Functional Polyglycidol-Based Block Copolymers for DNA Complexation
Abstract
:1. Introduction
2. Results and Discussion
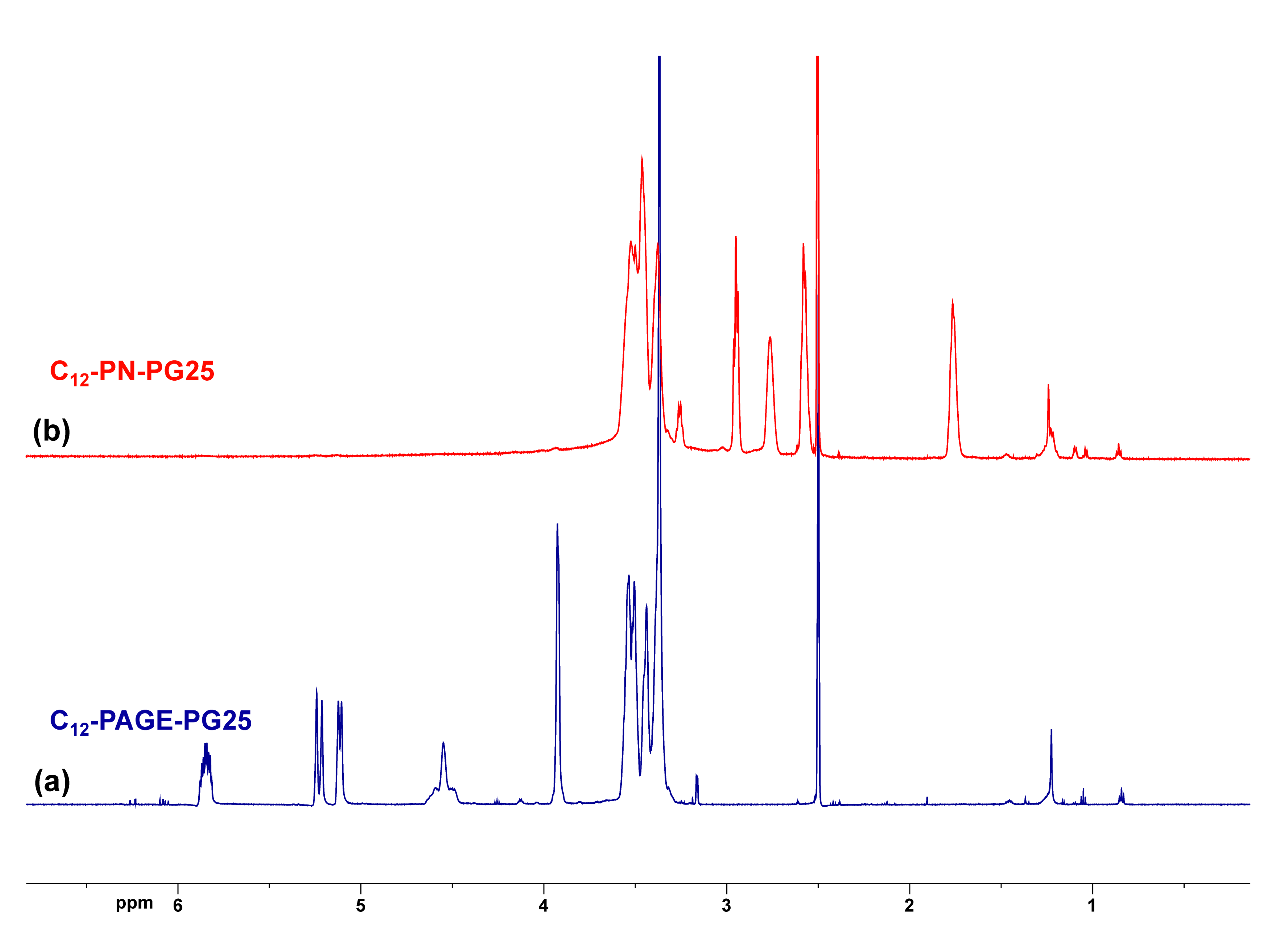
2.1. Synthesis and Characterization of Amine-Functionalized Block Copolyethers
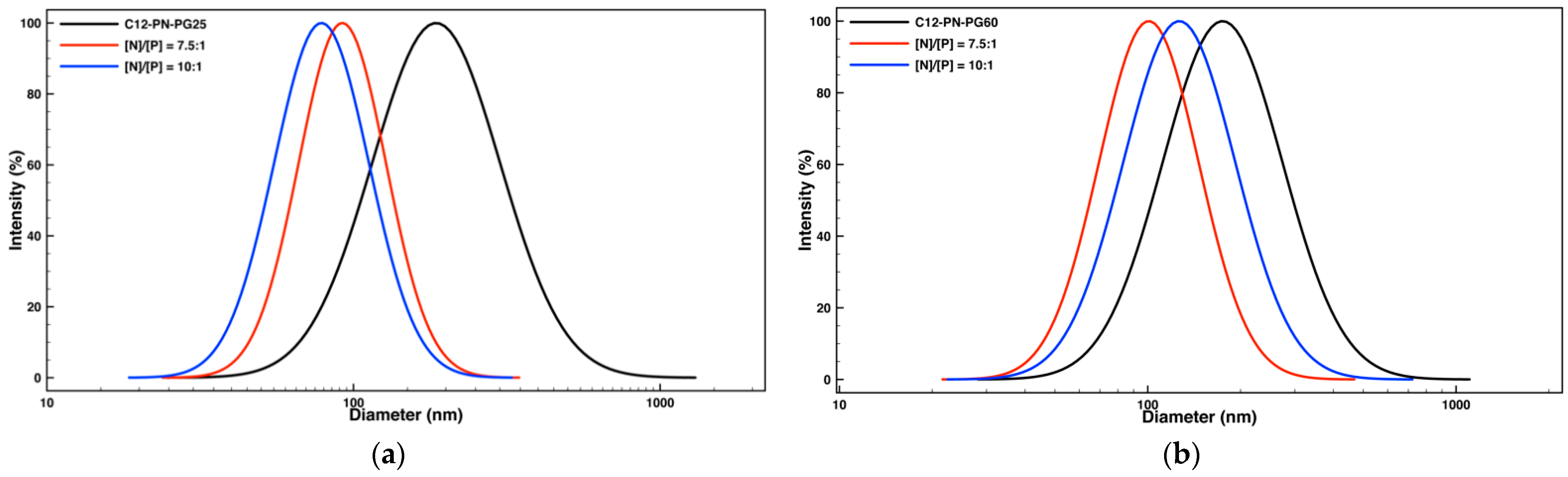
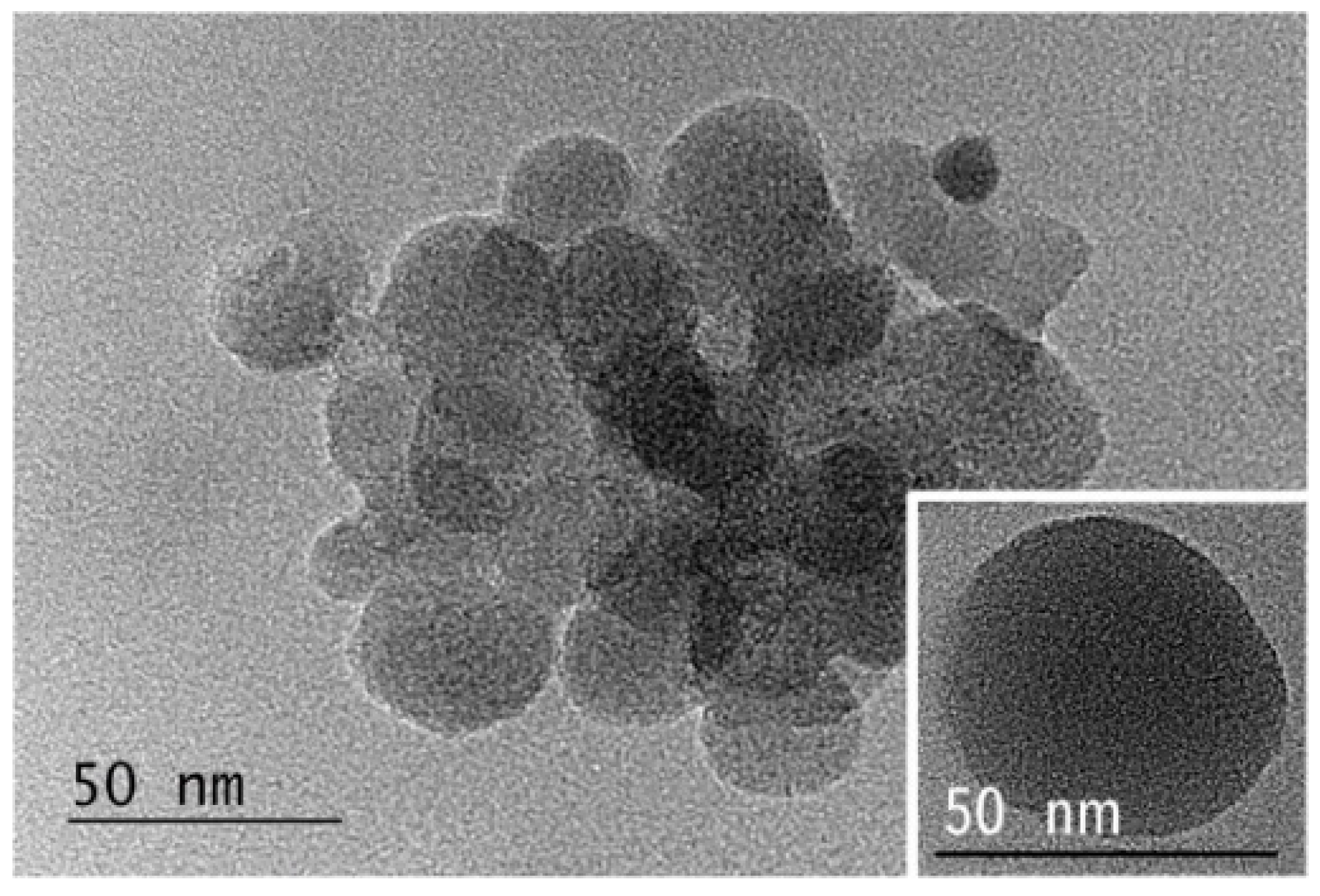
2.2. Self-Association of the Amine-Functional Block Copolymers and DNA Condensation
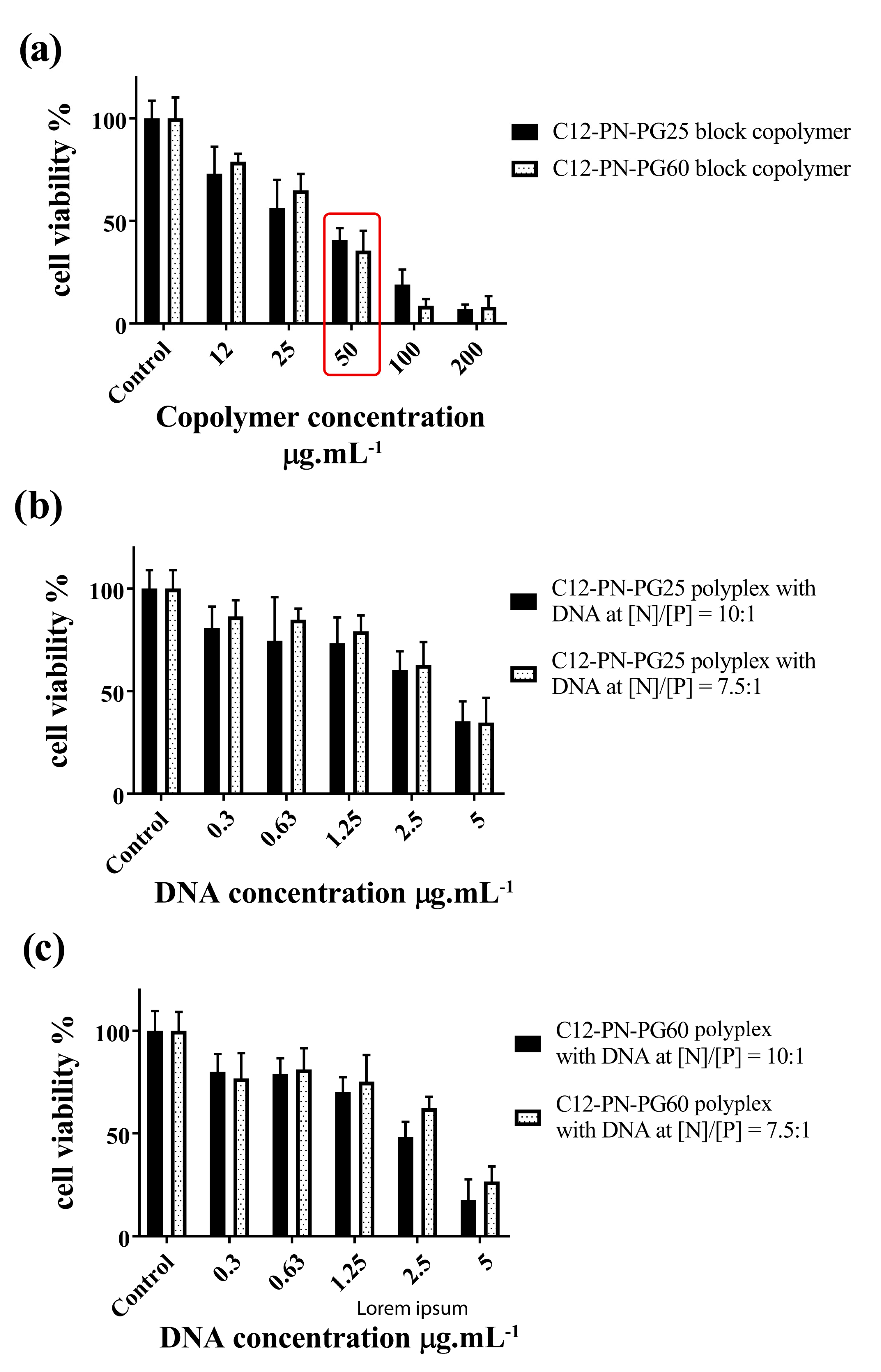
2.3. In Vitro Cytotoxicity Assessment of Block Copolymers and Polyplexes
2.4. Cell Internalization of Polyplexes
3. Materials and Methods
3.1. Materials and Reagents
3.2. Synthesis of Amine and Hydroxyl-Functional Copolyethers (C12-PN-PG)
3.3. Characterization
3.4. DNA Condensation
3.5. Cell Lines and Culture Conditions
3.6. Cytotoxicity Assessment (Trypan Blue Exclusion Test)
3.7. Cytotoxicity Assessment (MTT-Dye Reduction Assay)
3.8. Intracellular Localization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Miller, A.D. Human gene therapy comes of age. Nature 1992, 357, 455–460. [Google Scholar] [CrossRef]
- Burton, E.A.; Glorioso, J.C.; Fink, D.J. Gene therapy progress and prospects: Parkinson’s disease. Gene Ther. 2003, 10, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.; Burmester, J.K. Gene Therapy for cancer treatment: Past, present and future. Clin. Med. Res. 2006, 4, 218–227. [Google Scholar] [CrossRef] [Green Version]
- Jooss, K.; Yang, Y.; Fisher, K.J.; Wilson, J.M. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J. Virol. 1998, 72, 4212–4223. [Google Scholar] [CrossRef] [Green Version]
