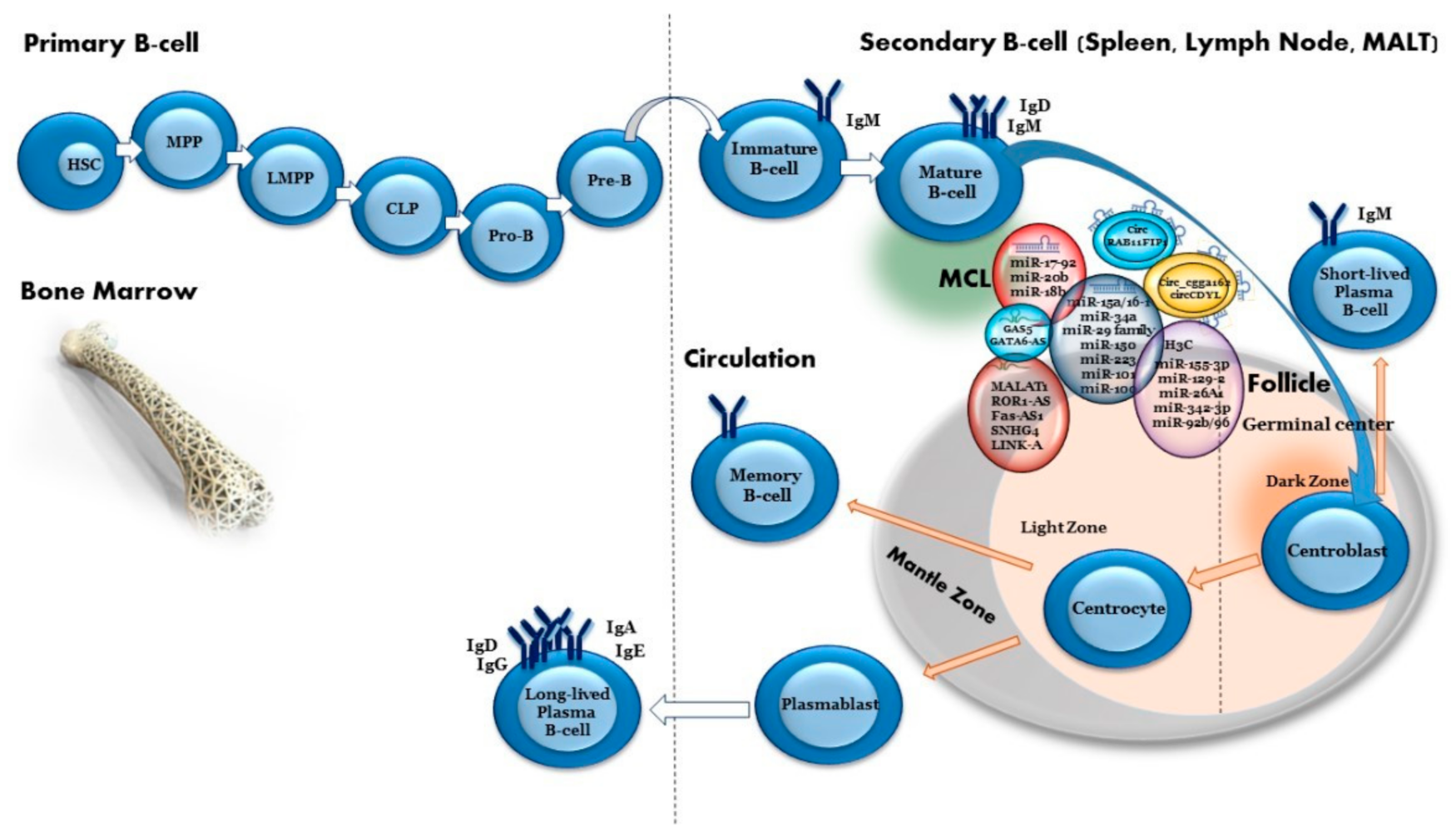
3. Mantle Cell Lymphoma
- Classical MCL is usually identified by the presence of immunoglobulin heavy-chain variable region gene (IGHV)-unmutated B cells, and SRY-box transcription factor 11 (SOX11) expression. This MCL type usually involves lymph nodes and extranodal sites;
- Leukemic non-nodal MCL is identified by the presence of IGHV-mutated genes without SOX11 expression. This type typically involves the bone marrow, peripheral blood and spleen and mostly has an indolent presentation.
4. MicroRNAs—Powerful Regulators of Gene Expression
5. MiRNAs in Normal B-Cell Development
5.1. MiR-150
5.2. MiR-17-92 Cluster
5.3. MiR-34a
5.4. MiR-126
5.5. MiR-212/132 Cluster
6. Germinal Center Regulation by microRNAs
6.1. MiR-155 and 181b
6.2. MiR-125b
6.3. MiR-9, MiR-30 and Let-7 Families
6.4. MiR-21
6.5. MiR-217
6.6. MiR-142
6.7. MiR-148a
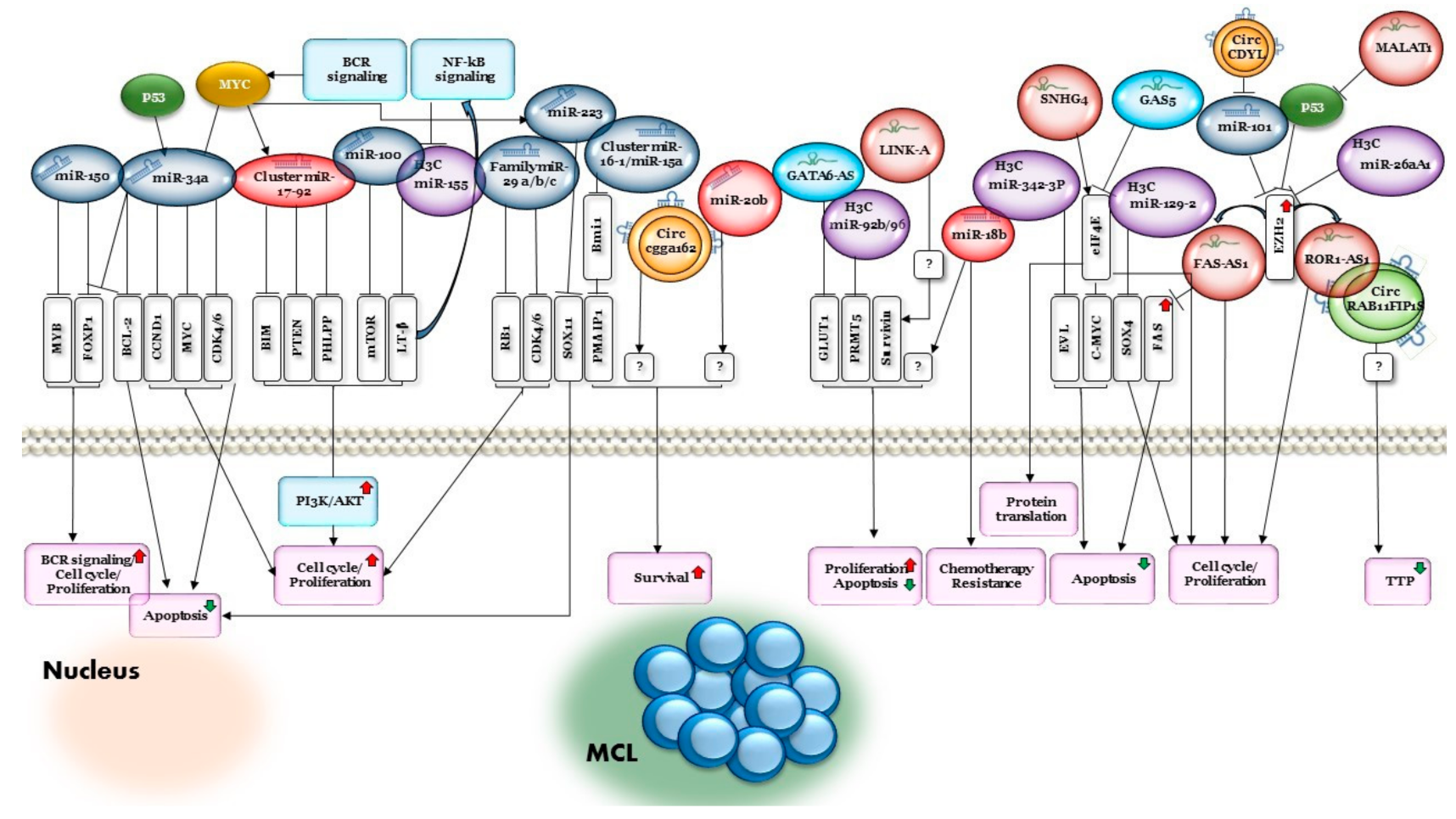
7. MiRNAs Related to MCL
7.1. The miRNA-17~92 Cluster
7.2. The miR-16-1/miR-15a Cluster
7.3. MiR-34a
7.4. MiR-29
7.5. MiR-150
7.6. MiR-18b
7.7. MiR-20b
7.8. MiR-223
7.9. MiR-101
7.10. MiR-100
8. Methylation of miRNA Genes in MCL
8.1. MiR-155-3p
8.2. MiR-129-2
8.3. MiR-26A1
8.4. MiR-342-3p
8.5. MiR-92b/96
9. Long Non-Coding RNAs in Normal Development
10. Long Non-Coding RNA in MCL
11. Circular RNAs
12. Circular RNA in B-Cell Development and Malignancies
13. Conclusions
| Type of ncRNA | ncRNA | Upregulated/Downregulated | Target | Role in B-Cell Development | References |
|---|---|---|---|---|---|
| miRNA | miR-150 | Upregulated | MYB/FOXP1 | MiR-150 is mainly expressed in the lymph nodes and spleen and is highly upregulated during the development of mature T and B cells. It regulates FOXP1 in mature B cells. Expression of miR-150 is sharply upregulated in immature B cells. Premature expression of miR-150 blocked the transition from pro-B to pre-B cells. In progenitor B cells, miR-150 targets include the transcription factor MYB. Precise levels of MYB are required for normal haematopoiesis and B-cell development. Loss of MYB leads to a dramatic loss in B cells. | [58,59,60,61] |
| miRNA | miR-17-92 cluster (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92-1) | Upregulated | PTEN/BIM | Overexpression of this cluster results in lympho-proliferative and autoimmune phenotypes due to reduced phosphatase and tensin homolog (PTEN) and Bcl-2-like 11 (Bim) protein expression. | [62,63,64,65,66] |
| miRNA | miR-34a | Upregulated | FOXP1 | MiR-34a regulates B-cell differentiation from pro-B cell to pre-B cell. Upregulation of miR-34a at the pro-B cell stage resulted in downregulation of Foxp1 protein, a transcriptional regulator required for B-cell differentiation. | [67,68] |
| miRNA | miR-126 | Upregulated | IRS1 | MiR-126 has a crucial role in in B-cell lineage commitment. Overexpression of miR-126 in an immature B-cell line and in mouse hematopoietic stem and progenitor cells (HSPCs) induced B-cell differentiation through the regulation of IRS1. | [69] |
| miRNA | miR 212/132 cluster | Upregulated | SOX4 | The miR-212/132 cluster is an important regulator of hematopoietic stem cell function, inflammation and proliferation during wound healing. It inhibits the pre-pro-B-cell to pro-B-cell transition by targeting SOX4, which is a crucial mediator of pro-B cell development. | [70,71,72,73,74] |
| miRNA | miR-155/181b | Upregulated | AID | AID expression is required during the B-cell somatic hypermutation process of immunoglobulin genes and its loss leads to the impairment of class switching. | [77,78,79,80,81,82,83,84,85] |
| miRNA | miR-155-5p | Upregulated/ Downregulated | CD10/PU.1 | Important to germinal-center (GC) regulation. MiR-155-5p negatively regulates the expression of CD10, a marker of GC cells. Loss of miR-155-5p leads to reduced IgG1 switching by targeting PU.1 transcription factor. This in turn leads to PAX5 downregulation, enabling plasma cell commitment. | [81,82,83] |
| miRNA | miR-125b-5p | Upregulated | PRDM1/IRF4 | MiR-125b promotes B lymphocyte diversification in GC by inhibiting premature utilization of essential transcription factors for plasma cell differentiation. | [86] |
| miRNA | miR-9, let-7 and miR-30 families | Downregulated/ Upregulated | PRDM1 BCL6 | These miRNA families regulate transcription factors that promote GC B-cell survival and differentiation. They also regulate GC formation and terminal B-cell differentiation. | [87,88,89] |
| miRNA | miR-21 | Downregulated | PRDM1 binds to primary miR-21 and represses its translation | The primary miR-21 transcript is a direct target of PRDM1-dependent repression; therefore, it is repressed during interleukin (IL)-21-driven plasma cell differentiation. | [90] |
| miRNA | miR-217 | Upregulated | BCL6 | A positive regulator of the GC reaction; promotes mature B-cell lymphomagenesis. | [91] |
| miRNA | miR-142-5p | Upregulated | BAFFR | Critical for the development and homeostasis of lymphocytes. | [92] |
| miRNA | miR-148a | Upregulated | BACH2/MITF/PTEN/BIM | An important player in the regulatory network controlling terminal plasma cell differentiation. | [93,94] |
| LncRNAs | LEF1-AS1, SMAD1AS1, MYB-AS, TC-436K13.6 | Upregulated/ Downregulated | RAG2/VPREB1/DNTT/LEF1/SMAD1/MYB | Early B-cell development-specific genes, expressed in pro-B, pre-B and immature B cells (e.g., RAG2, VPREB1, DNTT, LEF1, SMAD1, MYB), were associated with LEF1-AS1, SMAD1-AS1 and MYB-AS1 antisense transcripts, as well as with the intergenic transcript CTC-436K13.6. | [158] |
| LncRNAs | OIP5AS, MME-AS1, CRNDE | Upregulated/ Downregulated | KIF23/PLK4/CENPE | Specifically expressed in the proliferative stages of B-cell development, i.e., pro-B, pre-B, centroblasts and centrocytes. | [159,160,161] |
| LncRNAs | PVT1 and multiple uncharacterized lncRNAs: LINC00487, LINC00877, RP11-203B7.2, RP11-132N15.3 | Upregulated/ Downregulated | AID/SERPINA/ BCL6 | Specifically expressed in GC centroblasts and centrocytes. | [151,152,153] |
| lncRNA | GAS5 | Upregulated/ Downregulated | MYC/CDK6/CIAP2/SGK1/MICRORNA-21 | GAS5 mediates growth arrest. In proliferating cells, GAS5 is repressed by active mTOR, miR-21 and DNA methylation. In growth arrested cells, GAS5 levels are increased and negatively influence growth- and apoptosis-associated genes, such as MYC, CDK6, cIAP2 and SGK1 (anti-apoptotic), as well as the oncomiR microRNA-21. | [169] |
| Type of ncRNA | ncRNA | Expression in MCL | Target | Mechanism/Target/Pathway | References |
|---|---|---|---|---|---|
