Follistatin-Like-1 (FSTL1) Is a Fibroblast-Derived Growth Factor That Contributes to Progression of Chronic Kidney Disease
Abstract
:1. Introduction
2. Results
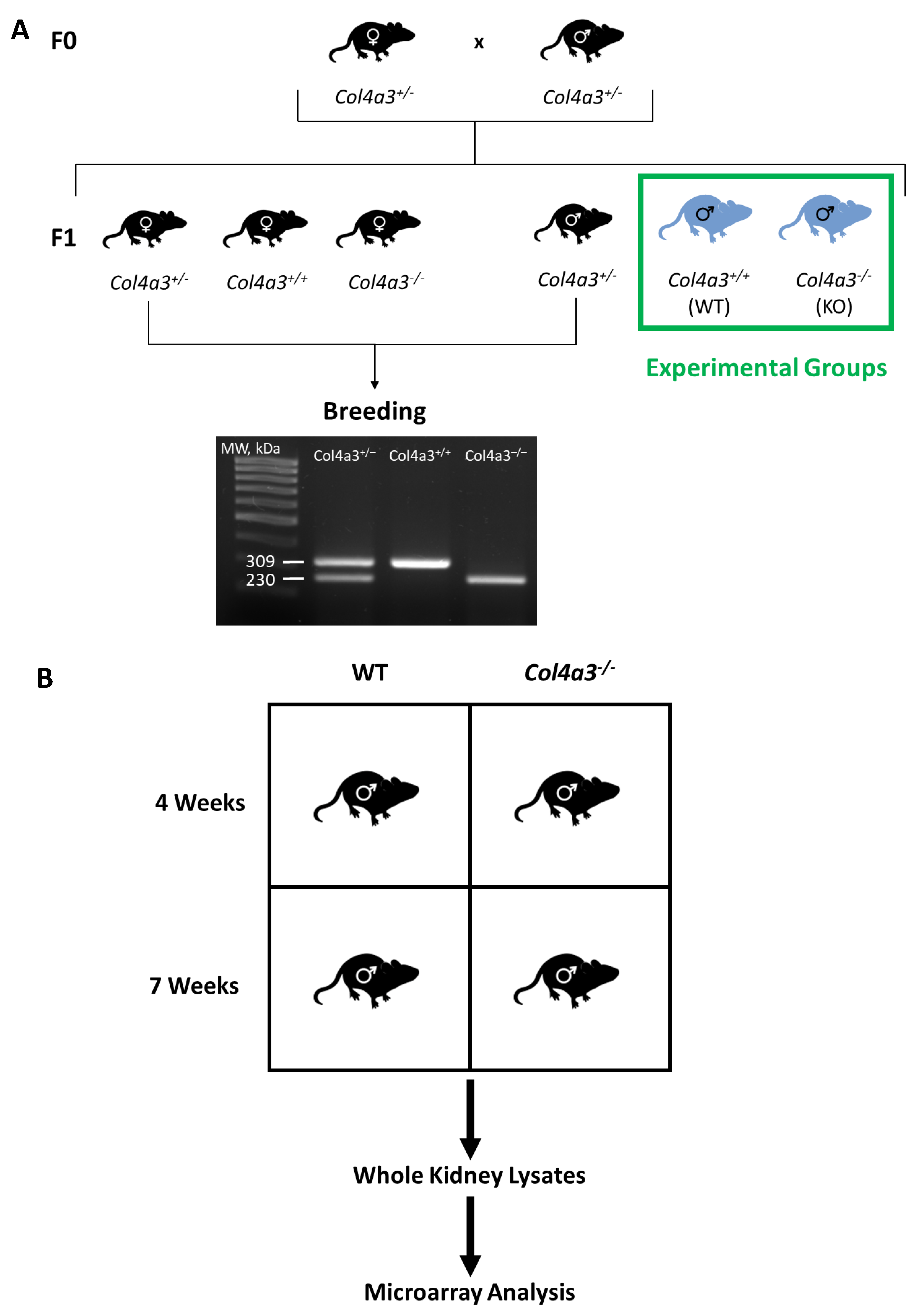
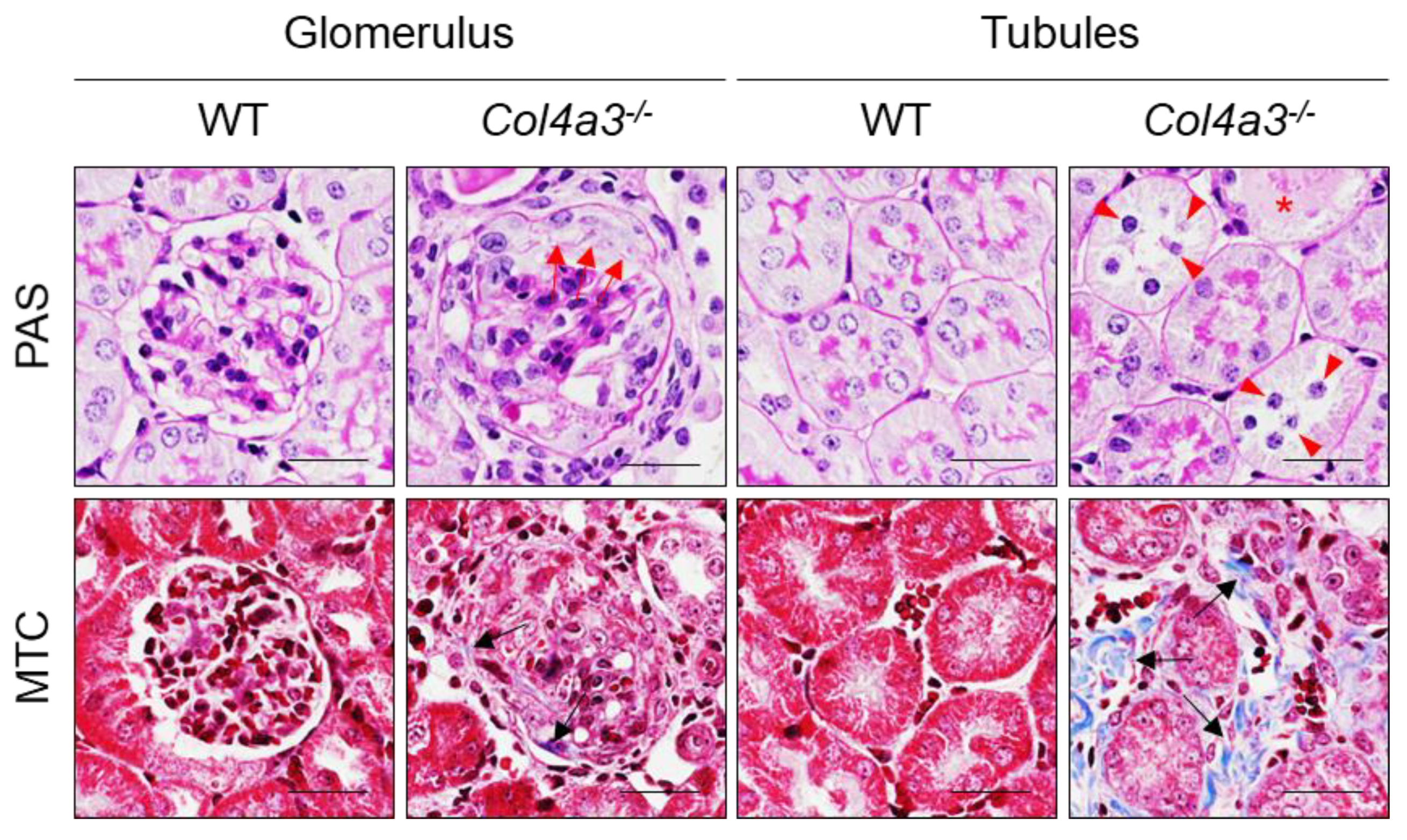
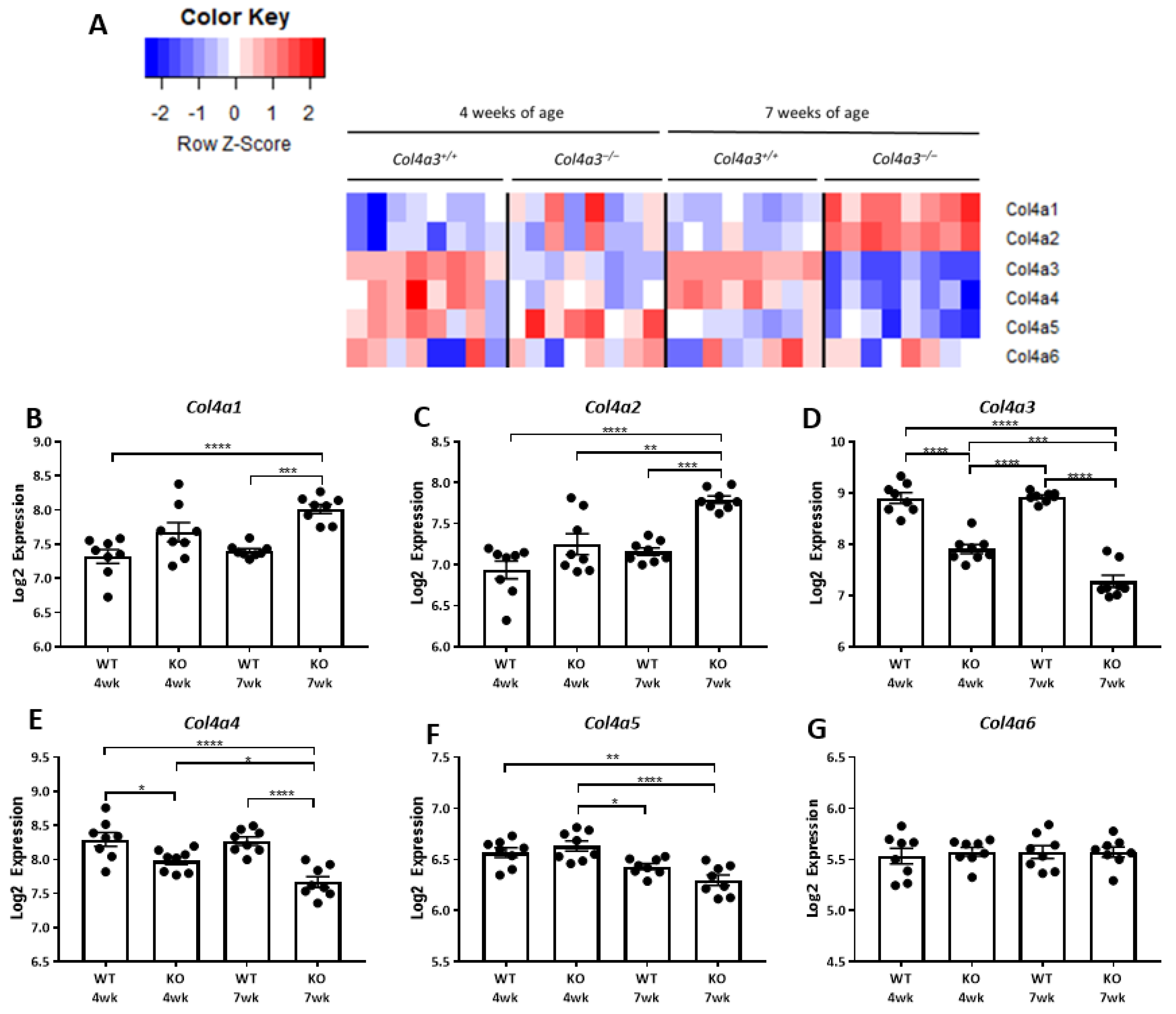
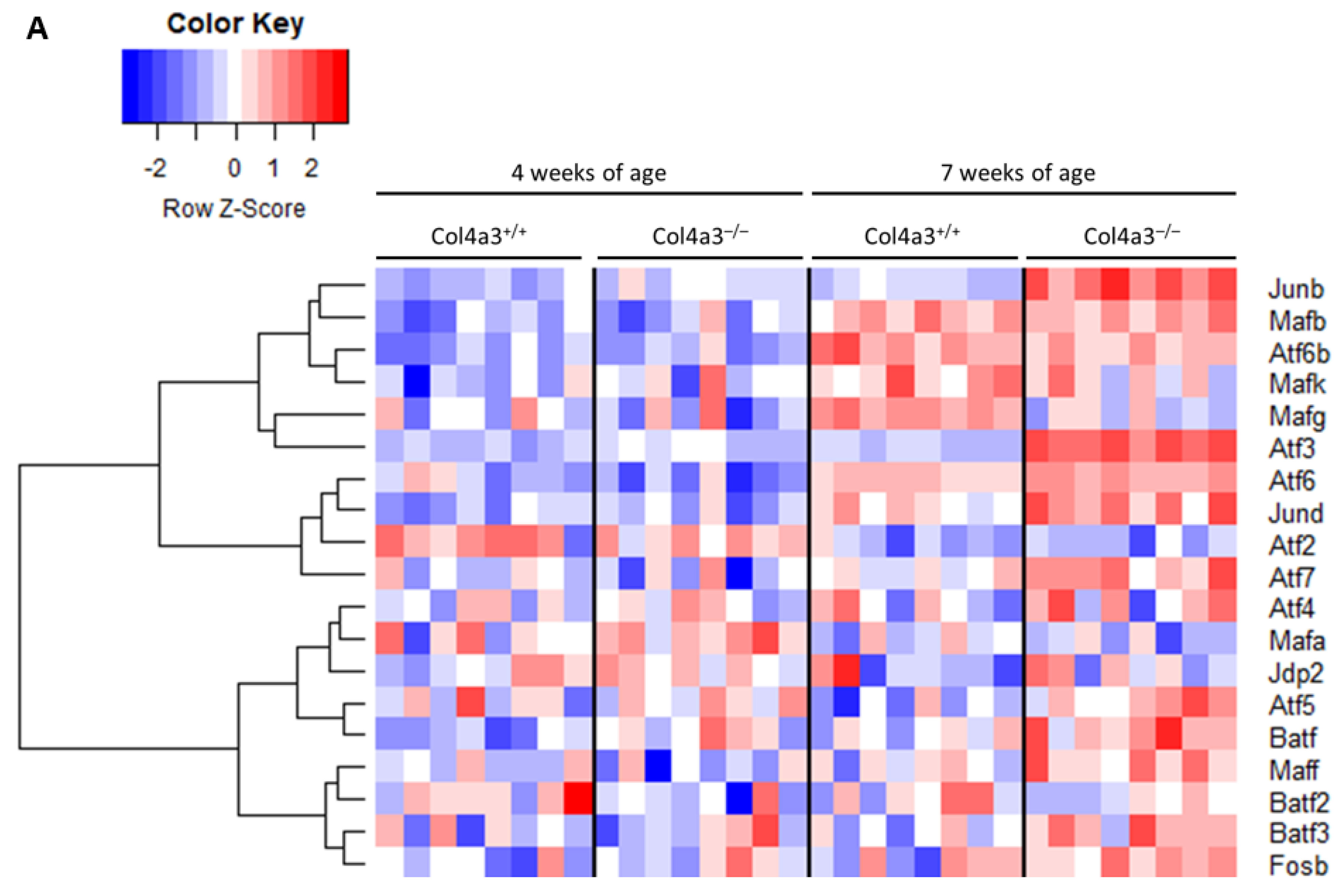
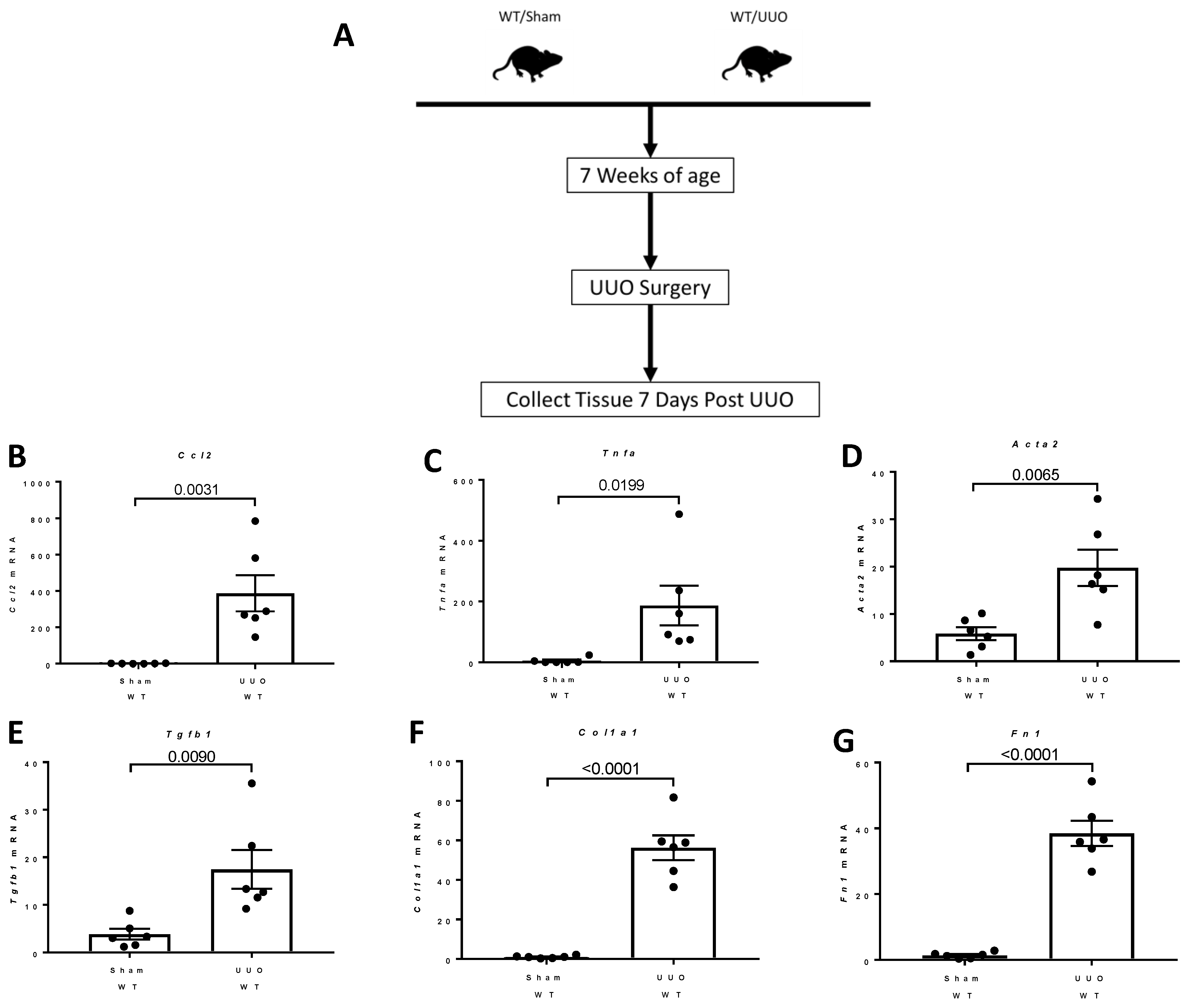
2.1. Breeding Strategy for Col4a3-/- Mice and Experimental Design
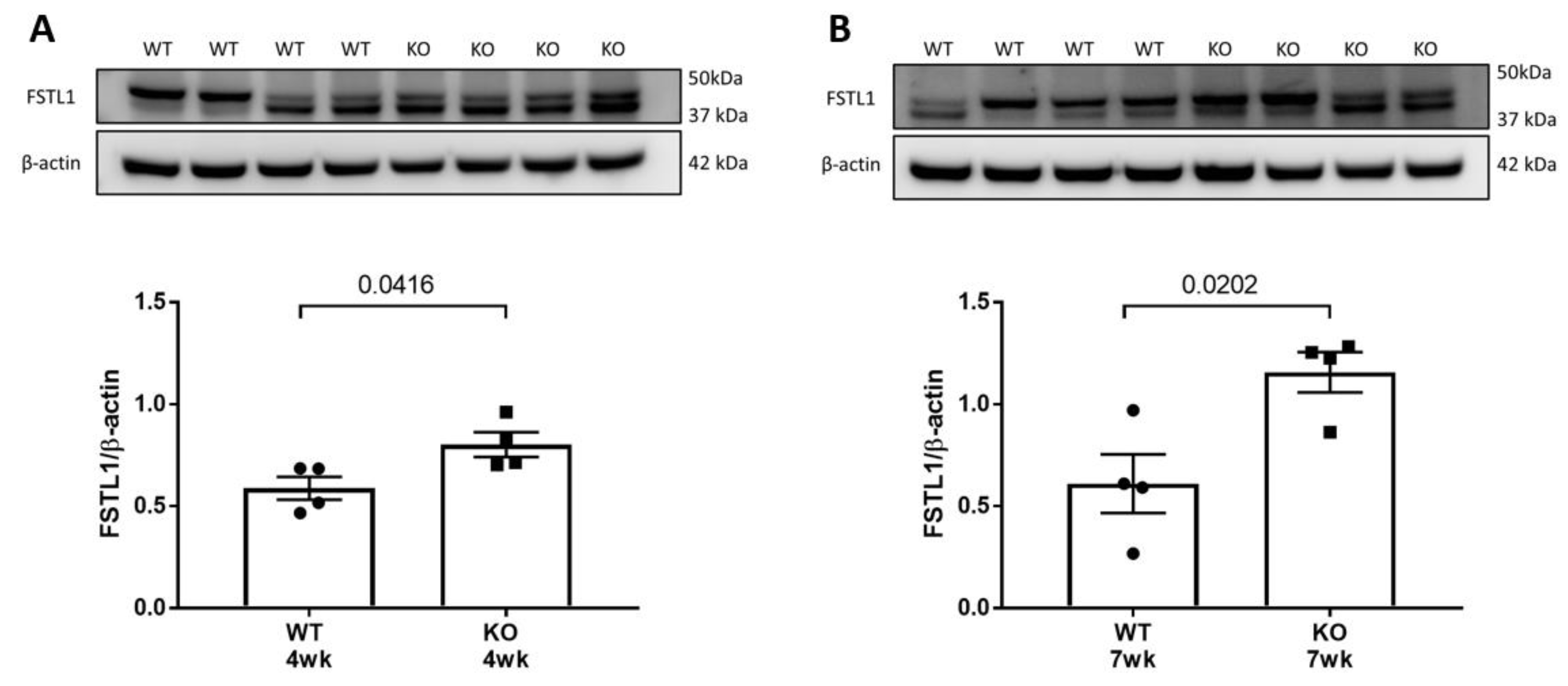
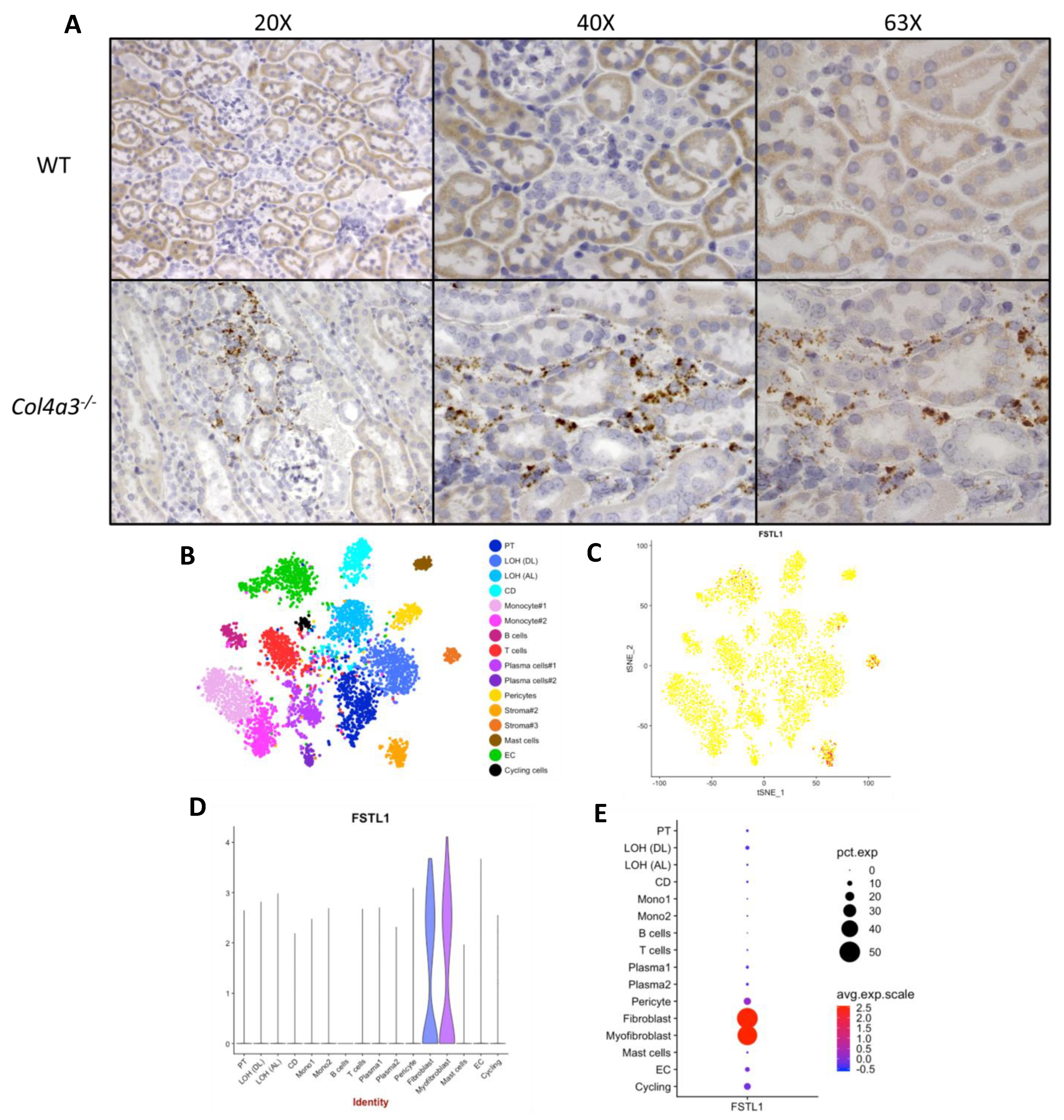
2.2. Localization of Fstl1 in the Kidney
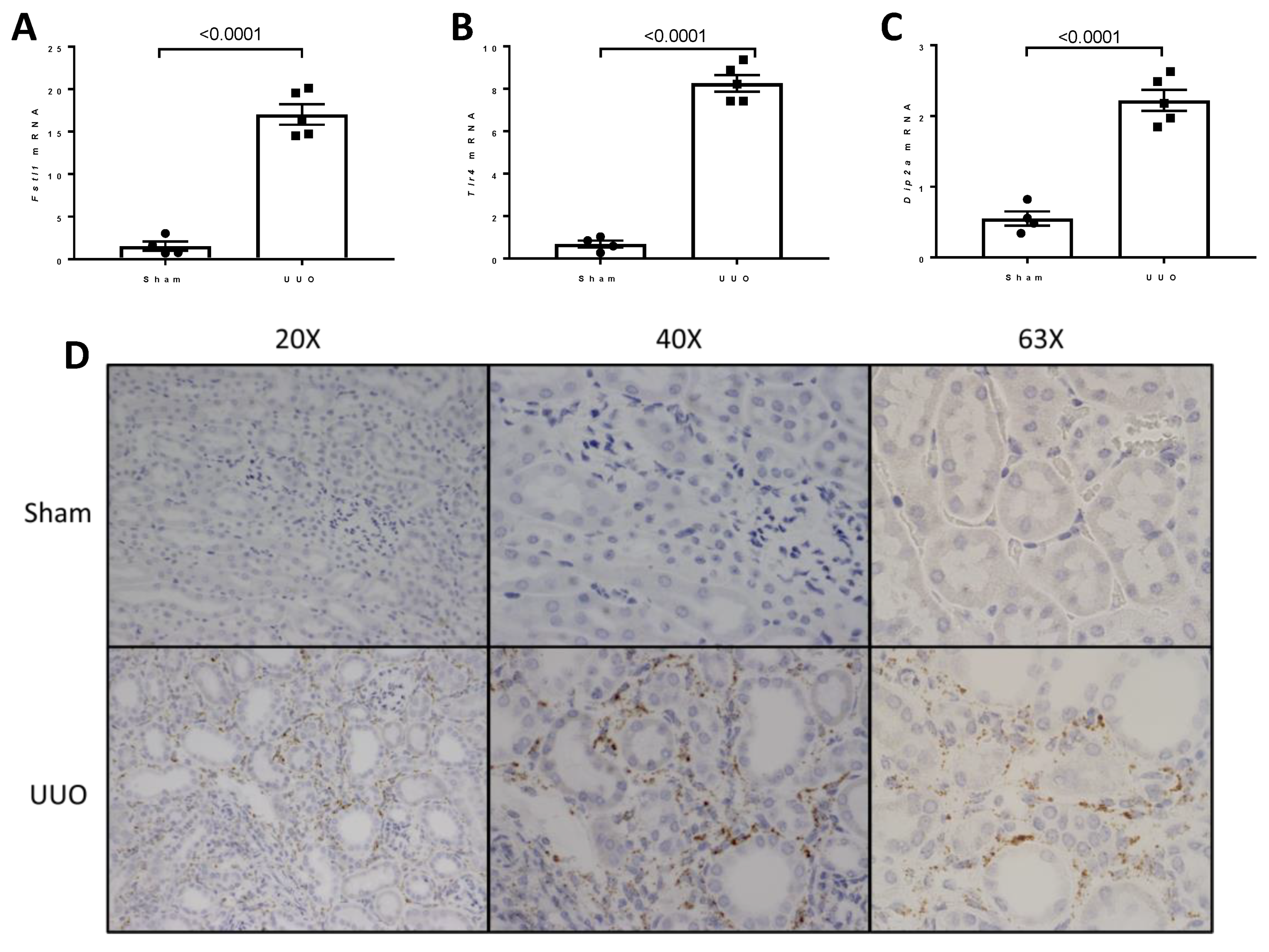
2.3. Expression of Cognate Receptors for FSTL1 in the Kidney
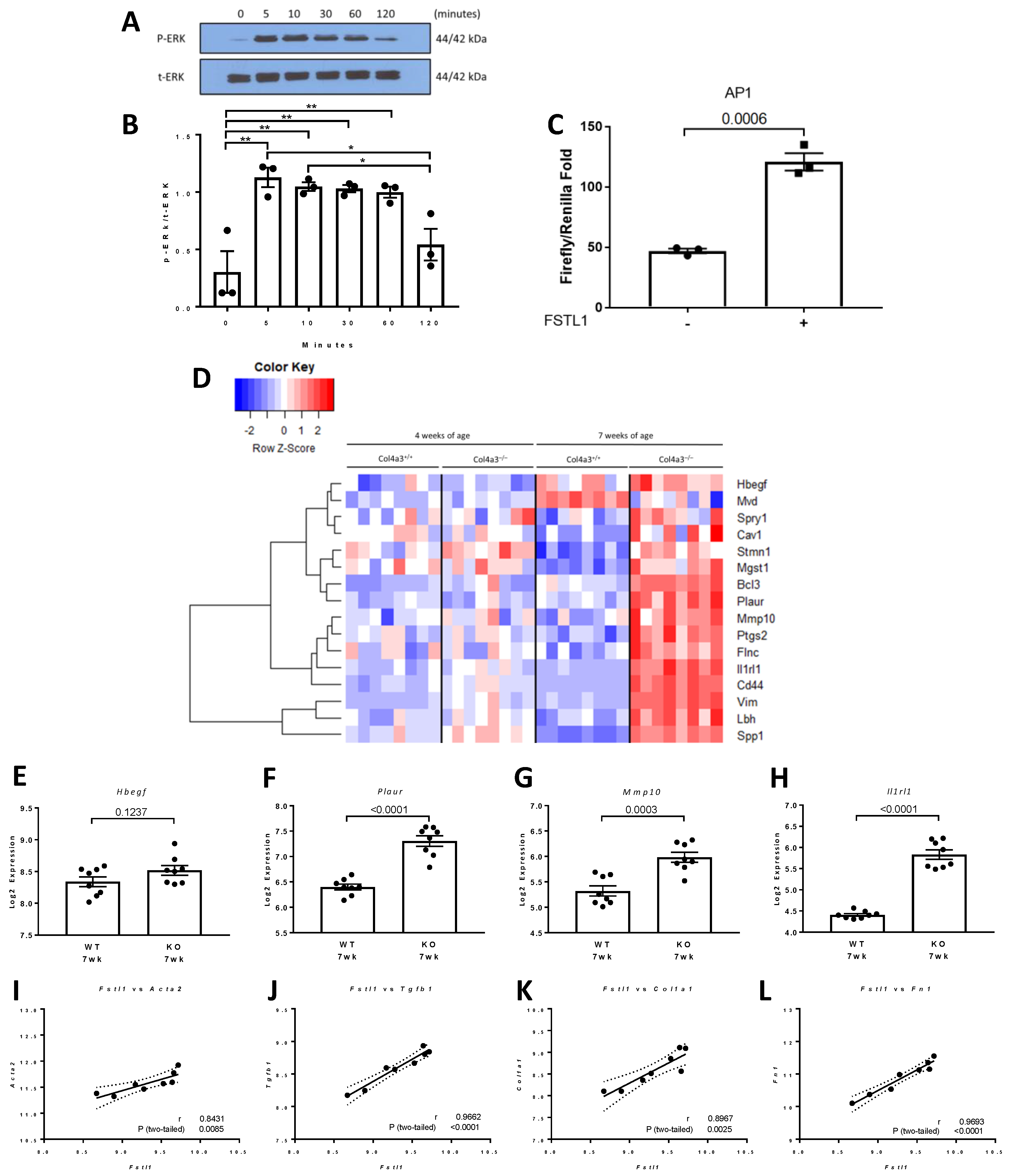
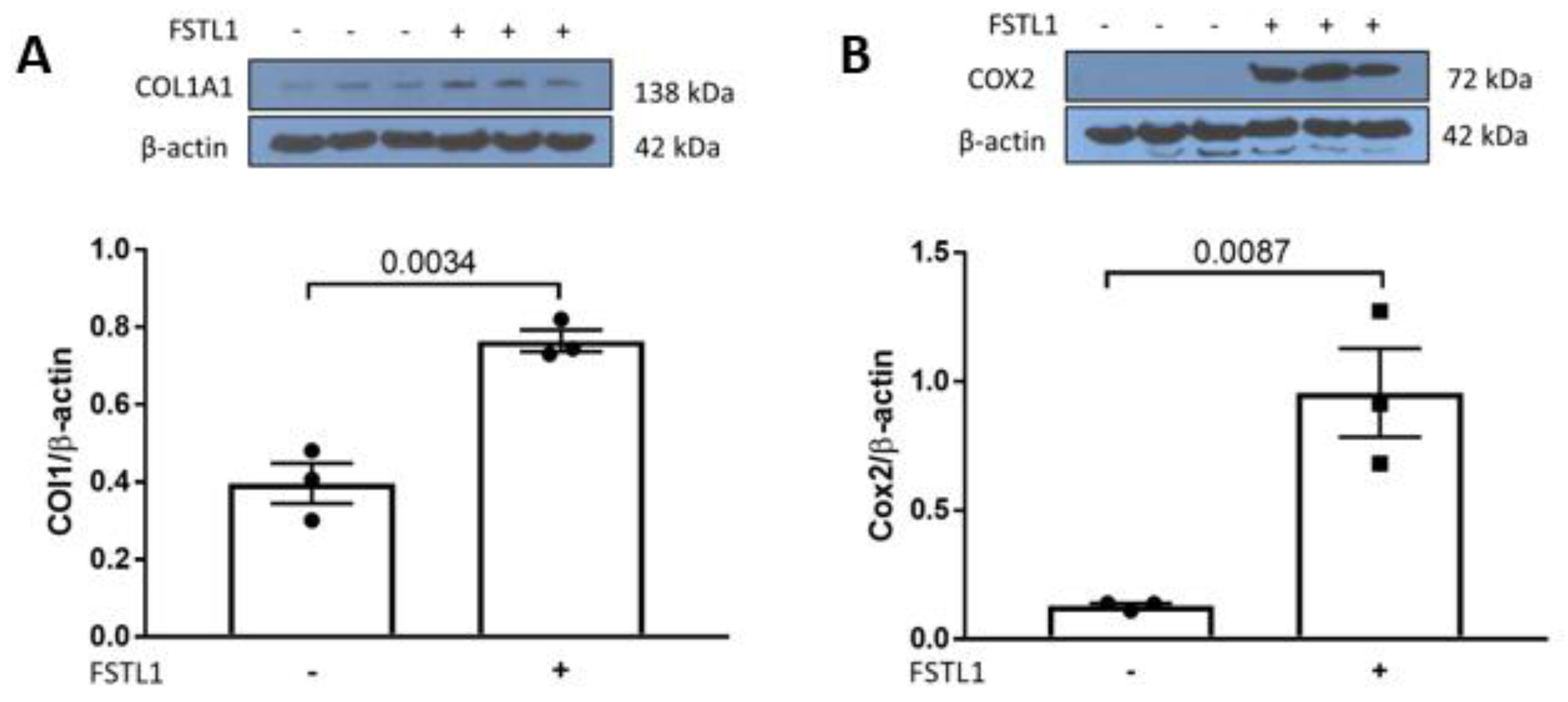
2.4. p42/44 MAPK and Activator Protein 1 (AP1) Signaling in Response to rhFSTL1
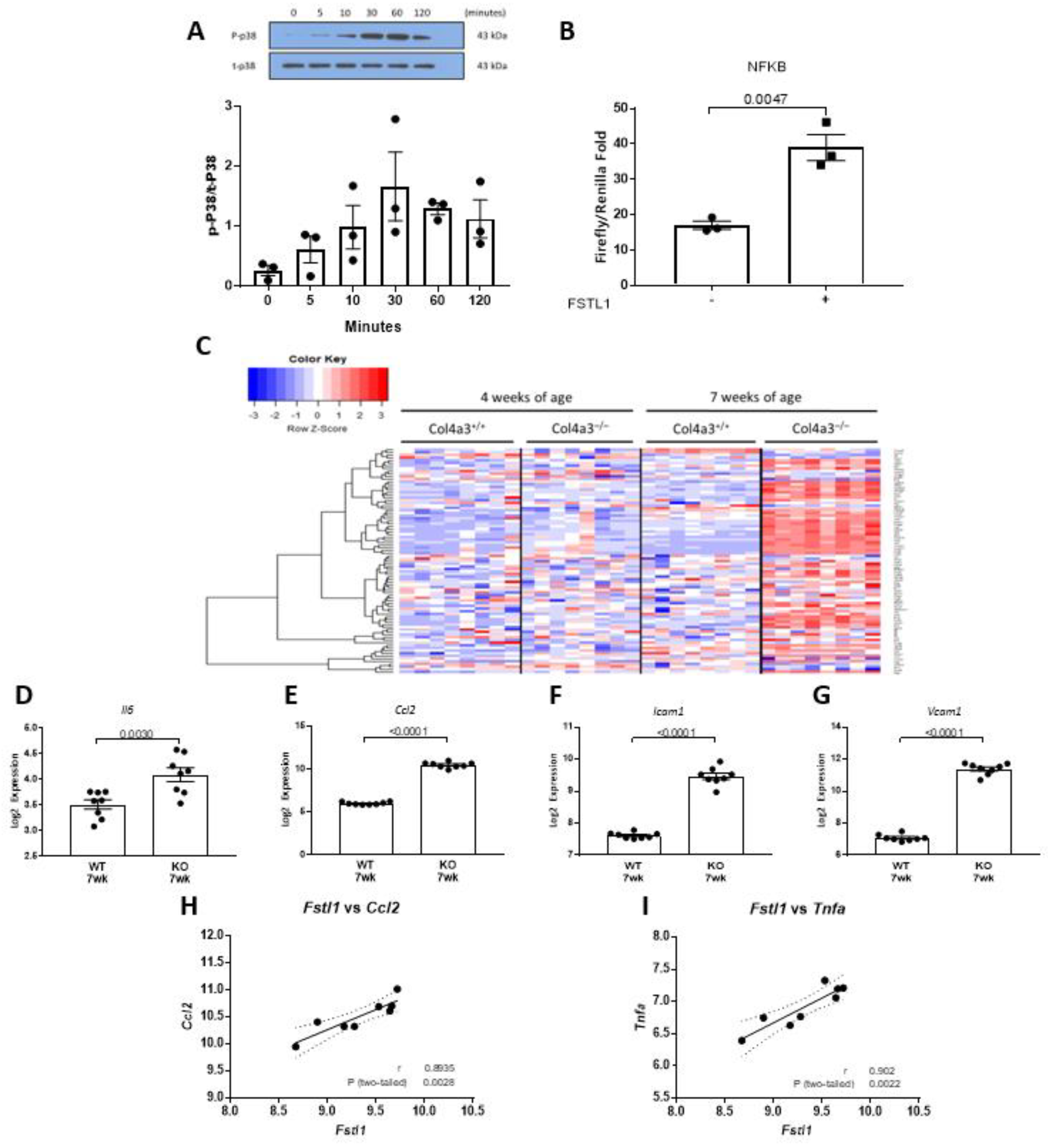
2.5. p38 MAPK and NFκB Signaling in Response to Fstl1
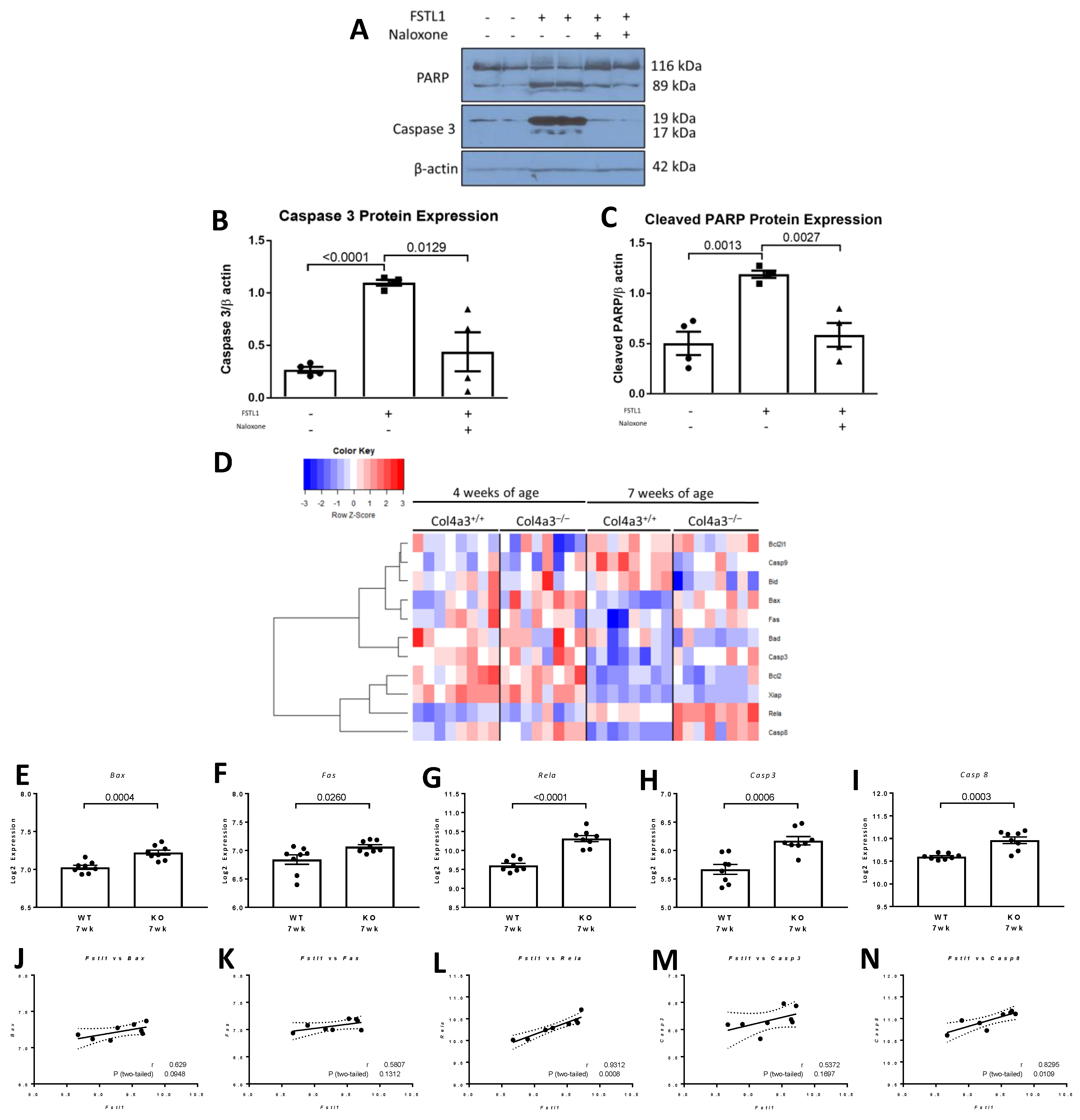
2.6. Apoptosis in Response to rhFSTL1
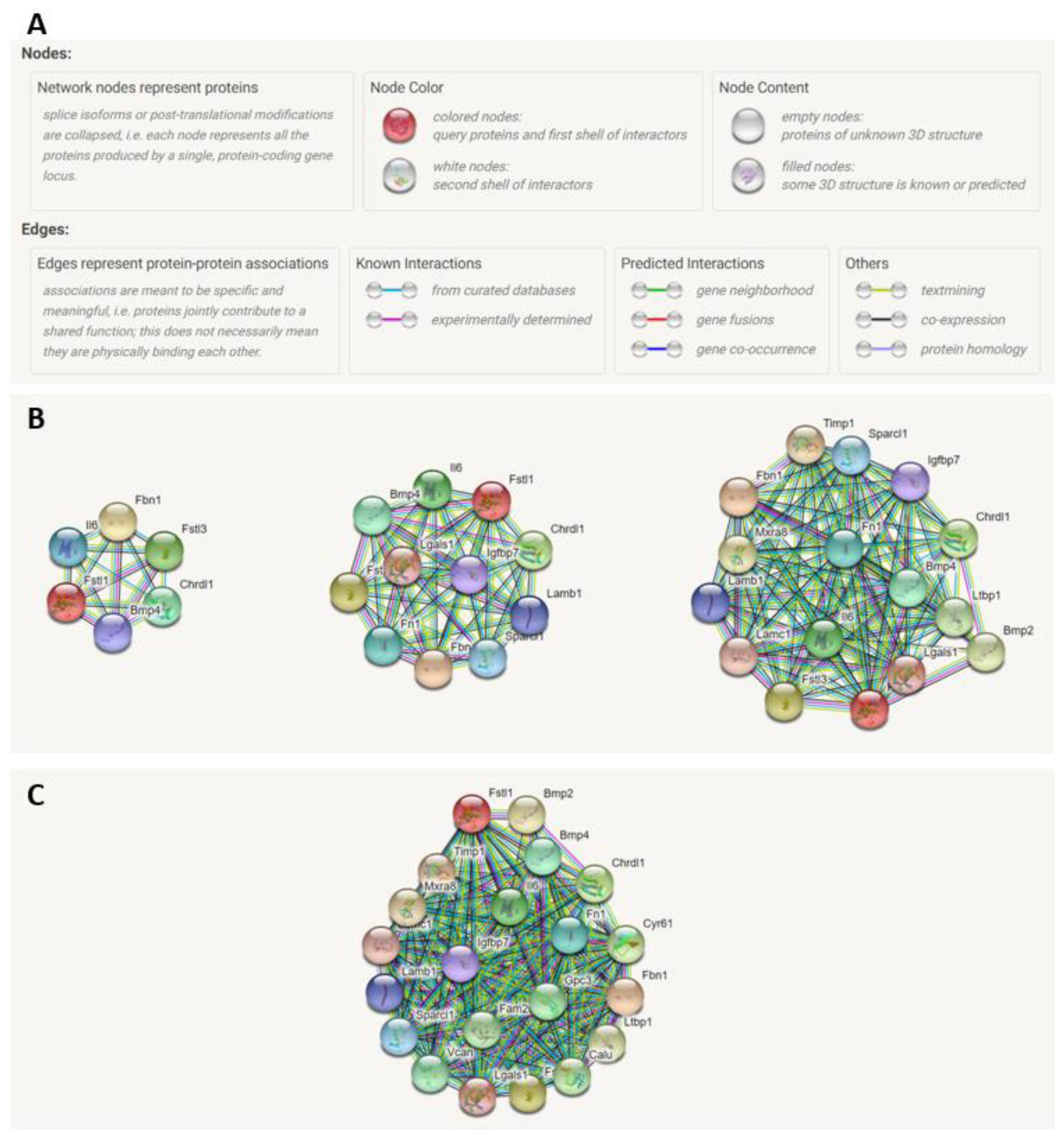
2.7. STRING Analysis of FSTL1 Protein–Protein Interactions
2.8. Studies of the Expression of Fstl1 and Its Cognate Receptors in Mice Subjected to Unilateral Ureteral Obstruction (UUO)
2.9. Studies of the Expression of Kidney Fstl1 Expression in the NEPTUNE Cohort
2.9.1. Patient Characteristics
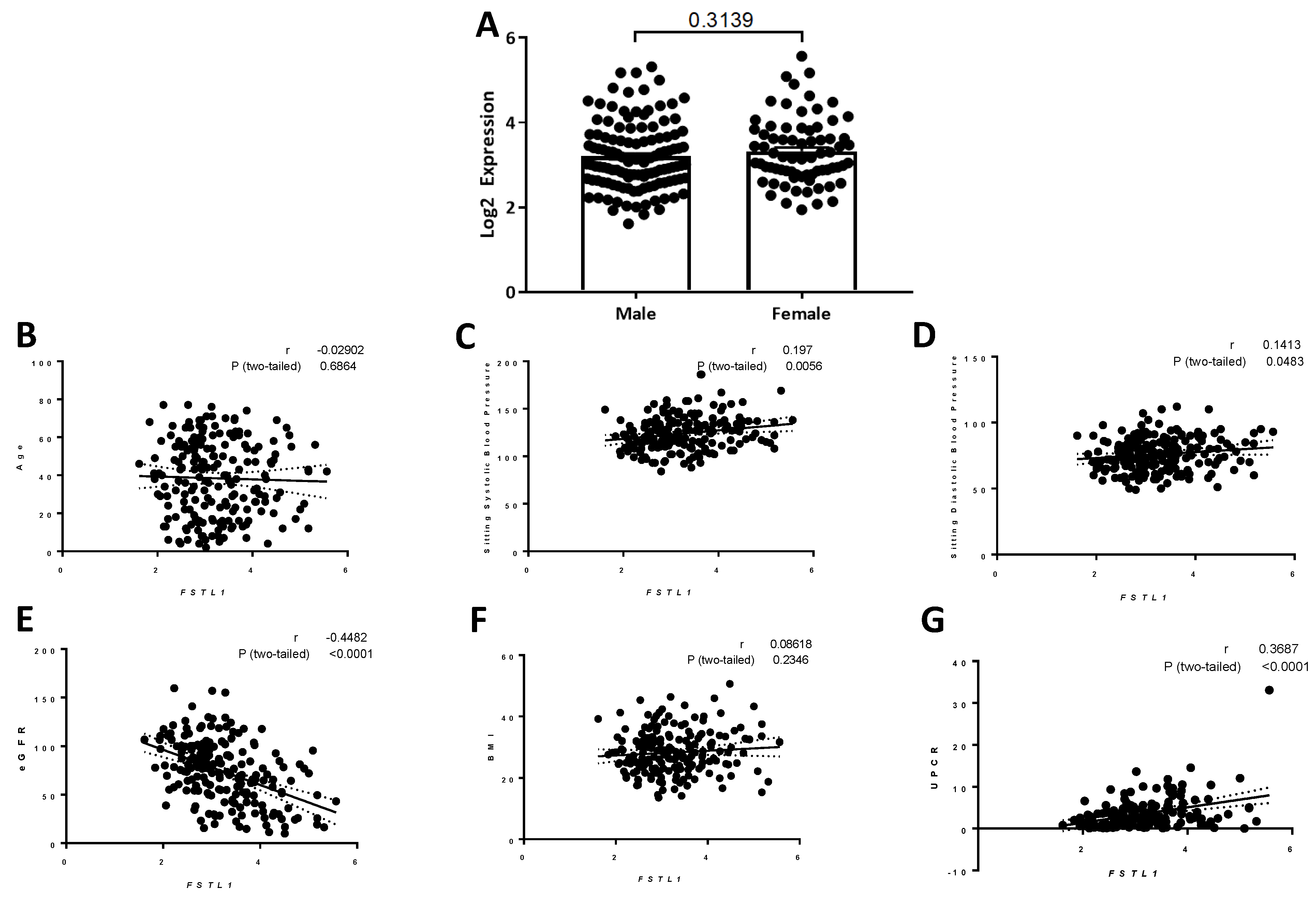
2.9.2. Correlation of FSTL1 mRNA Expression with Clinical Variables
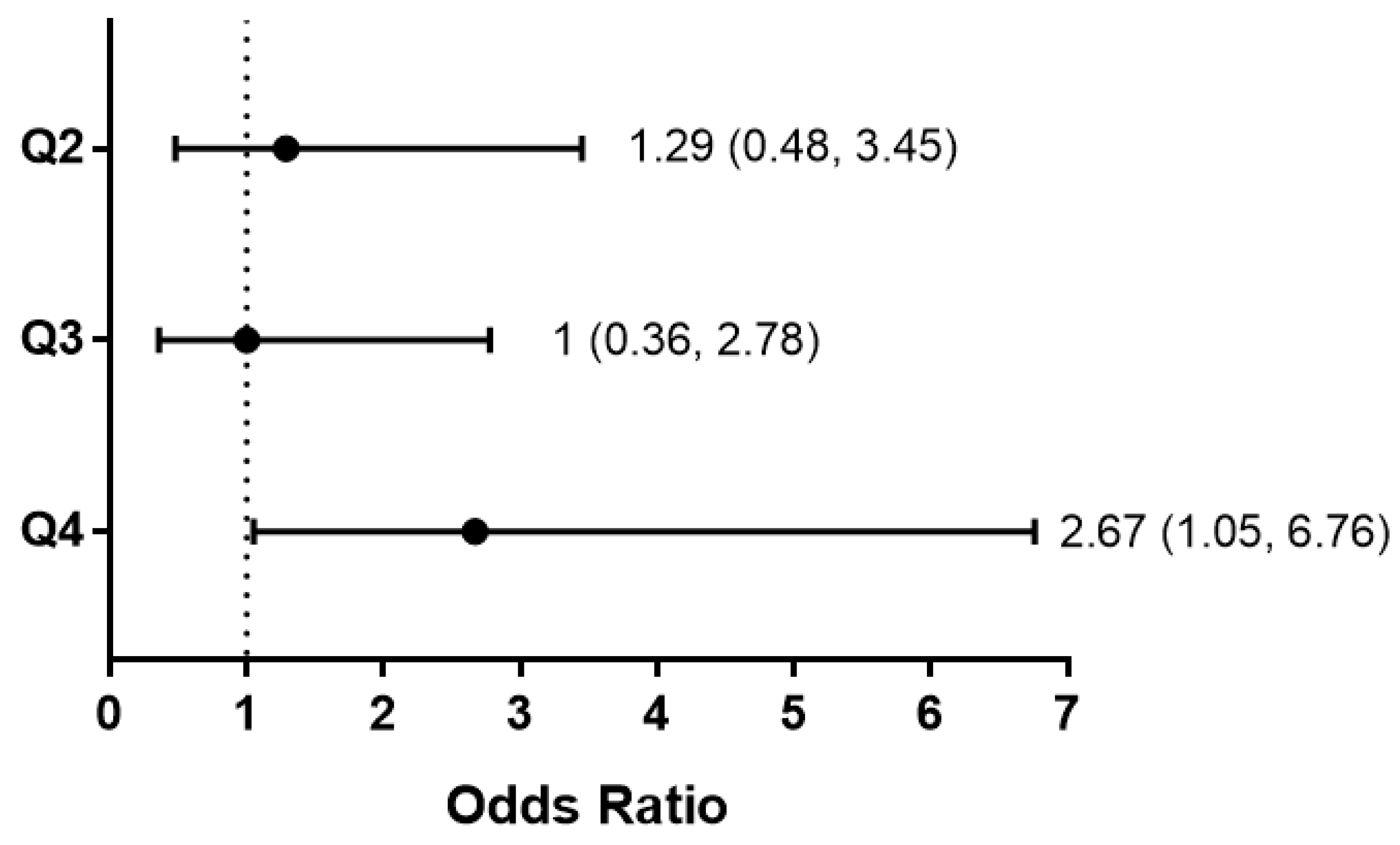
2.9.3. FSTL1 mRNA Levels and Kidney Outcomes
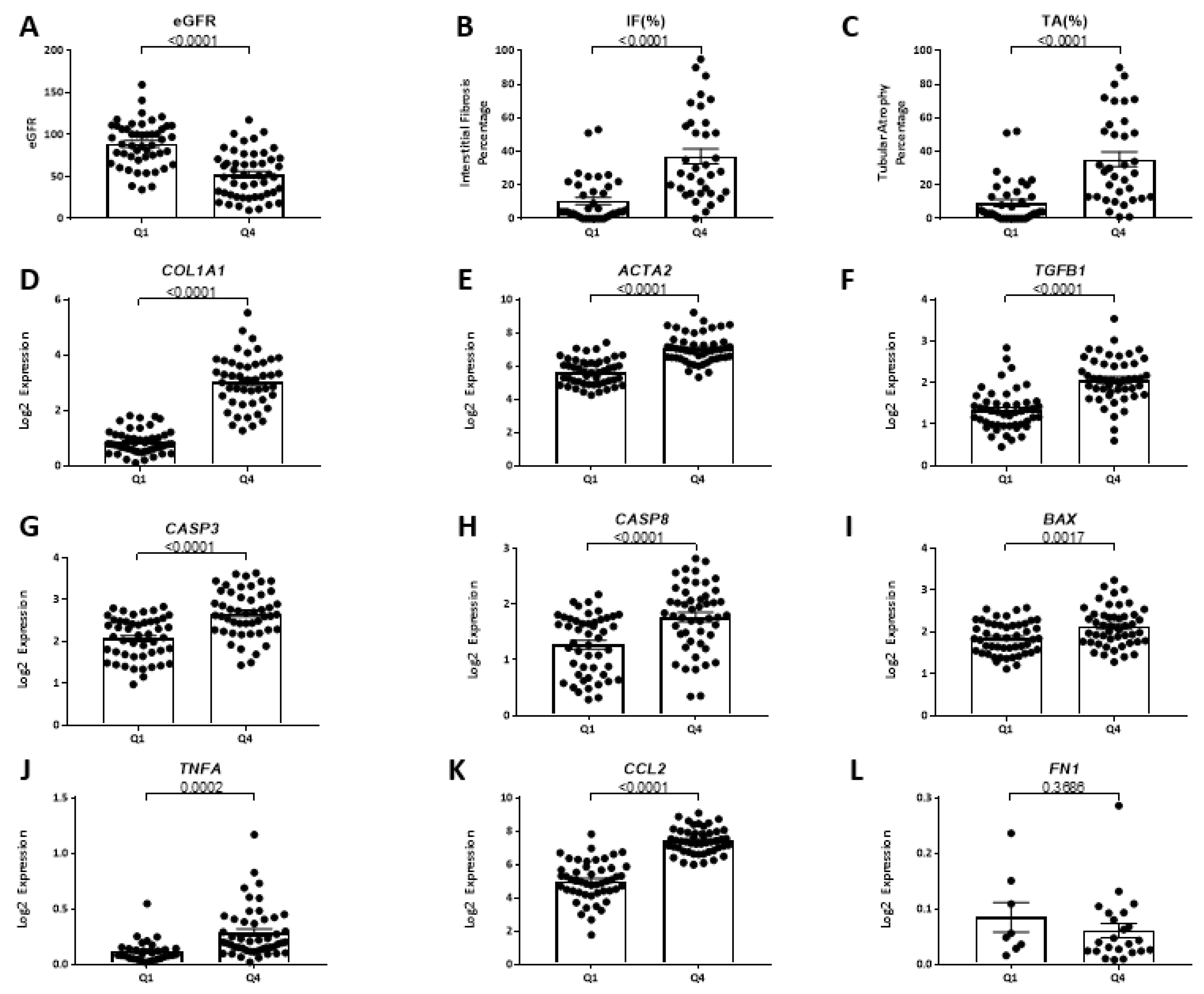
2.9.4. FSTL1 mRNA Levels and Kidney Structure and Function
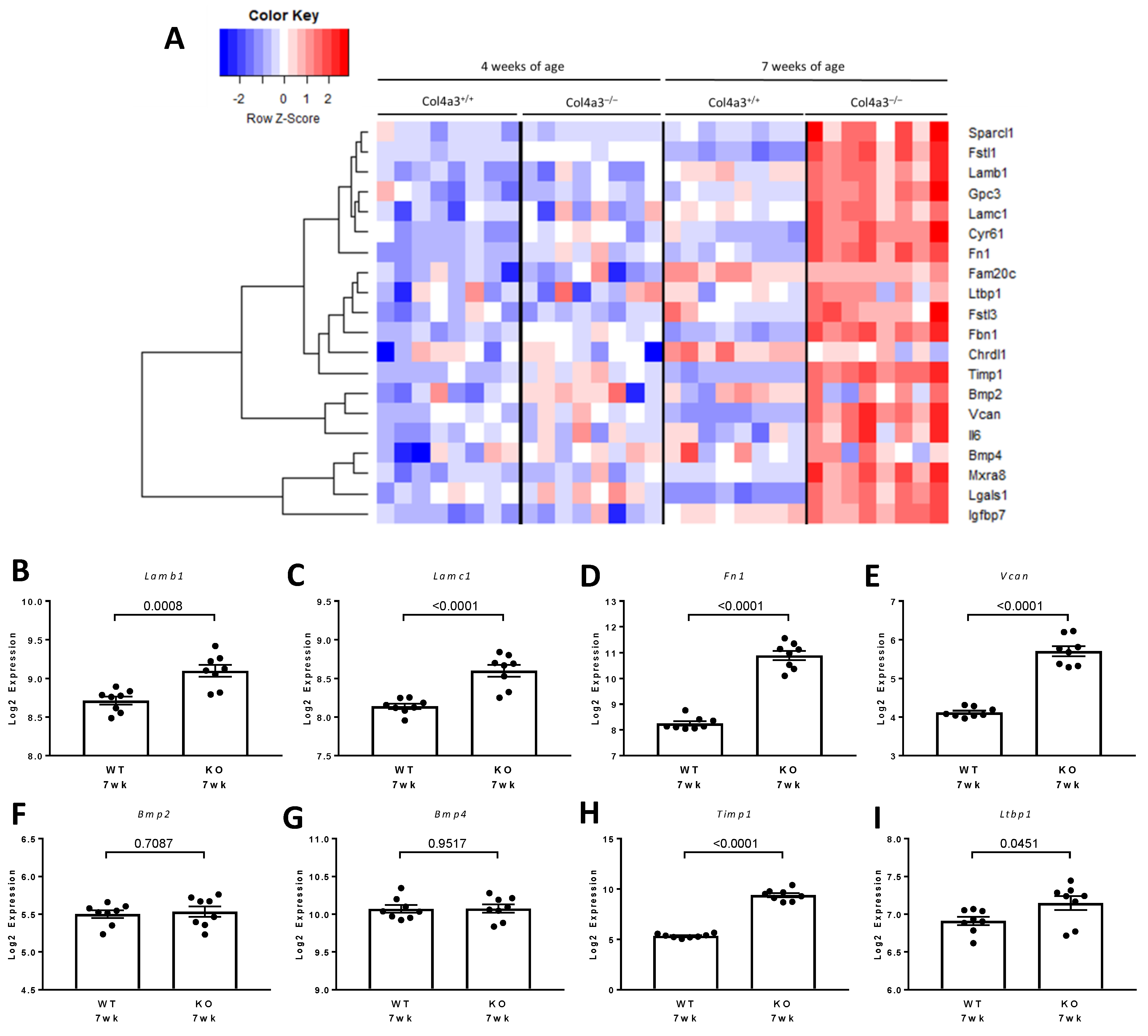
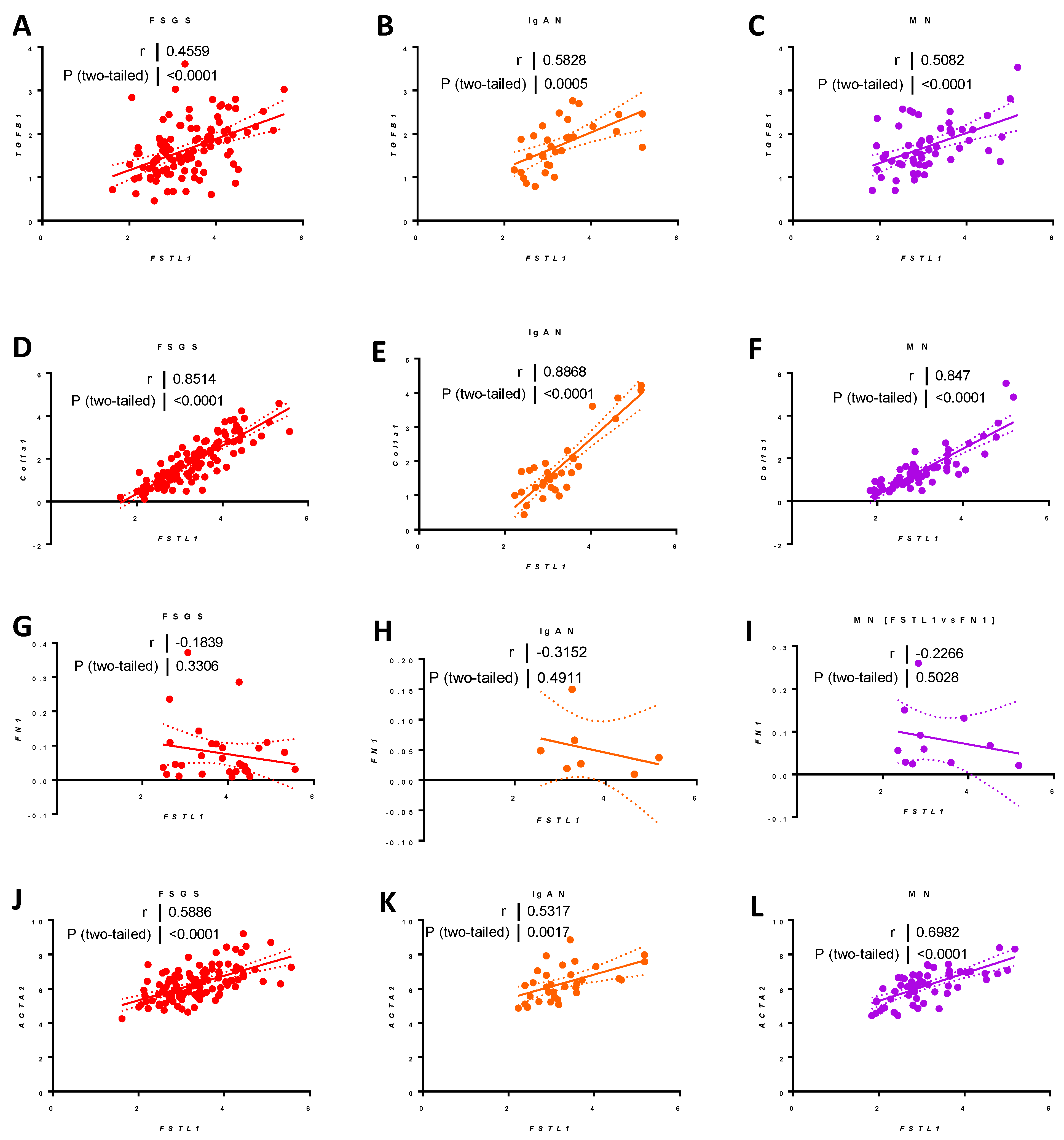
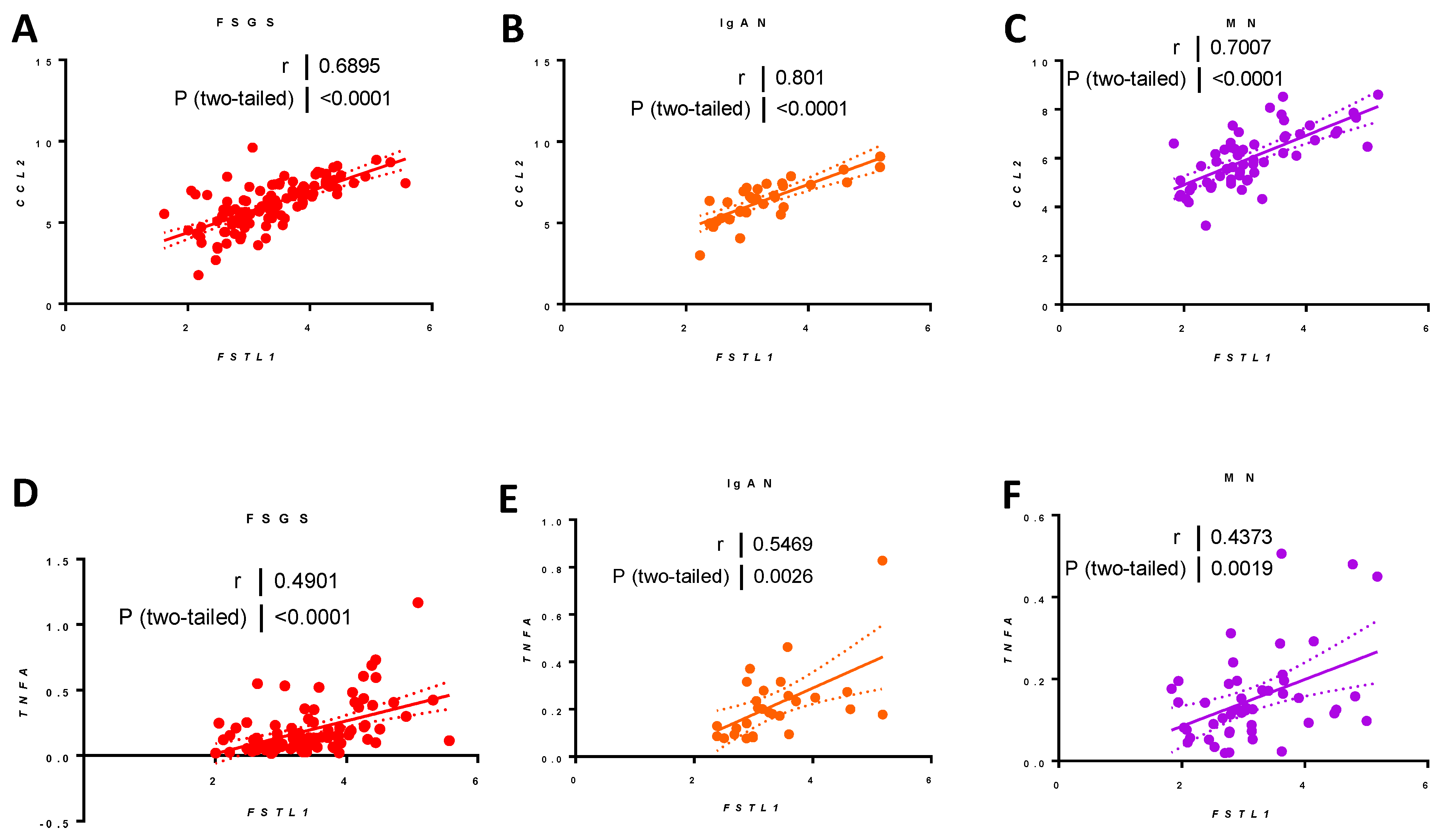
2.9.5. Associations between FSTL1 mRNA Levels and Genes Implicated in Fibrosis, Inflammation, and Apoptosis, in Each of the Three Cohorts
3. Discussion
4. Material and Methods
4.1. Animals
4.2. Cell Culture
4.3. Immunoblotting
4.4. RNAscope®
4.5. Luciferase
4.6. Quantitative PCR
4.7. Histological Staining
4.8. Heatmaps
4.9. Unilateral Ureteral Obstruction (UUO)
4.10. Data Collection and Study Cohort
4.11. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Saucedo, A.L.; Perales-Quintana, M.M.; Paniagua-Vega, D.; Sanchez-Martinez, C.; Cordero-Perez, P.; Minsky, N.W. Chronic Kidney Disease and the Search for New Biomarkers for Early Diagnosis. Curr. Med. Chem. 2018, 25, 3719–3747. [Google Scholar] [CrossRef]