- Marshall, E. Gene therapy death prompts review of adenovirus vector. Science 1999, 286, 2244–2245. [Google Scholar] [CrossRef]
- Check, E. A tragic setback. Nat. Cell Biol. 2002, 420, 116–118. [Google Scholar] [CrossRef]
- Sharma, D.; Arora, S.; Singh, J.; Layek, B. A review of the tortuous path of nonviral gene delivery and recent progress. Int. J. Biol. Macromol. 2021, 183, 2055–2073. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef]
- Jones, C.H.; Chen, C.-K.; Ravikrishnan, A.; Rane, S.; Pfeifer, B.A. Overcoming nonviral gene delivery barriers: Perspective and future. Mol. Pharm. 2013, 10, 4082–4098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trentin, D.; Hubbell, J.; Hall, H. Non-viral gene delivery for local and controlled DNA release. J. Control. Release 2005, 102, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Pelet, J.M.; Putnam, D. Polymer systems for gene delivery—Past, present, and future. Prog. Polym. Sci. 2007, 32, 799–837. [Google Scholar] [CrossRef]
- O’Rorke, S.; Keeney, M.; Pandit, A. Non-viral polyplexes: Scaffold mediated delivery for gene therapy. Prog. Polym. Sci. 2010, 35, 441–458. [Google Scholar] [CrossRef]
- Pichon, C.; Billiet, L.; Midoux, P. Chemical vectors for gene delivery: Uptake and intracellular trafficking. Curr. Opin. Biotechnol. 2010, 21, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, A.I.S.; Yun, C.-O.; Schiffelers, R.M.; Hennink, W.E. Polymeric delivery systems for nucleic acid therapeutics: Approaching the clinic. J. Control. Release 2021, 331, 121–141. [Google Scholar] [CrossRef]
