Novel Cold-Adapted Recombinant Laccase KbLcc1 from Kabatiella bupleuri G3 IBMiP as a Green Catalyst in Biotransformation
Abstract
1. Introduction
2. Results and Discussion
2.1. Expression of KbLcc1 Laccase from K. bupleuri G3 IBMiP
2.2. Purification of Recombinant Laccase
2.3. Characterization of Purified KbLcc1 Laccase
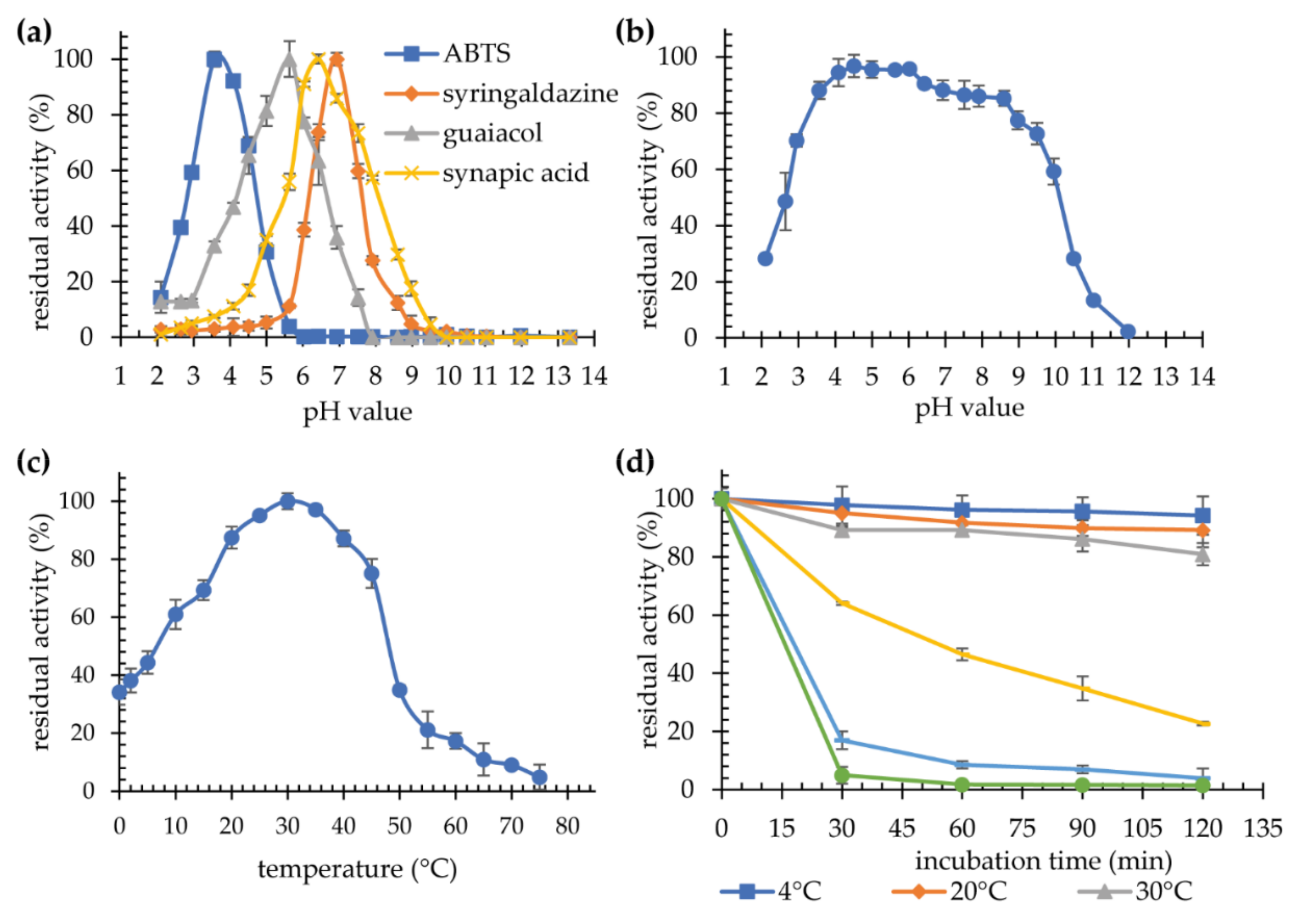
2.3.1. Influence of pH and Temperature on Purified Laccase KbLcc1 Activity and Stability
2.3.2. Substrate Specificity
2.3.3. Influence of Ions on Laccase KbLcc1 Activity
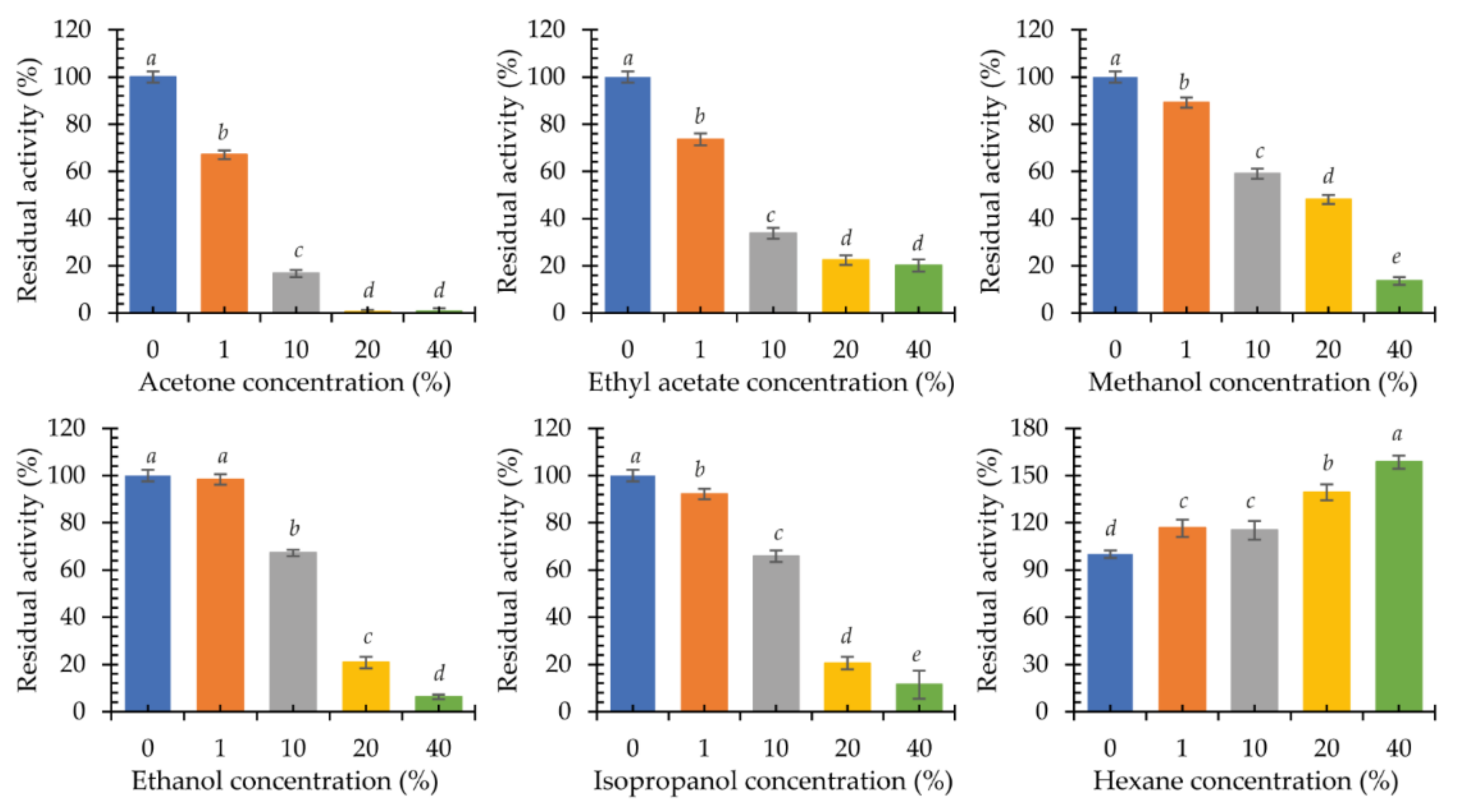
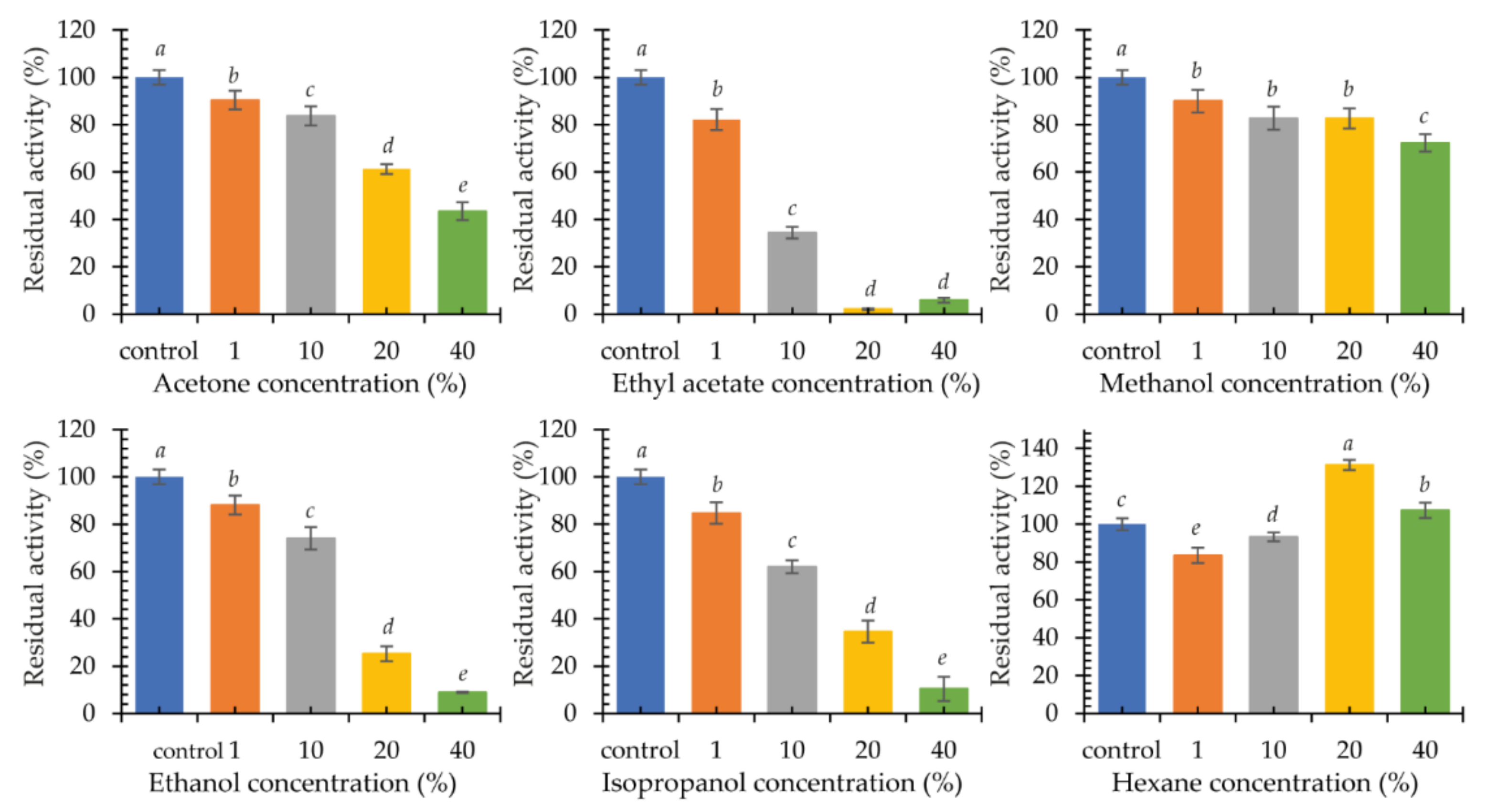
2.3.4. Influence of Organic Solvents on Laccase KbLcc1 Activity
2.4. Application of the Recombinant Laccase from K. bupleuri
2.4.1. Decolorization of Synthetic Dyes
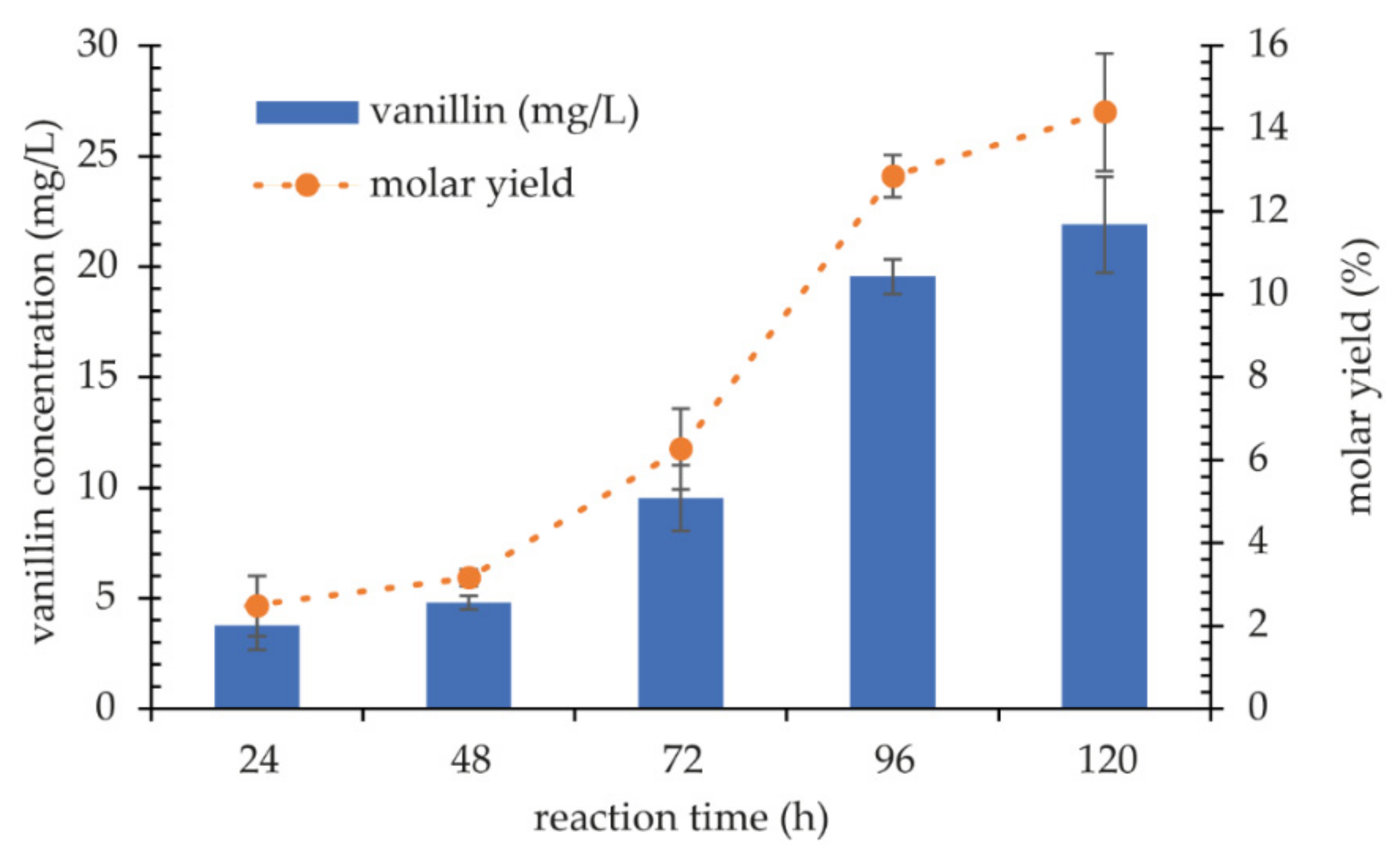
2.4.2. Biotransformation of Ferulic Acid into Vanillin
3. Materials and Methods
3.1. Strains, Plasmids and Media
3.2. Cloning of Laccase Gene from K. bupleuri G3 IBMiP
3.2.1. Genome Sequencing
3.2.2. Bioinformatics Analysis of Laccase Gene
3.2.3. KbLcc1 Gene Amplification and Cloning
3.2.4. Splicing In Vitro of KbLcc1 Gene
3.2.5. Cloning of KbLcc1 Coding Sequence to Expression Vector
3.2.6. Expression of the KbLcc1 Laccase in P. pastoris
3.2.7. Purification of Recombinant Laccase KbLcc1
3.3. Characterization of Purified Recombinant Laccase KbLcc1
3.3.1. Effect of Temperature and pH on Laccase Activity and Stability
3.3.2. Substrate Specificity of Recombinant Laccase
3.3.3. The Effects of Metal Cations and Chemicals on the Activity and Stability of the Laccase
3.4. Application of Laccase KbLcc1
3.4.1. Decolorization of Synthetic Dyes
3.4.2. Biotransformation of Ferulic Acid into Vanillin
3.5. Analytical Methods
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klonowska, A.; Gaudin, C.; Fournel, A.; Asso, M.; Le Petit, J.; Giorgi, M.; Tron, T. Characterization of low redox potential laccase from the basidiomycete C30. Eur. J. Biochem. 2002, 269, 6119–6125. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Hoyos, C.M.; Morales-Alvarez, E.D.; Poutou-Pinales, R.A.; Pedroza-Rodriguez, A.M.; Rodriguez-Vazquez, R.; Delgado-Boada, J.M. Fungal laccases. Fungal Biol. Rev. 2013, 37, 67–82. [Google Scholar] [CrossRef]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Pourmir, A.; Johannes, T.W. Directed evolution: Selection of the host organism. Comput. Struct. Biotechnol. J. 2012, 2, e201203012. [Google Scholar] [CrossRef] [PubMed]
- Piscitelli, A.; Pezzella, C.; Giardina, P.; Faraco, V.; Sannia, G. Heterologous laccase production and its role in industrial applications. Bioeng. Bugs 2010, 1, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Balakshin, M.; Chen, C.L.; Gratzl, J.S.; Kirkman, A.G.; Jacob, H. Biobleaching of pulp with dioxygen in laccase-mediator system—Effect of variables on the reaction kinetics. J. Mol. Catal. B Enzym. 2001, 16, 205–215. [Google Scholar] [CrossRef]