- Prakoura, N.; Chatziantoniou, C. Periostin and Discoidin Domain Receptor 1: New Biomarkers or Targets for Therapy of Renal Disease. Front. Med. 2017, 4, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.-Z.; Sasaki, K.; Li, Y.; Li, Z.; Pan, Y.; Jin, G.-N.; Wang, Y.; Niu, A.; Wang, S.; Fan, X.; et al. The Role of the EGF Receptor in Sex Differences in Kidney Injury. J. Am. Soc. Nephrol. 2019, 30, 1659–1673. [Google Scholar] [CrossRef] [PubMed]
- Escobar, C.; Aranda, U.; Palacios, B.; Capel, M.; Sicras, A.; Sicras, A.; Hormigo, A.; Alcázar, R.; Manito, N.; Botana, M. Epidemiology, clinical profile, management, and two-year risk complications among patients with chronic kidney disease in Spain. Nefrologia 2021, in press. [Google Scholar] [CrossRef]
- Bae, E.H.; Fang, F.; Williams, V.R.; Konvalinka, A.; Zhou, X.; Patel, V.B.; Song, X.; John, R.; Oudit, G.Y.; Pei, Y.; et al. Murine recombinant angiotensin-converting enzyme 2 attenuates kidney injury in experimental Alport syndrome. Kidney Int. 2017, 91, 1347–1361. [Google Scholar] [CrossRef]
- Williams, V.R.; Konvalinka, A.; Song, X.; Zhou, X.; John, R.; Pei, Y.; Scholey, J.W. Connectivity mapping of a chronic kidney disease progression signature identified lysine deacetylases as novel therapeutic targets. Kidney Int. 2020, 98, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Bae, E.H.; Konvalinka, A.; Fang, F.; Zhou, X.; Williams, V.; Maksimowski, N.; Song, X.; Zhang, S.-L.; John, R.; Oudit, G.Y.; et al. Characterization of the intrarenal renin-angiotensin system in experimental alport syndrome. Am. J. Pathol. 2015, 185, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Y.; Yang, Z.; Ning, W. Cell type specific expression of Follistatin-like 1 (Fstl1) in mouse embryonic lung development. J. Mol. Histol. 2018, 49, 399–409. [Google Scholar] [CrossRef]
- Chen, Z.; Fang, Y.; Zhang, S.; Li, L.; Wang, L.; Zhang, A.; Yuan, Z.; Wang, P.; Zhou, H.; Cui, W.; et al. Haplodeletion of Follistatin-Like 1 Attenuates Radiation-Induced Pulmonary Fibrosis in Mice. Int. J. Radiat. Oncol. Biol. Phys. 2019, 103, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, J.; Zhang, W.; Peng, M.; Chen, J.; Zheng, L.; Zhang, X.; Yang, H.; Liu, Y. Follistatin-Like 1 Protects against Doxorubicin-Induced Cardiomyopathy through Upregulation of Nrf2. Oxid. Med. Cell. Longev. 2020, 2020, 3598715. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, S.; Ohashi, K.; Shibata, R.; Kataoka, Y.; Miyabe, M.; Enomoto, T.; Joki, Y.; Shimizu, Y.; Kambara, T.; Uemura, Y.; et al. Cardiac myocyte-derived follistatin-like 1 prevents renal injury in a subtotal nephrectomy model. J. Am. Soc. Nephrol. 2015, 26, 636–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattiotti, A.; Prakash, S.; Barnett, P.; van den Hoff, M.J.B. Follistatin-like 1 in development and human diseases. Cell. Mol. Life Sci. 2018, 75, 2339–2354. [Google Scholar] [CrossRef] [PubMed]