| miRNA | miR-17-92 cluster (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, miR-92-1) | Upregulated | PTEN/PHLPP2/BIM | A mouse model demonstrated that c-Myc can both activate the expression of the miR-17-92 gene cluster and be regulated by it, suggesting negative regulation between c-Myc and the miR-17-92 gene cluster. Loss of regulation control of this cluster is involved in the development of B-cell non-Hodgkin lymphoma (B-NHL). MiR-17-92 can also inhibit phosphatase and tensin homolog (PTEN), PHLPP2, P21 and Bcl-2-like 11 (Bim) expression, promoting proliferation and suppressing cancer cell apoptosis. High expression levels of the miR-17-92 cluster are also related to poor survival rate in patients with MCL. | [30,96,97,98,99,100,101] |
| miRNA | miR-15a/16-1 | Loss/ downregulated | BMI1 | MiR-15a/16-1 inhibits cell proliferation, promotes apoptosis of cancer cells and suppresses tumorigenicity by targeting multiple oncogenes associated with short overall survival. MiR-15a/16-1 targets the proto-oncogene Bmi1, which directly regulates pro-apoptotic genes such as Bim and PMAIP1. MiR-15a/16-1 downregulation enhanced the anti-apoptotic potential of MCL. | [102,103,104,105,106,107,108,109,110,111,112] |
| miRNA | miR-34a | Downregulated | MYC/CDK4/6/CCND1/FOXP1/BCL2 | MCL patients with downregulated miR-34a had short overall survival associated with poor prognosis. Low expression of miR-34a was associated with high expression of MYC oncogene, which was co-regulated with CDK4/6 and CCND1 to further promote cell-cycle progression and the development of MCL. MiR-34a is involved in the regulation of the tumor suppressor gene TP53 via FOXP1 and BCL2, thus affecting the differentiation and apoptosis of B cells. | [113,114,115,116,117] |
| miRNA | miR-29a/b/c | Downregulated | CDK6/RB1 | MiR-29 family members are critical regulators of extracellular matrix (ECM) proteins and signaling pathways associated with fibrosis via targeting of collagens, fibrillins, and elastin. MiR-29 inhibition activated CDK4/CDK6 and RB1 in MCL. Low expression of miR-29 was associated with poor prognosis in MCL. | [104,118,119,120] |
| miRNA | miR-150 | Downregulated | MET/FOXP1 | MiR-150 inhibits the proliferation and promotes the apoptosis of MCL cells by negatively regulating MET and FOXP1 expression. Premature miR-150 expression severely impairs B-cell development due to a pro- to pre-B-cell transition block, whereas miR-150 deletion promotes B1-cell expansion and increases antibody production. HGF/MET and FOXP1 signaling pathways are considered as treatment targets for B-cell lymphoma, particularly in MCL. | [121,122] |
| miRNA | miR-18b | Upregulated | unknown | MiR-18b decreases the MCL cell line proliferation rate without inducing apoptosis, suggesting that it may render MCL cells resistant to chemotherapy by decelerating the cell proliferation. MiR-18b is associated with poor prognosis. | [123,124] |
| miRNA | miR-20b | Upregulated | unknown | MiR-20b plays a role in survival of patients with MCL. High expression levels of miR-20b are associated with a worse prognosis. | [97,121,125,126] |
| miRNA | miR-223 | Downregulated | SOX11 | MiR-223 expression is repressed in MCL. It inhibits cell proliferation and promotes G0/G1 accumulation and cell apoptosis. Low expression of miR-223 predicts poorer outcomes in MCL, probably due to its direct targeting of SOX11. | [50,127,128,129,130] |
| miRNA | miR-101 | Downregulated | EZH2 | Inhibits cell proliferation and induces cell apoptosis of MCL by targeting EZH2. Low miR-101 expression is associated with low overall survival rates. | [131] |
| miRNA | miR-100 | Downregulated | mTOR | Inhibits cell proliferation in MCL by targeting mTOR. | [132] |
| miRNA | miR-155-3p | Downregulated | LT-β/non-canonical NF-κB signaling | Overexpression of miR-155-3p led to increased sub-G1 apoptotic cells and reduced cellular viability, demonstrating its tumor suppressive properties. | [134,135,136,137] |
| miRNA | miR-129-2 | Downregulated | SOX4 | MiR-129-2 has been shown to be a tumor suppressor hypermethylated in epithelial cancers. MiR-129 overexpression inhibited cellular proliferation and enhanced cell death, with concomitant SOX4 mRNA downregulation. | [136] |
| miRNA | miR-26-A-1 | Downregulated | EZH2 | MiR-26-A-1 is a tumor suppressor. Epigenetic silencing of miR-26-A-1 leads to increased EZH2 levels, which, in turn, translate into a worse outcome. | [138] |
| miRNA | miR-342-3p | Downregulated | Co-regulated with its host gene, EVL | MiR-342-3p is a tumor suppressor co-regulated with its host gene, EVL, by promoter DNA methylation in B-cell lymphomas including MCL. Re-expression of miR-342-3p and EVL and the tumor suppressor function of miR-342-3p were demonstrated by the inhibition of cellular proliferation and increase of cell death. | [139] |
| miRNA | miR-92b/96 | Downregulated | PRMT5 | Low expression of miR-92b and miR-96 is associated with enhanced PRMT5 translation which is overexpressed in aggressive B-cell NHL, including MCL. Re-expression of miR-92b and miR-96 inhibits PRMT5 translation and alters the growth of MCL cells. | [140,141] |
| lncRNA | MALAT1 | Upregulated | MALAT1 is associated with cancer progression, acts through repression of TP53 promoter and increases EZH2 translation. EZH2, in turn, binds a ROR1-AS1 lncRNA, which leads to increased cell proliferation in MCL cell lines. | [164] | |
| lncRNA | ROR1-AS1 | Upregulated | SOX11/P16 | Overexpression of ROR1-AS1 lncRNA promoted growth of MCL cells and resistance to ibrutinib (BTK inhibitor) and dexamethasone treatment through regulation of SOX11 and P16 expression. | [162] |
| lncRNA | FAS-AS1 | Upregulated | The promoter region of FAS-AS1 is regulated by EZH2, which is upregulated in MCL. FAS-AS1 modulates alternative splicing of the FAS gene, a central inhibition molecule in the extrinsic apoptosis pathway, thereby downregulating FAS. | [7,165] | |
| lncRNA | SNHG4 | Upregulated | Co-regulated with eIF4 | SNHG4 plays a role in the regulation of gene expression and can bind with eIF4E and regulate protein translation in MCL. Knockdown SNHG4 expression through siRNA inhibits cell proliferation and global protein translation. | [167] |
| lncRNA | SNHG12, FTX, SNHG5, ZNFX1-AS1 | Upregulated | Associated with translation machinery via eIF4E/modulation of c-Myc translation | These lncRNAs are associated with translation machinery via eIF4E-RNA-binding motifs in MCL tumor cells. SNHG5 and SNHG12 can also modulate c-Myc translation in MCL cells. | [168] |