- Minussi, R.C.; Rossi, M.; Bologna, L.; Rotilio, D. Phenols removal in musts: Strategy for wine stabilization by laccase. J. Mol. Catal. B Enzym. 2007, 45, 102–107. [Google Scholar] [CrossRef]
- Wang, G.D.; Li, Q.J.; Luo, B.; Chen, X.Y. Ex planta phytoremediation of trichlorophenol and phenolic allelo chemicals via an engineered secretory laccase. Nat. Biotechnol. 2004, 22, 893–897. [Google Scholar] [CrossRef]
- Spina, F.; Cordero, C.; Schilliro, T.; Sgorbini, B.; Pignata, C.; Gilli, G.; Bicci, C.; Varese, G.C. Removal of micropollutants by fungal laccases in model solution and municipal wastewater: Evaluation of estrogenic activity and ecotoxicity. J. Clean Prod. 2015, 100, 185–194. [Google Scholar] [CrossRef]
- Watharkar, A.D.; Kadam, S.K.; Khandare, R.V.; Kolekar, P.D.; Jeon, B.H.; Jadhav, J.P.; Govindwar, S.P. Asparagus densiflorus in a vertical subsurface flow phytoreactor for treatment of real textile effluent: A lab to land approach for in situ soil remediation. Ecotoxicol. Environ. Saf. 2018, 161, 70–77. [Google Scholar] [CrossRef]
- Liu, H.; Cheng, Y.; Du, B.; Tong, C.; Liang, S.; Han, S.; Zheng, S.; Lin, Y. Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5 JP17-16 in Pichia pastoris and its application in synthetic dye decolorization. PLoS ONE 2015, 10, e0119833. [Google Scholar] [CrossRef]
- Huang, M.T.; Lu, Y.C.; Zhang, S.; Lu, F.; Yang, H. Rice (Oryza sativa) laccases involved in modification and detoxification of herbicides atrazine and izoproturon residues in plants. J. Agric. Food Chem. 2016, 64, 6397–6406. [Google Scholar] [CrossRef]
- Zheng, J.; Zhu, Q.; Wu, Y.; Lin, X. Oxidation of polycyclic aromatic hydrocarbons using Bacillus CotA with high laccase activity and copper independence. Chemosphere 2016, 148, 1–7. [Google Scholar] [CrossRef]
- Jahangiri, E.; Thomas, I.; Schulze, A.; Seiwert, B.; Cabana, H.; Schlosser, D. Characterization of electron beam irradiation-immobilised laccase for application in wastewater treatment. Sci. Total Environ. 2018, 624, 309–322. [Google Scholar] [CrossRef]
- Fabbrini, M.; Galli, C.; Gentili, P.; Macchitella, D. An oxidation of alcohols by oxygen with the enzyme laccase and mediation by TEMPO. Tetrahedron Lett. 2001, 42, 7551–7553. [Google Scholar] [CrossRef]
- Nicotra, S.; Cramaross, M.R.; Mucci, A.; Pagnoni, U.M.; Riva, S.; Forti, L. Biotransformation of resveratrol: Synthesis of trans-dehydrodimers catalyzed by laccases from Myceliophtora thermophyla and from Trametes pubescens. Tetrahedron 2004, 60, 595–600. [Google Scholar] [CrossRef]