- Garces de Los Fayos Alonso, I.; Liang, H.-C.; Turner, S.D.; Lagger, S.; Merkel, O.; Kenner, L. The Role of Activator Protein-1 (AP-1) Family Members in CD30-Positive Lymphomas. Cancers 2018, 10, 93. [Google Scholar] [CrossRef] [Green Version]
- Kassiri, Z.; Oudit, G.Y.; Kandalam, V.; Awad, A.; Wang, X.; Ziou, X.; Maeda, N.; Herzenberg, A.M.; Scholey, J.W. Loss of TIMP3 enhances interstitial nephritis and fibrosis. J. Am. Soc. Nephrol. 2009, 20, 1223–1235. [Google Scholar] [CrossRef] [Green Version]
- Bascands, J.-L.; Schanstra, J.P. Obstructive nephropathy: Insights from genetically engineered animals. Kidney Int. 2005, 68, 925–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevalier, R.L.; Forbes, M.S.; Thornhill, B.A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 2009, 75, 1145–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tinti, F.; Lai, S.; Noce, A.; Rotondi, S.; Marrone, G.; Mazzaferro, S.; Di Daniele, N.; Mitterhofer, A.P. Chronic Kidney Disease as a Systemic Inflammatory Syndrome: Update on Mechanisms Involved and Potential Treatment. Life 2021, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Peng, L.; Tian, W.; Li, Y.; Zhang, P.; Sun, K.; Yang, Y.; Li, X.; Li, G.; Zhu, X. Loss of phosphatidylserine flippase β-subunit Tmem30a in podocytes leads to albuminuria and glomerulosclerosis. Dis. Models Mech. 2021, 14, dmm048777. [Google Scholar] [CrossRef]
- Ammar, S.; Kanoun, H.; Kammoun, K.; Domingo-Gallego, A.; Ruiz, P.; Lorente-Grandoso, L.; Pybus, M.; Maalej, B.; Boudawara, T.; Kamoun, H.; et al. Next-generation sequencing in patients with familial FSGS: First report of collagen gene mutations in Tunisian patients. J. Hum. Genet. 2021, 66, 795–803. [Google Scholar] [CrossRef]
- Salem, R.M.; Todd, J.N.; Sandholm, N.; Cole, J.B.; Chen, W.-M.; Andrews, D.; Pezzolesi, M.G.; McKeigue, P.M.; Hiraki, L.T.; Qiu, C.; et al. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen. J. Am. Soc. Nephrol. 2019, 30, 2000–2016. [Google Scholar] [CrossRef] [Green Version]
- Maksimowski, N.A.; Scholey, J.W.; Williams, V.R. Nephrotic Syndrome Study Network (NEPTUNE) Sex and kidney ACE2 expression in primary focal segmental glomerulosclerosis: A NEPTUNE study. PLoS ONE 2021, 16, e0252758. [Google Scholar] [CrossRef] [PubMed]
- Maksimowski, N.; Williams, V.R.; Scholey, J.W. Kidney ACE2 expression: Implications for chronic kidney disease. PLoS ONE 2020, 15, e0241534. [Google Scholar] [CrossRef]