| lncRNA | GAS5 | Downregulated | eIF4E | Downregulation of the tumor suppressor GAS5, which interacts with translation initiation factor eIF4E to suppress the translation of c-MYC mRNA, resulted in decreased apoptosis levels in MCL cell lines. Overexpression of GAS5 constructs is also sufficient to induce growth arrest in normal and transformed human lymphocytes. | [169] |
| lncRNA | GATA6-AS | Downregulated | GLUT1 | GATA6-AS is involved in endothelial–mesenchymal transition. Overexpression of lncRNA GATA6-AS inhibits cancer cell proliferation by downregulating GLUT1 and therefore inhibits glucose uptake in MCL. | [170] |
| lncRNA | LINK-A | Upregulated | Survivin (not directly) | LINK-A lncRNA overexpression promotes cell proliferation, inhibits cell apoptosis and upregulates survivin expression. | [171] |
| circRNA | circCDYL | Upregulated | miR-101 | Overexpression of circCDYL in MCL cells promotes malignant proliferation, self-renewal and chemoresistance. Inhibition of circCDYL suppressed cell proliferation in vitro, consistent with its functions in regulating transcription, programmed cell death, cell death and apoptosis. CircCDYL might serve as a sponge for miR-101, targeting EZH2 and further regulating p21 and p27. | [183] |
| circRNA | circ_cgga162 | Upregulated | unknown | High circ_cgga162 expression was associated with low overall survival rate. | [184] |
| circRNA | circRAB11FIP1 | Downregulated | unknown | Low circRAB11FIP1 expression was significantly associated with TP53 mutations and with shorter median time to progression. | [180] |
Funding
Conflicts of Interest
References
- Pennisi, E. Genomics. ENCODE project writes eulogy for junk DNA. Science 2012, 337, 1159–1161. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Adelman, K. Emerging roles of non-coding RNA transcription. Trends Biochem. Sci. 2018, 43, 654–667. [Google Scholar] [CrossRef]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. 2010, 220, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, C.A.; Voinnet, O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009, 21, 416–425. [Google Scholar] [CrossRef]
- Zheng, B.; Xi, Z.; Liu, R.; Yin, W.; Sui, Z.; Ren, B.; Miller, H.; Gong, Q.; Liu, C. The function of microRNAs in B-cell development, lymphoma, and their potential in clinical practice. Front. Immunol. 2018, 9, 936. [Google Scholar] [CrossRef] [PubMed]
- Marques, S.C.; Laursen, M.B.; Bodker, J.S.; Kjeldsen, M.K.; Falgreen, S.; Schmitz, A.; Bogsted, M.; Johnsen, H.E.; Dybkaer, K. MicroRNAs in B-cells: From normal differentiation to treatment of malignancies. Oncotarget 2015, 6, 7–25. [Google Scholar] [CrossRef]
- Dahl, M.; Kristensen, L.S.; Gronbaek, K. Long non-coding RNAs guide the fine-tuning of gene regulation in B-cell development and malignancy. Int. J. Mol. Sci. 2018, 19, 2475. [Google Scholar] [CrossRef] [PubMed]
- Coffre, M.; Koralov, S.B. miRNAs in B Cell development and lymphomagenesis. Trends Mol. Med. 2017, 23, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Chen, Q.; Tao, S.; Shi, Y.; Chen, Y.; Shen, L.; Wang, C.; Yu, L. The research progress of circular RNAs in hematological malignancies. Hematology 2019, 24, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; Kluiver, J.L.; Diepstra, A.; van den Berg, A. Emerging roles for long noncoding RNAs in B-cell development and malignancy. Crit. Rev. Oncol. Hematol. 2017, 120, 77–85. [Google Scholar] [CrossRef]
- Glass, S.; Phan, A.; Williams, J.N.; Flowers, C.R.; Koff, J.L. Integrating understanding of epidemiology and genomics in B-cell non-Hodgkin lymphoma as a pathway to novel management strategies. Discov. Med. 2016, 21, 181–188. [Google Scholar] [PubMed]
- Wang, J.; Pan, Y.; Wu, J.; Zhang, C.; Huang, Y.; Zhao, R.; Cheng, G.; Liu, J.; Qin, C.; Shao, P.; et al. The association between abnormal long noncoding RNA MALAT-1 expression and cancer lymph node metastasis: A meta-analysis. Biomed. Res. Int. 2016, 2016, 1823482. [Google Scholar] [CrossRef]
- Nemazee, D. Mechanisms of central tolerance for B cells. Nat. Rev. Immunol. 2017, 17, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Balatti, V.; Pekarky, Y.; Croce, C.M. Role of microRNA in chronic lymphocytic leukemia onset and progression. J. Hematol. Oncol. 2015, 8, 12. [Google Scholar] [CrossRef]
- Wang, G.G.; Konze, K.D.; Tao, J. Polycomb genes, miRNA, and their deregulation in B-cell malignancies. Blood 2015, 125, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Musilova, K.; Mraz, M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia 2015, 29, 1004–1017. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, D.; Wei, W.; Chen, H.; Shi, H.; Zhou, N.; Wu, L.; Peng, R. Comprehensive profiling of circular RNA expressions reveals potential diagnostic and prognostic biomarkers in multiple myeloma. BMC Cancer 2020, 20, 40. [Google Scholar] [CrossRef]
- Wu, W.; Wu, Z.; Xia, Y.; Qin, S.; Li, Y.; Wu, J.; Liang, J.; Wang, L.; Zhu, H.; Fan, L.; et al. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging 2019, 11, 3561–3573. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, L.; Wu, J.; Khadka, B.; Fang, Z.; Gu, J.; Tang, B.; Xiao, R.; Pan, G.; Liu, J. CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 54. [Google Scholar] [CrossRef] [PubMed]
- Berindan-Neagoe, I.; Monroig Pdel, C.; Pasculli, B.; Calin, G.A. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef]
- Sole, C.; Larrea, E.; Di Pinto, G.; Tellaetxe, M.; Lawrie, C.H. miRNAs in B-cell lymphoma: Molecular mechanisms and biomarker potential. Cancer Lett. 2017, 405, 79–89. [Google Scholar] [CrossRef]
- Tan, H.; Gan, L.; Fan, X.; Liu, L.; Liu, S. Diagnostic value of circular RNAs as effective biomarkers for cancer: A systematic review and meta-analysis. Onco Targets Ther. 2019, 12, 2623–2633. [Google Scholar] [CrossRef]
- Hardy, R.R.; Hayakawa, K. B cell development pathways. Annu. Rev. Immunol. 2001, 19, 595–621. [Google Scholar] [CrossRef]
- Berkowska, M.A.; Driessen, G.J.; Bikos, V.; Grosserichter-Wagener, C.; Stamatopoulos, K.; Cerutti, A.; He, B.; Biermann, K.; Lange, J.F.; van der Burg, M.; et al. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood 2011, 118, 2150–2158. [Google Scholar] [CrossRef]
- Santos, P.; Arumemi, F.; Park, K.S.; Borghesi, L.; Milcarek, C. Transcriptional and epigenetic regulation of B cell development. Immunol. Res. 2011, 50, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Northrup, D.L.; Allman, D. Transcriptional regulation of early B cell development. Immunol. Res. 2008, 42, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Croce, C.M.; Calin, G.A. MicroRNAs in the ontogeny of leukemias and lymphomas. Leuk. Lymphoma 2009, 50, 160–170. [Google Scholar] [CrossRef]