- Kurisawa, M.; Chung, J.E.; Uyama, H.; Kobayashi, S. Laccase-catalyzed synthesis and antioxidant property of poly(catechin). Macromol. Biosci. 2003, 3, 758–764. [Google Scholar] [CrossRef]
- Kurisawa, M.; Chung, J.E.; Uyama, H.; Kobayashi, S. Oxidative coupling of epigallocatechin gallate amplifies antioxidant activity and inhibits xanthine oxidase activity. Chem. Commun. 2004, 3, 294–295. [Google Scholar] [CrossRef]
- Anderson, J.S. The chemistry of hair colorants. J. Soc. Dyers Colour. 2000, 116, 193–196. [Google Scholar]
- Kim, S.; Lopez, C.; Guebitz, G.; Cavaco-Paulo, A. Biological coloration of flax fabrics with flavonoids using laccase from Trametes hirsute. Eng. Life Sci. 2008, 8, 324–330. [Google Scholar] [CrossRef]
- Mustafa, R.; Muniglia, L.; Rovel, B.; Girardin, M. Phenolic colorants obtained by enzymatic synthesis using a fungal laccase in a hydro-organic biphasic system. Food Res. Int. 2015, 38, 995–1000. [Google Scholar] [CrossRef]
- Gallage, N.J.; Møller, B.L. Vanilla: The most popular flavour. In Biotechnology of Natural Products; Schwab, W., Lange, B., Wüst, M., Eds.; Springer: Cham, Switzerland, 2018; pp. 3–23. [Google Scholar] [CrossRef]
- Chattopadhyay, P.; Banerjee, G.; Sen, S.K. Cleaner production of vanillin through biotransformation of ferulic acid esters from agro residue by Streptomyces sannanensis. J. Clean Prod. 2018, 182, 272–279. [Google Scholar] [CrossRef]
- Tang, P.L.; Hassan, O. Bioconversion of ferulic acid attained from pineapple peels and pineapple crown leaves into vanilic acid and vanillin by Aspergillus niger I-1472. BMC Chem. 2020, 14, 7. [Google Scholar] [CrossRef]
- Chen, P.; Yan, L.; Zhang, S.; Wu, Z.; Li, S.; Yan, X.; Wang, N.; Liang, N.; Li, H. Optimizing bioconversion of ferulic acid to vanillin by Bacillus subtilis in the stirred packed reactor using Box-Behnken design and desirability function. Food Sci. Biotechnol. 2017, 26, 143–152. [Google Scholar] [CrossRef]
- Perez-Rodriguez, N.; de Souza Oliveira, R.P.; Agrasar, A.M.; Dominguez, J.M. Ferulic acid transformation into the main vanilla aroma compounds by Amycolatopsis sp. ATCC 39116. Appl. Microbiol. Biotechnol. 2016, 100, 1677–1689. [Google Scholar] [CrossRef]
- Falconnier, B.; Lapierre, C.; Lesage-Meessen, L.; Yonnet, G.; Brunerie, P.; Colonna-Ceccaldi, B.; Corrieu, G.; Asther, M. Vanillin as a product of ferulic acid biotransformation by the white-rot fungus Pycnoporus cinnabarinus I-937: Identification of metabolic pathways. J. Biotechnol. 1994, 37, 123–132. [Google Scholar] [CrossRef]
- Furuya, T.; Miura, M.; Kuroiwa, M.; Kino, K. High-yield production of vanillin from ferulic acid by a coenzyme-independent decarboxylase/oxygenase two-stage process. N. Biotechnol. 2015, 32, 335–339. [Google Scholar] [CrossRef]