- Madsen, B.S.; Thiele, M.; Detlefsen, S.; Sørensen, M.D.; Kjaergaard, M.; Møller, L.S.; Rasmussen, D.N.; Schlosser, A.; Holmskov, U.; Trebicka, J.; et al. Prediction of liver fibrosis severity in alcoholic liver disease by human microfibrillar-associated protein 4. Liver Int. 2020, 40, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-B.; Yang, J.; Shuai, W.; Yang, J.; Liu, L.-B.; Xu, M.; Tang, Q.-Z. Deletion of Microfibrillar-Associated Protein 4 Attenuates Left Ventricular Remodeling and Dysfunction in Heart Failure. J. Am. Heart Assoc. 2020, 9, e015307. [Google Scholar] [CrossRef]
- Pan, Z.; Yang, K.; Wang, H.; Xiao, Y.; Zhang, M.; Yu, X.; Xu, T.; Bai, T.; Zhu, H. MFAP4 deficiency alleviates renal fibrosis through inhibition of NF-κB and TGF-β/Smad signaling pathways. FASEB J. 2020, 34, 14250–14263. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Zhou, Z.; Ren, Y. Crucial genes associated with diabetic nephropathy explored by microarray analysis. BMC Nephrol. 2016, 17, 128. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Liang, W.; Li, J.; Long, J. Knockdown of FSTL1 inhibits oxLDL-induced inflammation responses through the TLR4/MyD88/NF-κB and MAPK pathway. Biochem. Biophys. Res. Commun. 2016, 478, 1528–1533. [Google Scholar] [CrossRef]
- Wang, B.; Li, J.; Tian, F. Downregulation of lncRNA SNHG14 attenuates osteoarthritis by inhibiting FSTL-1 mediated NLRP3 and TLR4/NF-κB pathway through miR-124-3p. Life Sci. 2021, 270, 119143. [Google Scholar] [CrossRef]
- Ni, S.; Li, C.; Xu, N.; Liu, X.; Wang, W.; Chen, W.; Wang, Y.; van Wijnen, A.J. Follistatin-like protein 1 induction of matrix metalloproteinase 1, 3 and 13 gene expression in rheumatoid arthritis synoviocytes requires MAPK, JAK/STAT3 and NF-κB pathways. J. Cell. Physiol. 2018, 234, 454–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, D.C.; Karolak, M.J.; Larman, B.W.; Liaw, L.; Nolin, J.D.; Oxburgh, L. Follistatin-like 1 regulates renal IL-1β expression in cisplatin nephrotoxicity. Am. J. Physiol. Renal Physiol. 2010, 299, F1320–F1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, D.; Ngov, C.; Henley, N.; Boufaied, N.; Gerarduzzi, C. Characterization of Matricellular Protein Expression Signatures in Mechanistically Diverse Mouse Models of Kidney Injury. Sci. Rep. 2019, 9, 16736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ramesh, G.; Uematsu, S.; Akira, S.; Reeves, W.B. TLR4 signaling mediates inflammation and tissue injury in nephrotoxicity. J. Am. Soc. Nephrol. 2008, 19, 923–932. [Google Scholar] [CrossRef]