- Rangelov, S.; Pispas, A. Polymer and Polymer-Hybrid Nanoparticles: From Synthesis to Biomedical Applications; CRC Press Taylor and Francis Group: Boca Raton, FL, USA, 2014. [Google Scholar]
- Yang, C.; Gao, S.; Dagnæs-Hansen, F.; Jakobsen, M.; Kjems, J. Impact of PEG chain length on the physical properties and bioactivity of PEGylated Chitosan/SiRNA nanoparticles In Vitro and In Vivo. ACS Appl. Mater. Interfaces 2017, 9, 12203–12216. [Google Scholar] [CrossRef]
- Venault, A.; Huang, Y.-C.; Lo, J.W.; Chou, C.-J.; Chinnathambi, A.; Higuchi, A.; Chen, W.-S.; Chen, W.-Y.; Chang, Y. Tunable PEGylation of branch-type PEI/DNA polyplexes with a compromise of low cytotoxicity and high transgene expression: In Vitro and In Vivo gene delivery. J. Mater. Chem. B 2017, 5, 4732–4744. [Google Scholar] [CrossRef] [PubMed]
- Santo, D.; Mendonça, P.V.; Lima, M.S.; Cordeiro, R.A.; Cabanas, L.; Serra, A.; Coelho, J.F.; Faneca, H. Poly(ethylene glycol)-block-poly(2-aminoethyl methacrylate hydrochloride)-Based Polyplexes as Serum-Tolerant Nanosystems for Enhanced Gene Delivery. Mol. Pharm. 2019, 16, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ye, M.; Xie, R.; Gong, S. Enhancing the In Vitro and In Vivo stabilities of polymeric nucleic acid delivery nanosystems. Bioconjug. Chem. 2019, 30, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Feijen, J.; Lok, M.C.; Hennink, W.E.; Christensen, L.V.; Yockman, J.W.; Kim, A.Y.