- Wiśniewska, K.M.; Twarda-Clapa, A.; Białkowska, A.M. Screening of novel laccase producers—Isolation and characterization of cold-adapted laccase from Kabatiella bupleuri G3 capable of synthetic dye decolorization. Biomolecules 2021, 11, 828. [Google Scholar] [CrossRef]
- Heckman, K.L.; Pease, L.R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007, 2, 924–932. [Google Scholar] [CrossRef]
- Viswanath, B.; Rajesh, B.; Janardhan, A.; Kumar, A.P.; Narasimha, G. Fungal laccases and their application in bioremediation. Enzyme Res. 2014, 2014, 163242. [Google Scholar] [CrossRef]
- Pinar, O.; Tamerler, C.; Yazgan Karatas, A. Heterologus expression and characterization of a high redox potential laccase from Coriolopsis polyzona MUCL 38443. Turk. J. Biol. 2017, 41, 278–291. [Google Scholar] [CrossRef]
- Bao, S.; Teng, Z.; Ding, S. Heterologous expression and characterization of a novel laccase isoenzyme with dyes decolorization potential from Coprinus comatus. Mol. Biol. Rep. 2013, 40, 1927–1936. [Google Scholar] [CrossRef]
- Li, Q.; Pei, J.; Zhao, L.; Xie, J.; Cao, F.; Wang, G. Overexpression and characterization of laccase from Trametes versicolor in Pichia pastoris. Appl. Biochem. Microbiol. 2014, 50, 140–147. [Google Scholar] [CrossRef]
- Wang, B.; Yan, Y.; Tian, Y.; Zhao, W.; Li, Z.; Gao, J.; Peng, R.; Yao, Q. Heterologous expression and characterization of a laccase from Colletotrichum lagenarium and decolourisation of different synthetic dyes. World J. Microbiol. Biotechnol. 2016, 32, 40. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gomez-Gil, X.; Gutierrez-Soto, G.; Hernandez-Luna, C.E.; de los Santos, M.H.; Levin, L.; Rojo-Dominguez, A.; Romero-Martinez, D.; Saparrat, M.C.; et al. Laccase: Structure, function, and potential application in water bioremediation. Microb. Cell Factories 2019, 18, 200. [Google Scholar] [CrossRef]
- Atalla, M.M.; Zeinab, H.K.; Eman, R.H.; Amani, A.Y.; Abeer, A.E. Characterization and kinetic properties of the purified Trematosphaeria mangrovei laccase enzyme. Saudi J. Biol. Sci. 2013, 20, 373–381. [Google Scholar] [CrossRef]
- Maestre-Reyna, M.; Liu, W.C.; Jeng, W.Y.; Lee, C.C.; Hsu, C.A.; Wen, T.N.; Wang, A.H.; Shyur, L.F. Structural and functional roles of glycosylation in fungal laccase from Lentinus sp. PLoS ONE 2015, 10, e0120601. [Google Scholar] [CrossRef]
- Rich, J.O.; Leathers, T.D.; Anderson, A.M.; Bischoff, K.M.; Manitchotpisit, P. Laccases from Aureobasidium pullulans. Enzyme Microb. Technol. 2013, 53, 33–37. [Google Scholar] [CrossRef]