- Calvo, J.; BenYoucef, A.; Baijer, J.; Rouyez, M.C.; Pflumio, F. Assessment of human multi-potent hematopoietic stem/progenitor cell potential using a single in vitro screening system. PLoS ONE 2012, 7, e50495. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, H. A close developmental relationship between the lymphoid and myeloid lineages. Trends Immunol. 2006, 27, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Notta, F.; Eppert, K.; Nguyen, L.T.; Ohashi, P.S.; Dick, J.E. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 2010, 11, 585–593. [Google Scholar] [CrossRef]
- Di Lisio, L.; Martinez, N.; Montes-Moreno, S.; Piris-Villaespesa, M.; Sanchez-Beato, M.; Piris, M.A. The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood 2012, 120, 1782–1790. [Google Scholar] [CrossRef]
- Mastio, J.; Saeed, M.B.; Wurzer, H.; Krecke, M.; Westerberg, L.S.; Thomas, C. Higher incidence of B cell malignancies in primary immunodeficiencies: A combination of intrinsic genomic instability and exocytosis defects at the immunological synapse. Front. Immunol. 2020, 11, 581119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, H.; Fang, W.; Romaguer, J.E.; Zhang, Y.; Delasalle, K.B.; Kwak, L.; Yi, Q.; Du, X.L.; Wang, M. Incidence trends of mantle cell lymphoma in the United States between 1992 and 2004. Cancer 2008, 113, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Howell, D.; Patmore, R.; Jack, A.; Roman, E. Incidence of haematological malignancy by sub-type: A report from the Haematological Malignancy Research Network. Br. J. Cancer 2011, 105, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Raffeld, M.; Jaffe, E.S. bcl-1, t(11;14), and mantle cell-derived lymphomas. Blood 1991, 78, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.E.; Meeker, T.C.; Swerdlow, S.H. Rearrangement of the chromosome 11 bcl-1 locus in centrocytic lymphoma: Analysis with multiple breakpoint probes. Blood 1991, 78, 493–498. [Google Scholar] [CrossRef]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Hershkovitz-Rokah, O.; Geva, P.; Salmon-Divon, M.; Shpilberg, O.; Liberman-Aronov, S. Network analysis of microRNAs, genes and their regulation in diffuse and follicular B-cell lymphomas. Oncotarget 2018, 9, 7928–7941. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.H.; Chen, L.; Fu, Y.; Wang, W.; Fu, S.W. Cell-free circulating miRNA biomarkers in cancer. J. Cancer 2012, 3, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef]
- Larsen, M.T.; Hager, M.; Glenthoj, A.; Asmar, F.; Clemmensen, S.N.; Mora-Jensen, H.; Borregaard, N.; Cowland, J.B. miRNA-130a regulates C/EBP-epsilon expression during granulopoiesis. Blood 2014, 123, 1079–1089. [Google Scholar] [CrossRef]
- Fazi, F.; Racanicchi, S.; Zardo, G.; Starnes, L.M.; Mancini, M.; Travaglini, L.; Diverio, D.; Ammatuna, E.; Cimino, G.; Lo-Coco, F.; et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 2007, 12, 457–466. [Google Scholar] [CrossRef]
- Katsaraki, K.; Karousi, P.; Artemaki, P.I.; Scorilas, A.; Pappa, V.; Kontos, C.K.; Papageorgiou, S.G. MicroRNAs: Tiny regulators of gene expression with pivotal roles in normal B-cell development and B-cell chronic lymphocytic leukemia. Cancers 2021, 13, 593. [Google Scholar] [CrossRef]
- Koralov, S.B.; Muljo, S.A.; Galler, G.R.; Krek, A.; Chakraborty, T.; Kanellopoulou, C.; Jensen, K.; Cobb, B.S.; Merkenschlager, M.; Rajewsky, N.; et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 2008, 132, 860–874. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, D.; Mecklenbrauker, I.; Das, P.P.; Santana, A.; Koenig, U.; Enright, A.J.; Miska, E.A.; Tarakhovsky, A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007, 21, 1999–2004. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Guo, K.; Zeng, Q.; Huo, J.; Lam, K.P. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood 2012, 119, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Coffre, M.; Benhamou, D.; Riess, D.; Blumenberg, L.; Snetkova, V.; Hines, M.J.; Chakraborty, T.; Bajwa, S.; Jensen, K.; Chong, M.M.W.; et al. miRNAs are essential for the regulation of the PI3K/AKT/FOXO pathway and receptor editing during B cell maturation. Cell Rep. 2016, 17, 2271–2285. [Google Scholar] [CrossRef][Green Version]
- Brandl, A.; Daum, P.; Brenner, S.; Schulz, S.R.; Yap, D.Y.; Bosl, M.R.; Wittmann, J.; Schuh, W.; Jack, H.M. The microprocessor component, DGCR8, is essential for early B-cell development in mice. Eur. J. Immunol. 2016, 46, 2710–2718. [Google Scholar] [CrossRef]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, S.; Mayr, C.; Bartel, D.P.; Lodish, H.F. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. USA 2007, 104, 7080–7085. [Google Scholar] [CrossRef]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef]
- Thompson, C.B.; Challoner, P.B.; Neiman, P.E.; Groudine, M. Expression of the c-myb proto-oncogene during cellular proliferation. Nature 1986, 319, 374–380. [Google Scholar] [CrossRef]
- Lahortiga, I.; De Keersmaecker, K.; Van Vlierberghe, P.; Graux, C.; Cauwelier, B.; Lambert, F.; Mentens, N.; Beverloo, H.B.; Pieters, R.; Speleman, F.; et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007, 39, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.T. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008, 133, 217–222. [Google Scholar] [CrossRef]
- Tanzer, A.; Stadler, P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M.; Kutok, J.L.; Rajewsky, K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008, 9, 405–414. [Google Scholar] [CrossRef]
- Lai, M.; Gonzalez-Martin, A.; Cooper, A.B.; Oda, H.; Jin, H.Y.; Shepherd, J.; He, L.; Zhu, J.; Nemazee, D.; Xiao, C. Regulation of B-cell development and tolerance by different members of the miR-17 approximately 92 family microRNAs. Nat. Commun. 2016, 7, 12207. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, B.; Borde, M.; Nardone, J.; Maika, S.; Allred, L.; Tucker, P.W.; Rao, A. Foxp1 is an essential transcriptional regulator of B cell development. Nat. Immunol. 2006, 7, 819–826. [Google Scholar] [CrossRef]
- Rao, D.S.; O’Connell, R.M.; Chaudhuri, A.A.; Garcia-Flores, Y.; Geiger, T.L.; Baltimore, D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity 2010, 33, 48–59. [Google Scholar] [CrossRef]