- Morrissey, J.; Guo, G.; McCracken, R.; Tolley, T.; Klahr, S. Induction of CD14 in tubular epithelial cells during kidney disease. J. Am. Soc. Nephrol. 2000, 11, 1681–1690. [Google Scholar] [CrossRef]
- Zhang, L.; Mabwi, H.A.; Palange, N.J.; Jia, R.; Ma, J.; Bah, F.B.; Sah, R.K.; Li, D.; Wang, D.; Bah, F.B.M.; et al. Expression Patterns and Potential Biological Roles of Dip2a. PLoS ONE 2015, 10, e0143284. [Google Scholar] [CrossRef]
- Tan, Y.; Cao, H.; Li, Q.; Sun, J. The role of transcription factor Ap1 in the activation of the Nrf2/ARE pathway through TET1 in diabetic nephropathy. Cell Biol. Int. 2021. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Liu, J.; Lawrence, A.; Dykstra, M.J.; Fannin, R.; Gerrish, K.; Tucker, C.J.; Scappini, E.; Dixon, D. Prolonged cadmium exposure alters benign uterine fibroid cell behavior, extracellular matrix components, and TGFB signaling. FASEB J. 2021, 35, e21738. [Google Scholar] [CrossRef]
- Surmiak, M.; Kosałka-Węgiel, J.; Polański, S.; Sanak, M. Endothelial cells response to neutrophil-derived extracellular vesicles miRNAs in anti-PR3 positive vasculitis. Clin. Exp. Immunol. 2021, 204, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Niño, M.D.; Poveda, J.; Sanz, A.B.; Mezzano, S.; Carrasco, S.; Fernandez-Fernandez, B.; Burkly, L.C.; Nair, V.; Kretzler, M.; Hodgin, J.B.; et al. Fn14 in podocytes and proteinuric kidney disease. Biochim. Biophys. Acta 2013, 1832, 2232–2243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arab, H.H.; Ashour, A.M.; Alqarni, A.M.; Arafa, E.-S.A.; Kabel, A.M. Camel Milk Mitigates Cyclosporine-Induced Renal Damage in Rats: Targeting p38/ERK/JNK MAPKs, NF-κB, and Matrix Metalloproteinases. Biology 2021, 10, 442. [Google Scholar] [CrossRef]
- Wilkening, A.; Krappe, J.; Mühe, A.M.; Lindenmeyer, M.T.; Eltrich, N.; Luckow, B.; Vielhauer, V. C-C chemokine receptor type 2 mediates glomerular injury and interstitial fibrosis in focal segmental glomerulosclerosis. Nephrol. Dial. Transplant. 2020, 35, 227–239. [Google Scholar] [CrossRef]
- Huang, J.; Wan, D.; Li, J.; Chen, H.; Huang, K.; Zheng, L. Histone acetyltransferase PCAF regulates inflammatory molecules in the development of renal injury. Epigenetics 2015, 10, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Zhu, J.; Wang, Y.; Zhu, Z.; Chen, Y.; Lu, L.; Jin, W.; Yan, X.; Zhang, R. Renal recruitment of B lymphocytes exacerbates tubulointerstitial fibrosis by promoting monocyte mobilization and infiltration after unilateral ureteral obstruction. J. Pathol. 2017, 241, 80–90. [Google Scholar] [CrossRef]