- Aung, T.; Jiang, H.; Chen, C.C.; Liu, G.L.; Hu, Z.; Chi, Z.M.; Chi, Z. Production, gene cloning and overexpression of a laccase in the marine-derived yeast Aureobasidium melanogenum strain 11-1 and characterization of the recombinant laccase. Mar. Biotechnol. 2019, 21, 76–87. [Google Scholar] [CrossRef]
- Rich, J.O.; Manitchotpisit, P.; Peterson, S.W.; Leathers, T.D. Laccase production by diverse phylogenetic clades of Aureobasidium pullulans. RJAS 2011, 1, 41–47. [Google Scholar]
- Mtibaà, R.; Barriuso, J.; de Eugenio, L.; Aranda, E.; Belbahri, L.; Nasri, M.; Martínez, M.J.; Mechichi, T. Purification and characterization of a fungal laccase from the ascomycete Thielavia sp. and its role in the decolorization of a recalcitrant dye. Int. J. Biol. Macromol. 2018, 120, 1744–1751. [Google Scholar] [CrossRef]
- Kumari, A.; Kishor, N.; Guptasarma, P. Characterization of a mildly alkalophilic and thermostable recombinant Thermus thermiphilus laccase with applications in decolourization of dyes. Biotechnol. Lett. 2018, 40, 285–295. [Google Scholar] [CrossRef]
- Baldrian, P. Fungal laccases—Occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef]
- Kiiskinen, L.L.; Viikari, L.; Kruus, K. Purification and characterization of a novel laccase from the ascomycete Melanocarpus albomyces. Appl. Microbiol. Biotechnol. 2002, 59, 198–204. [Google Scholar] [CrossRef]
- Chen, S.; Ge, W.; Buswell, J.A. Biochemical and molecular characterization of a laccase from the edible straw mushroom, Volvariella volvacea. Eur. J. Biochem. 2004, 271, 318–328. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, Y.; Liao, X.; Cai, Y. Purification and characterization of a new laccase from Shiraia sp. SUPER-H168. Process Biochem. 2013, 48, 351–357. [Google Scholar] [CrossRef]
- Singhal, A.; Choudhary, G.; Thakur, I.S. Characterization of laccase activity produced by Cryptococcus albidus. Prep. Biochem. Biotechnol. 2012, 42, 113–124. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, M.; Zhang, M.; Wang, C.; Liu, Y.; Fan, X.; Li, H. Characterization of a novel, cold-adapted and thermostable laccase-like enzyme with high tolerance for organic solvents and salt and potent dye decolorization ability, derived from a marine metagenomic library. Front. Microbiol. 2018, 9, 2998. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Mohd Omar, S.; Siddiqui, K.S.; Williams, T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef]
- You, L.F.; Liu, Z.M.; Lin, J.F.; Guo, L.Q.; Huang, X.L.; Yang, H.X. Molecular cloning of a laccase gene from Ganoderma lucidum and heterologous expression in Pichia pastoris. J. Basic Microbiol. 2014, 54, 134–141. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Xiao, T.; Fan, H.; Wei, D. Molecular characterization of a novel thermostable laccase PLLCC2 from the brown rot fungus Postia placenta MAD-698-R. Electron. J. Biotechnol. 2015, 18, 451–458. [Google Scholar] [CrossRef]