- Okuyama, K.; Ikawa, T.; Gentner, B.; Hozumi, K.; Harnprasopwat, R.; Lu, J.; Yamashita, R.; Ha, D.; Toyoshima, T.; Chanda, B.; et al. MicroRNA-126-mediated control of cell fate in B-cell myeloid progenitors as a potential alternative to transcriptional factors. Proc. Natl. Acad. Sci. USA 2013, 110, 13410–13415. [Google Scholar] [CrossRef]
- Mehta, A.; Zhao, J.L.; Sinha, N.; Marinov, G.K.; Mann, M.; Kowalczyk, M.S.; Galimidi, R.P.; Du, X.; Erikci, E.; Regev, A.; et al. The microRNA-132 and microRNA-212 cluster regulates hematopoietic stem cell maintenance and survival with age by buffering FOXO3 expression. Immunity 2015, 42, 1021–1032. [Google Scholar] [CrossRef]
- Mehta, A.; Mann, M.; Zhao, J.L.; Marinov, G.K.; Majumdar, D.; Garcia-Flores, Y.; Du, X.; Erikci, E.; Chowdhury, K.; Baltimore, D. The microRNA-212/132 cluster regulates B cell development by targeting Sox4. J. Exp. Med. 2015, 212, 1679–1692. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Saunders, N.J.; Soneji, S.; Palazzo, S.; Dunlop, H.M.; Cooper, C.D.; Brown, P.J.; Troussard, X.; Mossafa, H.; Enver, T.; et al. MicroRNA expression in lymphocyte development and malignancy. Leukemia 2008, 22, 1440–1446. [Google Scholar] [CrossRef]
- Pede, V.; Rombout, A.; Vermeire, J.; Naessens, E.; Mestdagh, P.; Robberecht, N.; Vanderstraeten, H.; Van Roy, N.; Vandesompele, J.; Speleman, F.; et al. CLL cells respond to B-Cell receptor stimulation with a microRNA/mRNA signature associated with MYC activation and cell cycle progression. PLoS ONE 2013, 8, e60275. [Google Scholar] [CrossRef]
- Tavolaro, S.; Colombo, T.; Chiaretti, S.; Peragine, N.; Fulci, V.; Ricciardi, M.R.; Messina, M.; Bonina, S.; Brugnoletti, F.; Marinelli, M.; et al. Increased chronic lymphocytic leukemia proliferation upon IgM stimulation is sustained by the upregulation of miR-132 and miR-212. Genes Chromosomes Cancer 2015, 54, 222–234. [Google Scholar] [CrossRef]
- McHeyzer-Williams, M.; Okitsu, S.; Wang, N.; McHeyzer-Williams, L. Molecular programming of B cell memory. Nat. Rev. Immunol. 2011, 12, 24–34. [Google Scholar] [CrossRef]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef]
- De Yebenes, V.G.; Belver, L.; Pisano, D.G.; Gonzalez, S.; Villasante, A.; Croce, C.; He, L.; Ramiro, A.R. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 2008, 205, 2199–2206. [Google Scholar] [CrossRef]
- Teng, G.; Hakimpour, P.; Landgraf, P.; Rice, A.; Tuschl, T.; Casellas, R.; Papavasiliou, F.N. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 2008, 28, 621–629. [Google Scholar] [CrossRef]
- Muramatsu, M.; Kinoshita, K.; Fagarasan, S.; Yamada, S.; Shinkai, Y.; Honjo, T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 2000, 102, 553–563. [Google Scholar] [CrossRef]
- Okazaki, I.M.; Hiai, H.; Kakazu, N.; Yamada, S.; Muramatsu, M.; Kinoshita, K.; Honjo, T. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 2003, 197, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.C.; Herscovitch, M.; Zhao, I.; Ford, T.J.; Gilmore, T.D. NF-kappaB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J. Biol. Chem. 2011, 286, 1675–1682. [Google Scholar] [CrossRef]
- Vigorito, E.; Perks, K.L.; Abreu-Goodger, C.; Bunting, S.; Xiang, Z.; Kohlhaas, S.; Das, P.P.; Miska, E.A.; Rodriguez, A.; Bradley, A.; et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847–859. [Google Scholar] [CrossRef]
- Lu, D.; Nakagawa, R.; Lazzaro, S.; Staudacher, P.; Abreu-Goodger, C.; Henley, T.; Boiani, S.; Leyland, R.; Galloway, A.; Andrews, S.; et al. The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J. Exp. Med. 2014, 211, 2183–2198. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xi, Y.; Gao, N.; Xu, E.; Chang, J.; Liu, J. Identification of miRNA-34a and miRNA-155 as prognostic markers for mantle cell lymphoma. J. Int. Med. Res. 2021, 49, 3000605211016390. [Google Scholar] [CrossRef] [PubMed]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013, 122, 1891–1899. [Google Scholar] [CrossRef]
- Gururajan, M.; Haga, C.L.; Das, S.; Leu, C.M.; Hodson, D.; Josson, S.; Turner, M.; Cooper, M.D. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int. Immunol. 2010, 22, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; Dalla-Favera, R. Germinal centres: Role in B-cell physiology and malignancy. Nat. Rev. Immunol. 2008, 8, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, A.L.; Rosenwald, A.; Staudt, L.M. Lymphoid malignancies: The dark side of B-cell differentiation. Nat. Rev. Immunol. 2002, 2, 920–932. [Google Scholar] [CrossRef]
- Lin, J.; Lwin, T.; Zhao, J.J.; Tam, W.; Choi, Y.S.; Moscinski, L.C.; Dalton, W.S.; Sotomayor, E.M.; Wright, K.L.; Tao, J. Follicular dendritic cell-induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin’s B-cell lymphomas. Leukemia 2011, 25, 145–152. [Google Scholar] [CrossRef]
- Barnes, N.A.; Stephenson, S.; Cocco, M.; Tooze, R.M.; Doody, G.M. BLIMP-1 and STAT3 counterregulate microRNA-21 during plasma cell differentiation. J. Immunol. 2012, 189, 253–260. [Google Scholar] [CrossRef] [PubMed]
- de Yebenes, V.G.; Bartolome-Izquierdo, N.; Nogales-Cadenas, R.; Perez-Duran, P.; Mur, S.M.; Martinez, N.; Di Lisio, L.; Robbiani, D.F.; Pascual-Montano, A.; Canamero, M.; et al. miR-217 is an oncogene that enhances the germinal center reaction. Blood 2014, 124, 229–239. [Google Scholar] [CrossRef]
- Kramer, N.J.; Wang, W.L.; Reyes, E.Y.; Kumar, B.; Chen, C.C.; Ramakrishna, C.; Cantin, E.M.; Vonderfecht, S.L.; Taganov, K.D.; Chau, N.; et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 2015, 125, 3720–3730. [Google Scholar] [CrossRef]
- Porstner, M.; Winkelmann, R.; Daum, P.; Schmid, J.; Pracht, K.; Corte-Real, J.; Schreiber, S.; Haftmann, C.; Brandl, A.; Mashreghi, M.F.; et al. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur. J. Immunol. 2015, 45, 1206–1215. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Adams, B.D.; Lai, M.; Shepherd, J.; Salvador-Bernaldez, M.; Salvador, J.M.; Lu, J.; Nemazee, D.; Xiao, C. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat. Immunol. 2016, 17, 433–440. [Google Scholar] [CrossRef]
- Schraders, M.; Jares, P.; Bea, S.; Schoenmakers, E.F.; van Krieken, J.H.; Campo, E.; Groenen, P.J. Integrated genomic and expression profiling in mantle cell lymphoma: Identification of gene-dosage regulated candidate genes. Br. J. Haematol. 2008, 143, 210–221. [Google Scholar] [CrossRef]
- Navarro, A.; Bea, S.; Fernandez, V.; Prieto, M.; Salaverria, I.; Jares, P.; Hartmann, E.; Mozos, A.; Lopez-Guillermo, A.; Villamor, N.; et al. MicroRNA expression, chromosomal alterations, and immunoglobulin variable heavy chain hypermutations in Mantle cell lymphomas. Cancer Res. 2009, 69, 7071–7078. [Google Scholar] [CrossRef]