-H.; Kim, S.W. Low Molecular Weight Linear Polyethylenimine-b-poly(ethylene glycol)-b-polyethylenimine Triblock Copolymers: Synthesis, Characterization, and In Vitro Gene Transfer Properties. Biomacromolecules 2005, 6, 3440–3448. [Google Scholar] [CrossRef]
- Kim, J.; Kang, Y.; Tzeng, S.Y.; Green, J.J. Synthesis and application of poly(ethylene glycol)-co-poly(β-amino ester) copolymers for small cell lung cancer gene therapy. Acta Biomater. 2016, 41, 293–301. [Google Scholar] [CrossRef]
- Haladjova, E.; Chrysostomou, V.; Petrova, M.; Ugrinova, I.; Pispas, S.; Rangelov, S. Physicochemical Properties and Biological Performance of Polymethacrylate Based Gene Delivery Vector Systems: Influence of Amino Functionalities. Macromol. Biosci. 2021, 21, e2000352. [Google Scholar] [CrossRef]
- Chroni, A.; Forys, A.; Trzebicka, B.; Alemayehu, A.; Tyrpekl, V.; Pispas, S. Poly[oligo(ethylene glycol) methacrylate]-b-poly[(vinyl benzyl trimethylammonium chloride)] Based Multifunctional Hybrid Nanostructures Encapsulating Magnetic Nanoparticles and DNA. Polymers 2020, 12, 1283. [Google Scholar] [CrossRef] [PubMed]
- Mendrek, B.; Fus-Kujawa, A.; Teper, P.; Botor, M.; Kubacki, J.; Sieroń, A.L.; Kowalczuk, A. Star polymer-based nanolayers with immobilized complexes of polycationic stars and DNA for deposition gene delivery and recovery of intact transfected cells. Int. J. Pharm. 2020, 589, 119823. [Google Scholar] [CrossRef]