- Mandic, M.; Djokic, L.; Nikolaivits, E.; Prodanovic, R.; O’Connor, K.; Jeremic, S.; Topakas, E.; Nikodinovic-Runic, J. Identification and characterization of new laccase biocatalysts from Pseudomonas species suitable for degradation of synthetic textile dyes. Catalysts 2019, 9, 699. [Google Scholar] [CrossRef]
- Kumar, V.P.; Naik, C.; Sridhar, M. Production, purification and characterization of novel laccase produced by Schizophyllum commune NI-07 with potential for delignification of crop residues. Appl. Biochem. Microbiol. 2015, 51, 432–441. [Google Scholar] [CrossRef]
- Younes, S.B.; Sayadi, S. Purification and characterization of a novel trimeric and thermotolerant laccase produced from the ascomycete Scytalidium thermophilum strain. J. Mol. Catal. B Enzym. 2011, 73, 35–42. [Google Scholar] [CrossRef]
- Sondhi, S.; Kaur, R.; Madan, J. Purification and characterization of a novel white highly thermo stable laccase from a novel Bacillus sp. MSK-01 having potential to be used as anticancer agent. Int. J. Biol. Macromol. 2021, 170, 232–238. [Google Scholar] [CrossRef]
- Ramirez-Cavazos, L.I.; Junghanns, C.; Ornelas-Soto, N.; Cardenas-Chaves, D.L.; Hernandez-Luna, C.; Demarche, P.; Enaud, E.; Garcia-Morales, R.; Agathos, S.N.; Parra, R. Purification and characterization of two thermostable laccases from Pycnoporus sanguineus and potential role in degradation of endocrine disrupting chemicals. J. Mol. Catal. B Enzym. 2014, 108, 32–42. [Google Scholar] [CrossRef]
- Chauhan, A.K.; Choudhury, B. Suitability of organic solvent and cholinium based ionic liquid activated novel lignolytic enzymes of H. aswanensis for enhanced Kalson lignin degradation. Int. J. Biol. Macromol. 2020, 165, 107–117. [Google Scholar] [CrossRef]
- Ezike, T.C.; Ezugwu, A.L.; Udeh, J.O.; Eze, S.O.; Chilaka, F.C. Purification and characterisation of new laccase from Trametes polyzona WRF03. Biotechnol. Rep. 2020, 28, e00566. [Google Scholar] [CrossRef]
- Jafari, M.; Mojtabavi, S.; Faramarzi, M.A.; Mehrnejad, F.; Soleimani, M.; Mirjani, R. Molecular level insight into stability, activity and structure of laccase in aqueous ionic liquid and organic solvents: An experimental and computational research. J. Mol. Liq. 2020, 317, 113925. [Google Scholar] [CrossRef]
- Benghazi, L.; Record, E.; Suarez, A.; Gomez-Vidal, J.A.; Martinez, J.; de la Rubia, T. Production of the Phanerochaete flavido-alba laccase in Aspergillus niger for synthetic dyes decolorization and biotransformation. World J. Microbiol. Biotechnol. 2014, 30, 201–211. [Google Scholar] [CrossRef]