- Guan, Y.-M.; Diao, Z.-L.; Huang, H.-D.; Zheng, J.-F.; Zhang, Q.-D.; Wang, L.-Y.; Liu, W.-H. Bioactive peptide apelin rescues acute kidney injury by protecting the function of renal tubular mitochondria. Amino Acids 2021, 53, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tan, X.; Liu, W.; Chen, X.; Hou, X.; Shen, D.; Ding, Y.; Yin, J.; Wang, L.; Zhang, H.; et al. Follistatin-like protein 1 plays a tumor suppressor role in clear-cell renal cell carcinoma. Chin. J. Cancer 2018, 37, 2. [Google Scholar] [CrossRef] [Green Version]
- Sundaram, G.M.; Ismail, H.M.; Bashir, M.; Muhuri, M.; Vaz, C.; Nama, S.; Ow, G.S.; Vladimirovna, I.A.; Ramalingam, R.; Burke, B.; et al. EGF hijacks miR-198/FSTL1 wound-healing switch and steers a two-pronged pathway toward metastasis. J. Exp. Med. 2017, 214, 2889–2900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundaram, G.M.; Common, J.E.A.; Gopal, F.E.; Srikanta, S.; Lakshman, K.; Lunny, D.P.; Lim, T.C.; Tanavde, V.; Lane, E.B.; Sampath, P. “See-saw” expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature 2013, 495, 103–106. [Google Scholar] [CrossRef]
- Von Mering, C. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2004, 33, D433–D437. [Google Scholar] [CrossRef]
- Hanai, S.; Uchimura, K.; Takahashi, K.; Ishii, T.; Mitsui, T.; Furuya, F. Hypoxia-induced thyroid hormone receptor expression regulates cell-cycle progression in renal tubule epithelial cells. Endocr. J. 2021. [Google Scholar] [CrossRef]
- Gaedeke, J.; Noble, N.A.; Border, W.A. Curcumin blocks multiple sites of the TGF-beta signaling cascade in renal cells. Kidney Int. 2004, 66, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Zhu, J.; Wang, R.; Chen, X.; Mi, L.; Walz, T.; Springer, T.A. Latent TGF-β structure and activation. Nature 2011, 474, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Dickson, L.E.; Wagner, M.C.; Sandoval, R.M.; Molitoris, B.A. The proximal tubule and albuminuria: Really! J. Am. Soc. Nephrol. 2014, 25, 443–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagami, S.; Border, W.A.; Miller, D.E.; Noble, N.A. Angiotensin II stimulates extracellular matrix protein synthesis through induction of transforming growth factor-beta expression in rat glomerular mesangial cells. J. Clin. Investig. 1994, 93, 2431–2437. [Google Scholar] [CrossRef] [Green Version]