- Vuoriluoto, M.; Orelma, H.; Johansson, L.-S.; Zhu, B.; Poutanen, M.; Walther, A.; Laine, J.; Rojas, O.J. Effect of Molecular Architecture of PDMAEMA–POEGMA Random and Block Copolymers on Their Adsorption on Regenerated and Anionic Nanocelluloses and Evidence of Interfacial Water Expulsion. J. Phys. Chem. B 2015, 119, 15275–15286. [Google Scholar] [CrossRef]
- Han, S.; Hagiwara, M.; Ishizone, T. Synthesis of Thermally Sensitive Water-Soluble Polymethacrylates by Living Anionic Polymerizations of Oligo(ethylene glycol) Methyl Ether Methacrylates. Macromolecules 2003, 36, 8312–8319. [Google Scholar] [CrossRef]
- Trzebicka, B.; Szweda, D.; Rangelov, S.; Kowalczuk, A.; Mendrek, B.; Utrata-Wesołek, A.; Dworak, A. (Co)polymers of oligo(ethylene glycol) methacrylates—temperature-induced aggregation in aqueous solution. J. Polym. Sci. Part A Polym. Chem. 2012, 51, 614–623. [Google Scholar] [CrossRef]
- Liu, M.; Leroux, J.-C.; Gauthier, M.A. Conformation–function relationships for the comb-shaped polymer pOEGMA. Prog. Polym. Sci. 2015, 48, 111–121. [Google Scholar] [CrossRef]
- Adams, N.; Schubert, U.S. Poly(2-oxazolines) in biological and biomedical application contexts. Adv. Drug Deliv. Rev. 2007, 59, 1504–1520. [Google Scholar] [CrossRef]
- Hoogenboom, R. Poly(2-oxazoline)s: A Polymer Class with Numerous Potential Applications. Angew. Chem. Int. Ed. 2009, 48, 7978–7994. [Google Scholar] [CrossRef] [PubMed]