- Lu, L.; Zhang, B.B.; Yu, S.Y.; Bian, X.J.; Wang, W.; Wang, Y. Purification and characterization of laccase from Pycnoporus sanguineus and decolorization of an anthraquinone dye by the enzyme. Appl. Microbiol. Biotechnol. 2007, 74, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhao, M.; Liang, S.C.; Zhao, L.Y.; Li, D.B.; Zhang, B.B. Production and synthetic dyes decolourization capacity of a recombinant laccase from Pichia pastoris. J. Appl. Microbiol. 2009, 107, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Priefert, H.; Rabenhorst, J.; Steinbuchel, A. Biotechnological production of vanillin. Appl. Microbiol. Biotechnol. 2001, 56, 296–314. [Google Scholar] [CrossRef]
- Wellington, K.W. Application of laccases in organic synthesis: A review. In Green Chemistry; Luque, R., Ed.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2011; pp. 1–44. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prijbelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Bourbonnais, R.; Leech, D.; Paice, M.G. Electrochemical analysis of the interactions of laccase mediators with lignin model compounds. Biochim. Biophys. Acta 1998, 1379, 381–390. [Google Scholar] [CrossRef]
- Majcherczyk, A.; Johannes, C.; Huttermann, A. Oxidation of polycyclic aromatic hydrocarbons (PAH) by laccase of Trametes versicolor. Enzyme Microb. Technol. 1998, 22, 335–341. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
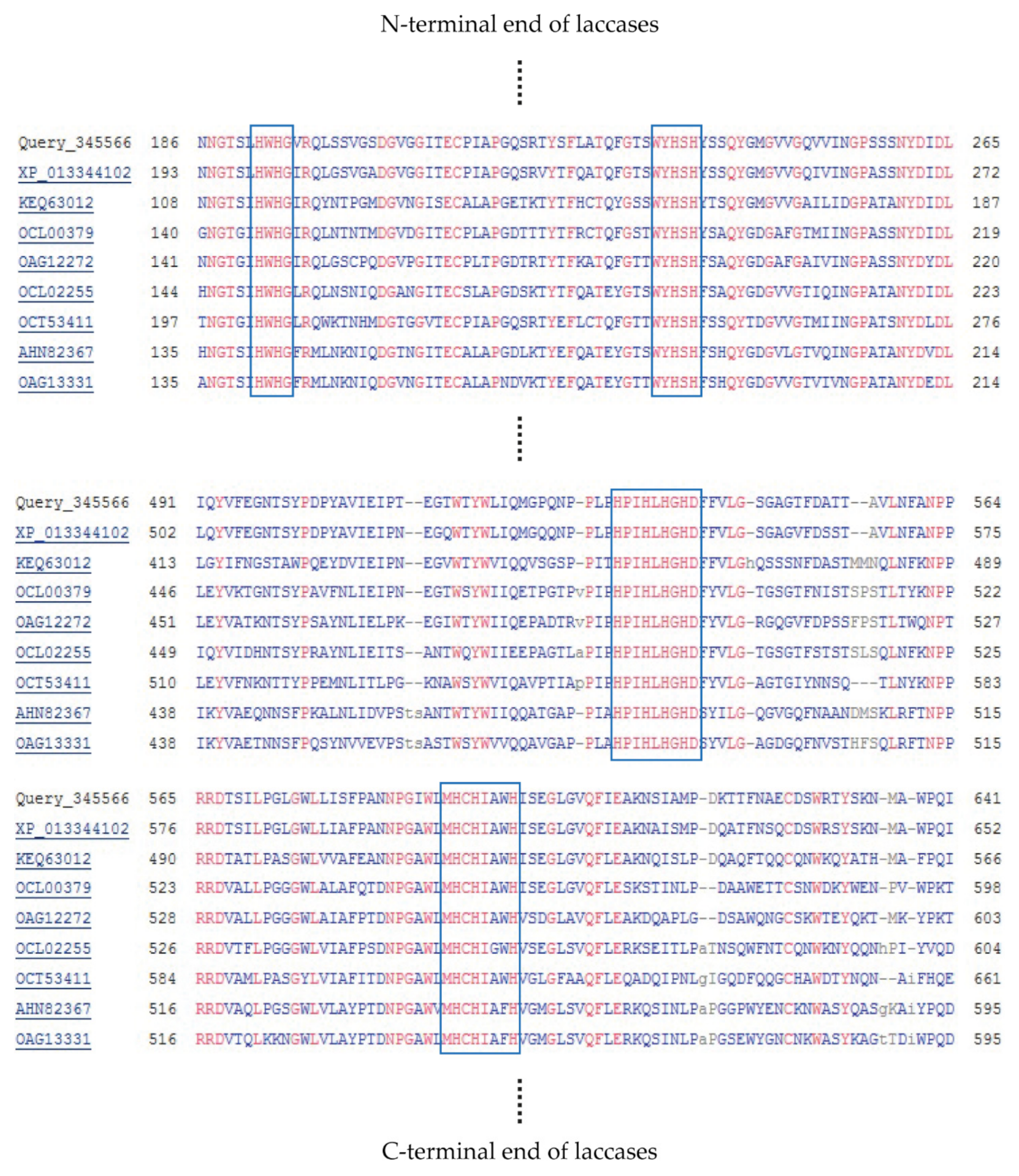


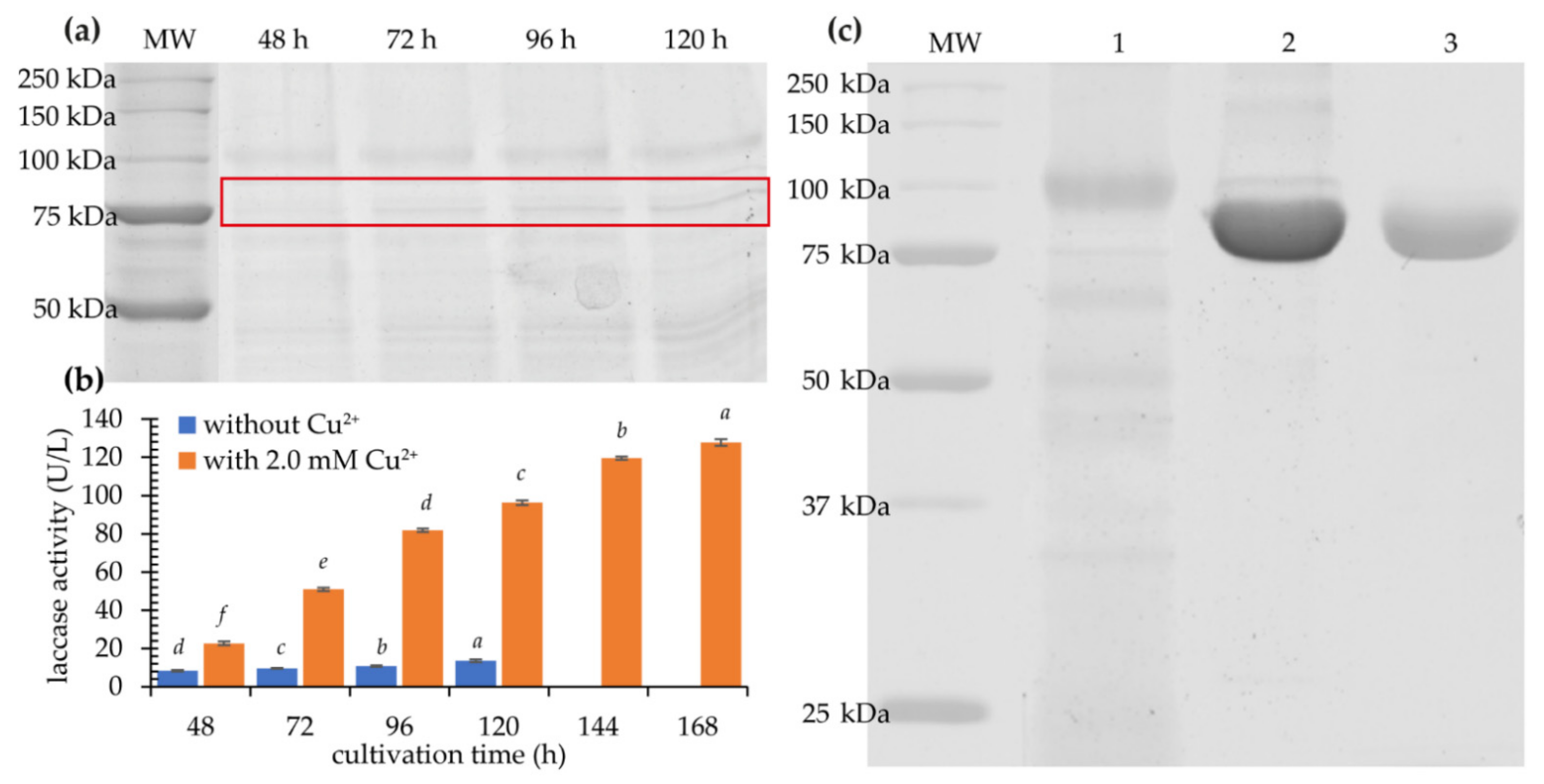


| Organism | GenBank Accession Number |
|---|---|
| Kabatiella bupleuri G3 IBMiP | MZ292708 |
| Aureobasidium subglaciale EXF–2481 | XP_013344102.1 |
| Aureobasidium melanogenum CBS 110374 | KEQ63012.1 |
| Cenococcum geophilum 1.58 | OCL00379.1 |
| Paraphaeosphaeria sporulosa | OAG12255.1 |
| Glonium stellatum | OCL02255.1 |
| Cladophialophora carrionii | OCT53411.1 |
| Shiraia sp. SUPER–H168 | AHN82367.1 |
| Alternaria alternata | OAG13331.1 |
| Purification Step | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 125.15 | 298.90 | 0.419 | 100% | 1.00 |
| Ultrafiltration | 105.26 | 139.15 | 0.756 | 84.11% | 1.81 |
| Ammonium sulfate precipitation | 65.96 | 54.24 | 1.216 | 52.71% | 2.90 |
| Mono S columnion exchange chromatography | 36.78 | 1.795 | 20.49 | 29.39% | 48.94 |
| Substrate | Topt (°C) | pHopt | KM (mM) | Vmax (U mg−1) | kcat * (s−1) | kcat/KM (mM−1 s−1) |
|---|---|---|---|---|---|---|
| ABTS | 30 | 3.5 | 0.580 | 34.18 | 39.64 | 68.35 |
| Syringaldazine | 30 | 7.0 | 0.021 | 33.03 | 38.31 | 1808.82 |
| Guaiacol | 30 | 5.5 | 13.28 | 4.70 | 5.45 | 0.41 |
| Sinapic acid | 30 | 6.5 | 0.015 | 1.39 | 1.61 | 109.26 |
| Dye | Group | Chemical Structure | % Decolorization | |
|---|---|---|---|---|
| Native | Recombinant | |||