- Iqbal, J.; Shen, Y.; Liu, Y.; Fu, K.; Jaffe, E.S.; Liu, C.; Liu, Z.; Lachel, C.M.; Deffenbacher, K.; Greiner, T.C.; et al. Genome-wide miRNA profiling of mantle cell lymphoma reveals a distinct subgroup with poor prognosis. Blood 2012, 119, 4939–4948. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.; Pastore, A.; Deshpande, A.J.; Zimmermann, Y.; Hutter, G.; Weinkauf, M.; Buske, C.; Hiddemann, W.; Dreyling, M. 3’UTR mediated regulation of the cyclin D1 proto-oncogene. Cell Cycle 2009, 8, 3592–3600. [Google Scholar] [CrossRef]
- Rao, E.; Jiang, C.; Ji, M.; Huang, X.; Iqbal, J.; Lenz, G.; Wright, G.; Staudt, L.M.; Zhao, Y.; McKeithan, T.W.; et al. The miRNA-17 approximately 92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia 2012, 26, 1064–1072. [Google Scholar] [CrossRef]
- Jiang, P.; Rao, E.Y.; Meng, N.; Zhao, Y.; Wang, J.J. MicroRNA-17-92 significantly enhances radioresistance in human mantle cell lymphoma cells. Radiat. Oncol. 2010, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Roisman, A.; Huaman Garaicoa, F.; Metrebian, F.; Narbaitz, M.; Kohan, D.; Garcia Rivello, H.; Fernandez, I.; Pavlovsky, A.; Pavlovsky, M.; Hernandez, L.; et al. SOXC and MiR17-92 gene expression profiling defines two subgroups with different clinical outcome in mantle cell lymphoma. Genes Chromosomes Cancer 2016, 55, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Chen, R.W.; Bemis, L.T.; Amato, C.M.; Myint, H.; Tran, H.; Birks, D.K.; Eckhardt, S.G.; Robinson, W.A. Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood 2008, 112, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S.; et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010, 115, 2630–2639. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Lin, J.; Lwin, T.; Wright, G.; Moscinski, L.C.; Dalton, W.S.; Seto, E.; Wright, K.; Sotomayor, E.; et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene 2012, 31, 3002–3008. [Google Scholar] [CrossRef]
- Teshima, K.; Nara, M.; Watanabe, A.; Ito, M.; Ikeda, S.; Hatano, Y.; Oshima, K.; Seto, M.; Sawada, K.; Tagawa, H. Dysregulation of BMI1 and microRNA-16 collaborate to enhance an anti-apoptotic potential in the side population of refractory mantle cell lymphoma. Oncogene 2014, 33, 2191–2203. [Google Scholar] [CrossRef]
- Leung, C.; Lingbeek, M.; Shakhova, O.; Liu, J.; Tanger, E.; Saremaslani, P.; Van Lohuizen, M.; Marino, S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 2004, 428, 337–341. [Google Scholar] [CrossRef]
- Behesti, H.; Bhagat, H.; Dubuc, A.M.; Taylor, M.D.; Marino, S. Bmi1 overexpression in the cerebellar granule cell lineage of mice affects cell proliferation and survival without initiating medulloblastoma formation. Dis. Model. Mech. 2013, 6, 49–63. [Google Scholar] [CrossRef]
- Hoenerhoff, M.J.; Chu, I.; Barkan, D.; Liu, Z.Y.; Datta, S.; Dimri, G.P.; Green, J.E. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene 2009, 28, 3022–3032. [Google Scholar] [CrossRef]
- Wu, Z.; Min, L.; Chen, D.; Hao, D.; Duan, Y.; Qiu, G.; Wang, Y. Overexpression of BMI-1 promotes cell growth and resistance to cisplatin treatment in osteosarcoma. PLoS ONE 2011, 6, e14648. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, F.; Jiao, Y.; Feng, J.; Tang, W.; Yao, H.; Gong, C.; Chen, J.; Su, F.; Zhang, Y.; et al. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin. Cancer Res. 2011, 17, 7105–7115. [Google Scholar] [CrossRef]
- Arakawa, F.; Kimura, Y.; Yoshida, N.; Miyoshi, H.; Doi, A.; Yasuda, K.; Nakajima, K.; Kiyasu, J.; Niino, D.; Sugita, Y.; et al. Identification of miR-15b as a transformation-related factor in mantle cell lymphoma. Int. J. Oncol. 2016, 48, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Christoffersen, N.R.; Shalgi, R.; Frankel, L.B.; Leucci, E.; Lees, M.; Klausen, M.; Pilpel, Y.; Nielsen, F.C.; Oren, M.; Lund, A.H. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010, 17, 236–245. [Google Scholar] [CrossRef]
- Chen, F.; Hu, S.J. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: A review. J. Biochem. Mol. Toxicol. 2012, 26, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.; Clot, G.; Prieto, M.; Royo, C.; Vegliante, M.C.; Amador, V.; Hartmann, E.; Salaverria, I.; Bea, S.; Martin-Subero, J.I.; et al. microRNA expression profiles identify subtypes of mantle cell lymphoma with different clinicobiological characteristics. Clin. Cancer Res. 2013, 19, 3121–3129. [Google Scholar] [CrossRef]
- Hartmann, E.; Fernandez, V.; Moreno, V.; Valls, J.; Hernandez, L.; Bosch, F.; Abrisqueta, P.; Klapper, W.; Dreyling, M.; Hoster, E.; et al. Five-gene model to predict survival in mantle-cell lymphoma using frozen or formalin-fixed, paraffin-embedded tissue. J. Clin. Oncol. 2008, 26, 4966–4972. [Google Scholar] [CrossRef]
- Kienle, D.; Katzenberger, T.; Ott, G.; Saupe, D.; Benner, A.; Kohlhammer, H.; Barth, T.F.; Holler, S.; Kalla, J.; Rosenwald, A.; et al. Quantitative gene expression deregulation in mantle-cell lymphoma: Correlation with clinical and biologic factors. J. Clin. Oncol. 2007, 25, 2770–2777. [Google Scholar] [CrossRef] [PubMed]
- Bowen, T.; Jenkins, R.H.; Fraser, D.J. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J. Pathol. 2013, 229, 274–285. [Google Scholar] [CrossRef]
- Filkowski, J.N.; Ilnytskyy, Y.; Tamminga, J.; Koturbash, I.; Golubov, A.; Bagnyukova, T.; Pogribny, I.P.; Kovalchuk, O. Hypomethylation and genome instability in the germline of exposed parents and their progeny is associated with altered miRNA expression. Carcinogenesis 2010, 31, 1110–1115. [Google Scholar] [CrossRef]
- van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed]
- Di Lisio, L.; Gomez-Lopez, G.; Sanchez-Beato, M.; Gomez-Abad, C.; Rodriguez, M.E.; Villuendas, R.; Ferreira, B.I.; Carro, A.; Rico, D.; Mollejo, M.; et al. Mantle cell lymphoma: Transcriptional regulation by microRNAs. Leukemia 2010, 24, 1335–1342. [Google Scholar] [CrossRef]
- Zhang, W.J.; Chen, F.F.; Wang, Y.Q.; Yu, W.; Dong, F.L.; Zhuang, H.X. MiR-150 inhibits proliferation of mantle-cell lymphoma cells via regulation of MET. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12063–12072. [Google Scholar] [CrossRef]
- Husby, S.; Ralfkiaer, U.; Garde, C.; Zandi, R.; Ek, S.; Kolstad, A.; Jerkeman, M.; Laurell, A.; Raty, R.; Pedersen, L.B.; et al. miR-18b overexpression identifies mantle cell lymphoma patients with poor outcome and improves the MIPI-B prognosticator. Blood 2015, 125, 2669–2677. [Google Scholar] [CrossRef][Green Version]