- Schlaad, H.; Diehl, C.; Gress, A.; Meyer, M.; Demirel, A.L.; Nur, Y.; Bertin, A. Poly(2-oxazoline)s as Smart Bioinspired Polymers. Macromol. Rapid Commun. 2010, 31, 511–525. [Google Scholar] [CrossRef]
- Luxenhofer, R.; Han, Y.; Schulz, A.; Tong, J.; He, Z.; Kabanov, A.; Jordan, R. Poly(2-oxazoline)s as Polymer Therapeutics. Macromol. Rapid Commun. 2012, 33, 1613–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brissault, B.; Kichler, A.; Guis, C.; Leborgne, C.; Danos, O.; Cheradame, H. Synthesis of Linear Polyethylenimine Derivatives for DNA Transfection. Bioconjug. Chem. 2003, 14, 581–587. [Google Scholar] [CrossRef]
- Lambermont-Thijs, H.M.L.; van der Woerdt, F.S.; Baumgaertel, A.; Bonami, L.; Du Prez, F.E.; Schubert, U.S.; Hoogenboom, R. Linear Poly(ethylene imine)s by Acidic Hydrolysis of Poly(2-oxazoline)s: Kinetic Screening, Thermal Properties, and Temperature-Induced Solubility Transitions. Macromolecules 2010, 43, 927–933. [Google Scholar] [CrossRef]
- Bludau, H.; Czapar, A.E.; Pitek, A.S.; Shukla, S.; Jordan, R.; Steinmetz, N.F. POxylation as an alternative stealth coating for biomedical applications. Eur. Polym. J. 2017, 88, 679–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haladjova, E.; Rangelov, S.; Tsvetanov, C. Thermoresponsive Polyoxazolines as Vectors for Transfection of Nucleic Acids. Polymers 2020, 12, 2609. [Google Scholar] [CrossRef] [PubMed]
- Halacheva, S.; Rangelov, S.; Tsvetanov, C. Poly(glycidol)-Based Analogues to Pluronic Block Copolymers. Synthesis and Aqueous Solution Properties. Macromolecules 2006, 39, 6845–6852. [Google Scholar] [CrossRef]