- Le, Y.; Zheng, Z.; Xue, J.; Cheng, M.; Guan, M.; Xue, Y. Effects of exendin-4 on the intrarenal renin-angiotensin system and interstitial fibrosis in unilateral ureteral obstruction mice: Exendin-4 and unilateral ureteral obstruction. J. Renin-Angiotensin-Aldosterone Syst. 2016, 17. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Geng, Y.; Li, L.; Li, X.; Yan, X.; Fang, Y.; Li, X.; Dong, S.; Liu, X.; Li, X.; et al. Blocking follistatin-like 1 attenuates bleomycin-induced pulmonary fibrosis in mice. J. Exp. Med. 2015, 212, 235–252. [Google Scholar] [CrossRef]
- Barisoni, L.; Nast, C.C.; Jennette, J.C.; Hodgin, J.B.; Herzenberg, A.M.; Lemley, K.V.; Conway, C.M.; Kopp, J.B.; Kretzler, M.; Lienczewski, C.; et al. Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE). Clin. J. Am. Soc. Nephrol. 2013, 8, 1449–1459. [Google Scholar] [CrossRef] [Green Version]
- Barisoni, L.; Troost, J.P.; Nast, C.; Bagnasco, S.; Avila-Casado, C.; Hodgin, J.; Palmer, M.; Rosenberg, A.; Gasim, A.; Liensziewski, C.; et al. Reproducibility of the NEPTUNE descriptor-based scoring system on whole-slide images and histologic and ultrastructural digital images. Mod. Pathol. 2016, 29, 671–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariani, L.H.; Martini, S.; Barisoni, L.; Canetta, P.A.; Troost, J.P.; Hodgin, J.B.; Palmer, M.; Rosenberg, A.Z.; Lemley, K.V.; Chien, H.-P.; et al. Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol. Dial. Transplant. 2018, 33, 310–318. [Google Scholar] [CrossRef] [Green Version]







| 4 Weeks | 7 Weeks | |||
|---|---|---|---|---|
| WT (n = 8) | KO (n = 8) | WT (n = 8) | KO (n = 8) | |
| Body weight (g) | 16.63 ± 0.74 | 17.60 ± 0.46 | 21.17 ± 0.50 | 19.51 ± 0.93 |
| LKW(g)/BW (Kg) | 7.08 ± 0.17 | 7.61 ± 0.33 | 7.43 ± 0.18 | 8.82 ± 0.17 |
| RKW(g)/BW (Kg) | 7.08 ± 0.17 | 7.69 ± 0.31 | 7.69 ± 0.07 | 8.84 ± 0.21 |
| PCr (μMol/L) | 16.80 ± 1.32 | 16.00 ± 1.00 | 18.25 ± 1.65 | 35.80 ± 14.06 |
| UalbV (mg/dl) | 20.45 ± 2.25 | 65.66 ± 6.33 | 20.14 ± 1.45 | 144.31 ± 12.84 |
| ALL | FSGS | IgAN | MN | |
|---|---|---|---|---|
| Age | 38.54 ± 1.364 | 32.62 ± 1.957 | 36.08 ± 2.664 | 50.89 ± 1.792 |
| Sex (male/female) | (133/78) | (66/45) | (28/11) | (39/22) |
| BMI | 28.51 ± 0.4938 | 27.39 ± 0.7284 | 28.24 ± 0.9308 | 30.72 ± 0.8484 |
| Sitting Systolic | 123.6 ± 1.209 | 122.7 ± 1.558 | 123.1 ± 2.67 | 125.5 ± 2.575 |
| Sitting Diastolic | 76 ± 0.8592 | 74.96 ± 1.212 | 75.85 ± 1.936 | 77.97 ± 1.557 |
| Hematocrit % | 39.08 ± 0.389 | 38.83 ± 0.5635 | 38.54 ± 0.8987 | 39.88 ± 0.654 |
| eGFR | 74.06 ± 2.214 | 72.93 ± 3.247 | 67.46 ± 5.559 | 80.32 ± 3.255 |
| Centrally measured timed urine protein | 243.8 ± 23.2 | 199.3 ± 27.74 | 119.9 ± 18.73 | 385.6 ± 52.7 |
| Centrally measured timed urine creatinine | 69.01 ± 3.536 | 69.07 ± 5.286 | 62.35 ± 5.869 | 72.86 ± 6.701 |