- Murakami, Y.; Tamori, A.; Itami, S.; Tanahashi, T.; Toyoda, H.; Tanaka, M.; Wu, W.; Brojigin, N.; Kaneoka, Y.; Maeda, A.; et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer 2013, 13, 99. [Google Scholar] [CrossRef]
- Xue, T.M.; Tao, L.D.; Zhang, M.; Xu, G.C.; Zhang, J.; Zhang, P.J. miR-20b overexpression is predictive of poor prognosis in gastric cancer. Onco Targets Ther. 2015, 8, 1871–1876. [Google Scholar] [CrossRef]
- Szymczyk, A.; Macheta, A.; Podhorecka, M. Abnormal microRNA expression in the course of hematological malignancies. Cancer Manag. Res. 2018, 10, 4267–4277. [Google Scholar] [CrossRef]
- Sun, W.; Shen, W.; Yang, S.; Hu, F.; Li, H.; Zhu, T.H. miR-223 and miR-142 attenuate hematopoietic cell proliferation, and miR-223 positively regulates miR-142 through LMO2 isoforms and CEBP-beta. Cell Res. 2010, 20, 1158–1169. [Google Scholar] [CrossRef]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef]
- Yuan, J.Y.; Wang, F.; Yu, J.; Yang, G.H.; Liu, X.L.; Zhang, J.W. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J. Cell Mol. Med. 2009, 13, 4551–4559. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Feng, X.; Wang, Y.; Liu, Y.; Tian, L.; Zuo, W.; Yi, S.; Wei, X.; Song, Y.; Qiu, L. miR-223 is repressed and correlates with inferior clinical features in mantle cell lymphoma through targeting SOX11. Exp. Hematol. 2017, 58, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Zou, Z.K.; Su, H.Y.; Huang, Y.Q. Expression of MiR101 and EZH2 in Patients with Mantle Cell Lymphoma and Its Clinical Significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2019, 27, 820–826. [Google Scholar] [CrossRef]
- Lin, L.; Huang, Y.; Zhuang, W.; Lin, P.; Ma, X. miR-100 inhibits cell proliferation in mantle cell lymphoma by targeting mTOR. Exp. Hematol. Oncol. 2020, 9, 25. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef]
- Chim, C.S.; Wong, K.Y.; Leung, C.Y.; Chung, L.P.; Hui, P.K.; Chan, S.Y.; Yu, L. Epigenetic inactivation of the hsa-miR-203 in haematological malignancies. J. Cell Mol. Med. 2011, 15, 2760–2767. [Google Scholar] [CrossRef]
- Wong, K.Y.; So, C.C.; Loong, F.; Chung, L.P.; Lam, W.W.; Liang, R.; Li, G.K.; Jin, D.Y.; Chim, C.S. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS ONE 2011, 6, e19027. [Google Scholar] [CrossRef]
- Wong, K.Y.; Yim, R.L.; Kwong, Y.L.; Leung, C.Y.; Hui, P.K.; Cheung, F.; Liang, R.; Jin, D.Y.; Chim, C.S. Epigenetic inactivation of the MIR129-2 in hematological malignancies. J. Hematol. Oncol. 2013, 6, 16. [Google Scholar] [CrossRef]
- Yim, R.L.; Wong, K.Y.; Kwong, Y.L.; Loong, F.; Leung, C.Y.; Chu, R.; Lam, W.W.; Hui, P.K.; Lai, R.; Chim, C.S. Methylation of miR-155-3p in mantle cell lymphoma and other non-Hodgkin’s lymphomas. Oncotarget 2014, 5, 9770–9782. [Google Scholar] [CrossRef][Green Version]
- Kopparapu, P.K.; Bhoi, S.; Mansouri, L.; Arabanian, L.S.; Plevova, K.; Pospisilova, S.; Wasik, A.M.; Croci, G.A.; Sander, B.; Paulli, M.; et al. Epigenetic silencing of miR-26A1 in chronic lymphocytic leukemia and mantle cell lymphoma: Impact on EZH2 expression. Epigenetics 2016, 11, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Calin, G.A.; Yuen, K.S.; Jin, D.Y.; Chim, C.S. Epigenetic silencing of miR-342-3p in B cell lymphoma and its impact on autophagy. Clin. Epigenet. 2020, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Karkhanis, V.; Alinari, L.; Ozer, H.G.; Chung, J.; Zhang, X.; Sif, S.; Baiocchi, R.A. Protein arginine methyltransferase 5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1 and c-MYC expression in aggressive B-cell lymphoma. J. Biol. Chem. 2020, 295, 1165–1180. [Google Scholar] [CrossRef]
- Pal, S.; Baiocchi, R.A.; Byrd, J.C.; Grever, M.R.; Jacob, S.T.; Sif, S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007, 26, 3558–3569. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.S.; Sunwoo, H.; Zhang, B.; Spector, D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011, 13, 95–101. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018, 46, 3742–3752. [Google Scholar] [CrossRef]
- Ulitsky, I. Evolution to the rescue: Using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016, 17, 601–614. [Google Scholar] [CrossRef]
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2013, 2, e01749. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Liu, J.; Chen, C.H.; Liao, Q.; Xu, P.; Xu, H.; Xiao, T.; Cao, Z.; Peng, J.; et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 2016, 34, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Bonnal, R.J.; Ranzani, V.; Arrigoni, A.; Curti, S.; Panzeri, I.; Gruarin, P.; Abrignani, S.; Rossetti, G.; Pagani, M. De novo transcriptome profiling of highly purified human lymphocytes primary cells. Sci. Data 2015, 2, 150051. [Google Scholar] [CrossRef] [PubMed]
- Casero, D.; Sandoval, S.; Seet, C.S.; Scholes, J.; Zhu, Y.; Ha, V.L.; Luong, A.; Parekh, C.; Crooks, G.M. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 2015, 16, 1282–1291. [Google Scholar] [CrossRef]
- Petri, A.; Dybkaer, K.; Bogsted, M.; Thrue, C.A.; Hagedorn, P.H.; Schmitz, A.; Bodker, J.S.; Johnsen, H.E.; Kauppinen, S. Long noncoding RNA expression during human B-cell development. PLoS ONE 2015, 10, e0138236. [Google Scholar] [CrossRef]
- Ranzani, V.; Rossetti, G.; Panzeri, I.; Arrigoni, A.; Bonnal, R.J.; Curti, S.; Gruarin, P.; Provasi, E.; Sugliano, E.; Marconi, M.; et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015, 16, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Tayari, M.M.; Winkle, M.; Kortman, G.; Sietzema, J.; de Jong, D.; Terpstra, M.; Mestdagh, P.; Kroese, F.G.; Visser, L.; Diepstra, A.; et al. Long noncoding RNA expression profiling in normal B-cell subsets and hodgkin lymphoma reveals hodgkin and reed-sternberg cell-specific long noncoding RNAs. Am. J. Pathol. 2016, 186, 2462–2472. [Google Scholar] [CrossRef]
- Brazao, T.F.; Johnson, J.S.; Muller, J.; Heger, A.; Ponting, C.P.; Tybulewicz, V.L. Long noncoding RNAs in B-cell development and activation. Blood 2016, 128, e10–e19. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Esposti, D.D.; Hernandez-Vargas, H.; Voegele, C.; Fernandez-Jimenez, N.; Forey, N.; Bancel, B.; Le Calvez-Kelm, F.; McKay, J.; Merle, P.; Herceg, Z. Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget 2016, 7, 31862–31877. [Google Scholar] [CrossRef]
- Song, H.; Han, L.M.; Gao, Q.; Sun, Y. Long non-coding RNA CRNDE promotes tumor growth in medulloblastoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2588–2597. [Google Scholar]