- Halacheva, S.; Rangelov, S.; Garamus, V.M. Structure and Interactions in Large Compound Particles Formed by Polyglycidol-Based Analogues to Pluronic Copolymers in Aqueous Solution. Macromolecules 2007, 40, 8015–8021. [Google Scholar] [CrossRef]
- Bakardzhiev, P.; Rangelov, S.; Trzebicka, B.; Momekova, D.; Lalev, G.; Garamus, V.M. Nanostructures by self-assembly of polyglycidol-derivatized lipids. RSC Adv. 2014, 4, 37208–37219. [Google Scholar] [CrossRef] [Green Version]
- Stoyanova, B.; Novakov, C.; Tsvetanov, C.B.; Rangelov, S. Synthesis and Aqueous Solution Properties of Block Copolyethers with Latent Chemical Functionality. Macromol. Chem. Phys. 2016, 217, 2380–2390. [Google Scholar] [CrossRef]
- Valchanova, M.; Yordanov, Y.; Tzankova, V.; Yoncheva, K.; Turmanova, S.; Rangelov, S. Functional amphiphilic block copolyethers as carriers of caffeic acid phenethyl ester. Polym. Int. 2019, 68, 1881–1890. [Google Scholar] [CrossRef]
- Yordanov, Y.; Aluani, D.; Tzankova, V.; Rangelov, S.; Odzhakov, F.; Apostolov, A.; Yoncheva, K. Safety assessment of a newly synthesized copolymer for micellar delivery of hydrophobic caffeic acid phenethyl ester. Pharm. Dev. Technol. 2020, 25, 1271–1280. [Google Scholar] [CrossRef]
- Hu, S.; Horii, F.; Odani, H. 1H NMR study of the solvation and gelation in a poly (vinyl alcohol)/DMSO-d6/H2O system. Bull. Inst. Chem. Res. Kyoto Univ. 1990, 67, 239–248. Available online: http://hdl.handle.net/2433/77317 (accessed on 12 March 2021).