| Centrally measured timed urine albumin | 1778 ± 169 | 1492 ± 208.3 | 927 ± 146.7 | 2737 ± 382 |
| Interstitial fibrosis (%) | 18.71 ± 1.658 | 20.98 ± 2.496 | 20.65 ± 3.155 | 12.38 ± 2.179 |
| Tubular atrophy (%) | 17.48 ± 1.654 | 19.44 ± 2.489 | 19.62 ± 3.175 | 11.71 ± 2.22 |
| Patient reached ESKD or 40% loss of eGFR (and eGFR<90) | 50 | 27 | 10 | 13 |
| Model: y = β0 + β1x1 + β2x2 + β3x3 + β4x4 + β5x5 + β6x6 | |||||
|---|---|---|---|---|---|
| Predictor | Coefficient | Estimate | Standard Error | t-Statistic | p-Value |
| FSTL1 | β0 | 4.0903 | 0.4391 | 9.3144 | <0.001 |
| Age | β1 | −0.0099 | 0.0029 | −3.4147 | 0.0008 |
| Sex | β2 | 0.0976 | 0.1078 | 0.9054 | 0.3667 |
| BPSS | β3 | 0.0035 | 0.0041 | 0.8616 | 0.3903 |
| BPSD | β4 | −0.0031 | 0.0057 | −0.5522 | 0.5817 |
| eGFR | β5 | −0.0134 | 0.0018 | −7.4402 | <0.001 |
| UPCR | β6 | 0.071 | 0.0131 | 5.4027 | <0.001 |
| Quartile Analysis | ||||
|---|---|---|---|---|
| Quartile | 1 | 2 | 3 | 4 |
| Percentage (%) | 18% | 22% | 18% | 38% |
| n | 9/49 | 11/49 | 9/49 | 18/48 |
| Range | 1.6–2.7 | 2.7–3.1 | 3.1–3.7 | 3.7–5.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maksimowski, N.A.; Song, X.; Bae, E.H.; Reich, H.; John, R.; Pei, Y.; Scholey, J.W.; Nephrotic Syndrome Study Network (NEPTUNE). Follistatin-Like-1 (FSTL1) Is a Fibroblast-Derived Growth Factor That Contributes to Progression of Chronic Kidney Disease. Int. J. Mol. Sci. 2021, 22, 9513. https://doi.org/10.3390/ijms22179513
Maksimowski NA, Song X, Bae EH, Reich H, John R, Pei Y, Scholey JW, Nephrotic Syndrome Study Network (NEPTUNE). Follistatin-Like-1 (FSTL1) Is a Fibroblast-Derived Growth Factor That Contributes to Progression of Chronic Kidney Disease. International Journal of Molecular Sciences. 2021; 22(17):9513. https://doi.org/10.3390/ijms22179513
Chicago/Turabian StyleMaksimowski, Nicholas A., Xuewen Song, Eun Hui Bae, Heather Reich, Rohan John, York Pei, James W. Scholey, and Nephrotic Syndrome Study Network (NEPTUNE). 2021. "Follistatin-Like-1 (FSTL1) Is a Fibroblast-Derived Growth Factor That Contributes to Progression of Chronic Kidney Disease" International Journal of Molecular Sciences 22, no. 17: 9513. https://doi.org/10.3390/ijms22179513
APA StyleMaksimowski, N. A., Song, X., Bae, E. H., Reich, H., John, R., Pei, Y., Scholey, J. W., & Nephrotic Syndrome Study Network (NEPTUNE). (2021). Follistatin-Like-1 (FSTL1) Is a Fibroblast-Derived Growth Factor That Contributes to Progression of Chronic Kidney Disease. International Journal of Molecular Sciences, 22(17), 9513. https://doi.org/10.3390/ijms22179513