- Meng, Y.; Li, Q.; Li, L.; Ma, R. The long non-coding RNA CRNDE promotes cervical cancer cell growth and metastasis. Biol. Chem. 2017, 399, 93–100. [Google Scholar] [CrossRef]
- Shao, K.; Shi, T.; Yang, Y.; Wang, X.; Xu, D.; Zhou, P. Highly expressed lncRNA CRNDE promotes cell proliferation through Wnt/beta-catenin signaling in renal cell carcinoma. Tumour Biol. 2016, 37, 15997–16004. [Google Scholar] [CrossRef]
- Hu, G.; Gupta, S.K.; Troska, T.P.; Nair, A.; Gupta, M. Long non-coding RNA profile in mantle cell lymphoma identifies a functional lncRNA ROR1-AS1 associated with EZH2/PRC2 complex. Oncotarget 2017, 8, 80223–80234. [Google Scholar] [CrossRef]
- Visser, H.P.; Gunster, M.J.; Kluin-Nelemans, H.C.; Manders, E.M.; Raaphorst, F.M.; Meijer, C.J.; Willemze, R.; Otte, A.P. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br. J. Haematol. 2001, 112, 950–958. [Google Scholar] [CrossRef]
- Wang, X.; Sehgal, L.; Jain, N.; Khashab, T.; Mathur, R.; Samaniego, F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016, 14, 346. [Google Scholar] [CrossRef]
- Sehgal, L.; Mathur, R.; Braun, F.K.; Wise, J.F.; Berkova, Z.; Neelapu, S.; Kwak, L.W.; Samaniego, F. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia 2014, 28, 2376–2387. [Google Scholar] [CrossRef] [PubMed]
- Inamdar, K.V.; Romaguera, J.E.; Drakos, E.; Knoblock, R.J.; Garcia, M.; Leventaki, V.; Medeiros, L.J.; Rassidakis, G.Z. Expression of eukaryotic initiation factor 4E predicts clinical outcome in patients with mantle cell lymphoma treated with hyper-CVAD and rituximab, alternating with rituximab, high-dose methotrexate, and cytarabine. Cancer 2009, 115, 4727–4736. [Google Scholar] [CrossRef]
- Hu, G.; Witzig, T.E.; Gupta, M. A novel long non-coding RNA, SNHG4 complex with eukaryotic initiation factor-4E and regulate aberrant protein translation in mantle cell lymphoma: Implications for novel biomarker. Blood 2013, 122, 81. [Google Scholar] [CrossRef]
- Hu, G.; Zhang, Y.; Gupta, M. RIP sequencing in mantle cell lymphoma identifies functional long non-coding RNAs associated with translation machinery. Blood Cancer J. 2019, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Lou, Z.; Gupta, M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS ONE 2014, 9, e107016. [Google Scholar] [CrossRef]
- Fan, Z.; Wang, X.; Li, P.; Mei, C.; Zhang, M.; Zhao, C. Overexpression of lncRNA GATA6-AS inhibits cancer cell proliferation in mantle cell lymphoma by downregulating GLUT1. Oncol. Lett. 2019, 18, 2443–2447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, P.; Du, H.; Zhang, L. LINK-A lncRNA promotes proliferation and inhibits apoptosis of mantle cell lymphoma cell by upregulating survivin. Med. Sci. Monit. 2019, 25, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.S.; Pan, F.; Mao, X.D.; Liu, C.; Chen, Y.J. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am. J. Transl. Res. 2019, 11, 1–15. [Google Scholar] [PubMed]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Tillo, T.H.; Giurini, J.M.; Habershaw, G.M.; Chrzan, J.S.; Rowbotham, J.L. Review of metatarsal osteotomies for the treatment of neuropathic ulcerations. J. Am. Podiatr. Med. Assoc. 1990, 80, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Ye, B.; Du, Y.; Li, C.; Xiong, Z.; Qu, Y.; Fan, Z. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 2018, 48, 688–701.e7. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 2016, 165, 289–302. [Google Scholar] [CrossRef]
- Dzikiewicz-Krawczyk, A.; Kok, K.; Slezak-Prochazka, I.; Robertus, J.L.; Bruining, J.; Tayari, M.M.; Rutgers, B.; de Jong, D.; Koerts, J.; Seitz, A.; et al. ZDHHC11 and ZDHHC11B are critical novel components of the oncogenic MYC-miR-150-MYB network in Burkitt lymphoma. Leukemia 2017, 31, 1470–1473. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Husby, S.; Eskelund, C.W.; Côme, C.; Ek, S.; Kolstad, A.; Räty, R.; Jerkeman, M.; Geisler, C.H.; Kristensen, L.S.; et al. A Circular RNA molecule, circRAB11FIP1, is associated with TP53 mutations and is of potential prognostic and functional significance in mantle cell lymphoma: Data from the Nordic MCL2 and MCL3 studies. Blood 2019, 134, 1495. [Google Scholar] [CrossRef]
- Dahl, M.; Daugaard, I.; Andersen, M.S.; Hansen, T.B.; Gronbaek, K.; Kjems, J.; Kristensen, L.S. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Investig. 2018, 98, 1657–1669. [Google Scholar] [CrossRef]
- Mei, M.; Wang, Y.; Wang, Q.; Liu, Y.; Song, W.; Zhang, M. CircCDYL serves as a new biomarker in mantle cell lymphoma and promotes cell proliferation. Cancer Manag. Res. 2019, 11, 10215–10221. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, S.; Ma, W.; Ke, W.; Zhang, C.; Liu, S.; Zhang, Y.; Pei, F.; Li, S.; Yi, M.; et al. CDYL suppresses epileptogenesis in mice through repression of axonal Nav1.6 sodium channel expression. Nat. Commun. 2017, 8, 355. [Google Scholar] [CrossRef]
- Li, Y.; Jia, Z.W.; Liu, J.X.; Ye, J.W.; Kong, X.Y.; Yang, Z.F.; Liu, X.Y.; Luo, J.M. Relationship of expression of Circ_cgga162 with the prognosis of patients with mantle cell lymphoma. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2020, 28, 876–880. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kersy, O.; Salmon-Divon, M.; Shpilberg, O.; Hershkovitz-Rokah, O. Non-Coding RNAs in Normal B-Cell Development and in Mantle Cell Lymphoma: From Molecular Mechanism to Biomarker and Therapeutic Agent Potential. Int. J. Mol. Sci. 2021, 22, 9490. https://doi.org/10.3390/ijms22179490
Kersy O, Salmon-Divon M, Shpilberg O, Hershkovitz-Rokah O. Non-Coding RNAs in Normal B-Cell Development and in Mantle Cell Lymphoma: From Molecular Mechanism to Biomarker and Therapeutic Agent Potential. International Journal of Molecular Sciences. 2021; 22(17):9490. https://doi.org/10.3390/ijms22179490
Chicago/Turabian StyleKersy, Olga, Mali Salmon-Divon, Ofer Shpilberg, and Oshrat Hershkovitz-Rokah. 2021. "Non-Coding RNAs in Normal B-Cell Development and in Mantle Cell Lymphoma: From Molecular Mechanism to Biomarker and Therapeutic Agent Potential" International Journal of Molecular Sciences 22, no. 17: 9490. https://doi.org/10.3390/ijms22179490
APA StyleKersy, O., Salmon-Divon, M., Shpilberg, O., & Hershkovitz-Rokah, O. (2021). Non-Coding RNAs in Normal B-Cell Development and in Mantle Cell Lymphoma: From Molecular Mechanism to Biomarker and Therapeutic Agent Potential. International Journal of Molecular Sciences, 22(17), 9490. https://doi.org/10.3390/ijms22179490