| Methylene blue | Heterocyclic/thiazine dye | 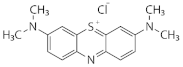 | 18.2 ± 2.5% b | 23.8 ± 1.2% a |
| Alkaline fuchsin | Triphenylmethane dye and aniline dye | 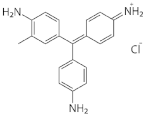 | 31.7 ± 3.3% b | 36.6 ± 1.8% a |
| Crystal violet | Triphenylmethane dye | 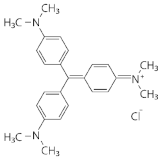 | 40.4 ± 7.0% a | 48.1 ± 4.3% a |
| Coomassie Brilliant Blue R-250 | Triphenylmethane dye | 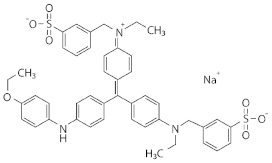 | 19.8 ± 5.8% a | 24.4 ± 3.9% a |
| Ferulic Acid (mM) | 20 °C | 30 °C | ||
|---|---|---|---|---|
| Vanillin (mg/L) | Molar Yield (%) | Vanillin (mg/L) | Molar Yield (%) | |
| 1.0 | 0.136 | 0.089 | 0.082 | 0.054 |
| 10.0 | 0.291 | 0.019 | 0.119 | 0.008 |
| Name of Primer | Sequence (5′–3′) |
|---|---|
| L1 LF | ATGCATCTACAGAAGCTCAGTGGCCTCT |
| L1 LR | CTAAAGTCCACTGTCGATTTGAGGCCAAGC |
| IntR | AAATCGTGACCGTGCAAATGGATGGGATGA |
| IntF | CATTTGCACGGTCACGATTTCTTCGTCCTC |
| L1 SF | ATGCATCTACAGAAGCTCAGTG |
| L1 SR | CTAAAGTCCACTGTCGATTTGAG |
| L1 StuIF | AGGCCTATGCATCTACAGAAGCTC |
| L1 FseIR | GGCCGGCCTAAAGTCCACT |
| 5′AOX1 | GACTGGTTCCAATTGACAAGC |
| 3′CYC1 | GCGTGAATGTAAGCGTGAC |
| Cycle Step | Temperature (°C) | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 98 | 30 s | |
| Denaturation | 98 | 10 s | 5 |
| Annealing/Extension | 72 | 60 s | |
| Extension | 72 | 5 min | |
| Denaturation | 98 | 10 s | 20 |
| Annealing | 72 (−1/cycle) | 30 s | |
| Extention | 72 | 60 s | |
| Final extension | 72 | 10 min | |
| Cooling | 4 | hold |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiśniewska, K.M.; Twarda-Clapa, A.; Białkowska, A.M. Novel Cold-Adapted Recombinant Laccase KbLcc1 from Kabatiella bupleuri G3 IBMiP as a Green Catalyst in Biotransformation. Int. J. Mol. Sci. 2021, 22, 9593. https://doi.org/10.3390/ijms22179593
Wiśniewska KM, Twarda-Clapa A, Białkowska AM. Novel Cold-Adapted Recombinant Laccase KbLcc1 from Kabatiella bupleuri G3 IBMiP as a Green Catalyst in Biotransformation. International Journal of Molecular Sciences. 2021; 22(17):9593. https://doi.org/10.3390/ijms22179593
Chicago/Turabian StyleWiśniewska, Katarzyna M., Aleksandra Twarda-Clapa, and Aneta M. Białkowska. 2021. "Novel Cold-Adapted Recombinant Laccase KbLcc1 from Kabatiella bupleuri G3 IBMiP as a Green Catalyst in Biotransformation" International Journal of Molecular Sciences 22, no. 17: 9593. https://doi.org/10.3390/ijms22179593
APA StyleWiśniewska, K. M., Twarda-Clapa, A., & Białkowska, A. M. (2021). Novel Cold-Adapted Recombinant Laccase KbLcc1 from Kabatiella bupleuri G3 IBMiP as a Green Catalyst in Biotransformation. International Journal of Molecular Sciences, 22(17), 9593. https://doi.org/10.3390/ijms22179593