- Burchard, W. Static and dynamic light scattering from branched polymers and biopolymers. Light Scatt. Polym. 2007, 48, 1–124. [Google Scholar] [CrossRef]
- Thurn, A.; Burchard, W.; Niki, R. Structure of casein micelles I. Small angle neutron scattering and light scattering from β- and χ-casein. Colloid Polym. Sci. 1987, 265, 653–666. [Google Scholar] [CrossRef]
- Agirre, M.; Zarate, J.; Puras, G.; Ojeda, E.; Pedraz, J.L. Improving transfection efficiency of ultrapure oligochitosan/DNA polyplexes 614 by medium acidification. Drug Deliv. 2015, 22, 100–110. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.; Firestein, B.L. DNA Transfection: Calcium Phosphate Method. Adv. Struct. Saf. Stud. 2013, 1018, 107–110. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. DNA Transfection Mediated by Cationic Lipid Reagents. Cold Spring Harb. Protoc. 2019, 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]




| Precursors | Aggregates of Modified Block Copolymers | |||||||
|---|---|---|---|---|---|---|---|---|
| Code | DPaPAGE | DPaPG | Mna (g·mol−1) | ÐMb | Code | dhc (nm) | PdIc | ζ c (mV) |
| C12-PAGE-PG25 | 44 | 16 | 6400 | 1.08 | C12-PN-PG25 | 184.2 | 0.293 | 41.1 |
| C12-PAGE-PG60 | 44 | 66 | 10,100 | 1.06 | C12-PN-PG60 | 173.9 | 0.232 | 40.7 |
| [N]/[P] | dha (nm) | PdI | ζa (mV) |
|---|---|---|---|
| Polyplexes from C12-PN-PG25 and DNA | |||
| 1:1 2.5:1 5:1 7.5:1 10:1 | 127.6 | 0.108 | −25.7 |
| 112.8 | 0.153 | 37.4 | |
| 95.4 | 0.073 | 35.3 | |
| 92.0 | 0.113 | 34.2 | |
| 78.7 | 0.135 | 35.7 | |
| Polyplexes from C12-PN-PG60 and DNA | |||
| 1:1 2.5:1 5:1 7.5:1 10:1 | 181.1 | 0.090 | −27.9 |
| 131.2 | 0.090 | 36.8 | |
| 119.5 | 0.187 | 40.7 | |
| 100.8 | 0.148 | 38.9 | |
| 126.6 | 0.191 | 41.8 | |
| [N]/[P] | A549 | MCF-7 | HeLa |
|---|---|---|---|
| Copolymer aggregates and polyplexes from C12-PN-PG25 and DNA | |||
| Initial C12-PN-PG25 aggregates 1:1 2.5:1 5:1 7.5:1 10:1 | 32.2 ± 5.1 | 29.8 ± 3.5 | 27.7 ± 2.8 |
| 20.7 ± 1.8 | 26.2 ± 3.1 | 31.5 ± 3.6 | |
| 24.7 ± 2.2 | 28.8 ± 2.6 | 24.6 ± 5.1 | |
| 37.2 ± 3.4 | 47.2 ± 3.3 | 53.7 ± 4.7 | |
| 46.5 ± 2.8 | 39.8 ± 4.1 | 52.8 ± 3.4 | |
| 49.8 ± 3.1 | 48.2 ± 2.5 | 43.7 ± 2.9 | |
| Copolymer aggregates and polyplexes from C12-PN-PG60 and DNA | |||
| Initial C12-PN-PG60 aggregates 1:1 2.5:1 5:1 7.5:1 10:1 | 22.0 ± 4.4 * | 24.6 ± 4.2 * | 28.7 ± 3.2 |
| 35.5 ± 2.7 | 19.8 ± 4.6 * | 32.6 ± 3.5 | |
| 40.3 ± 1.9 | 25.6 ± 2.8 | 39.5 ± 4.8 | |
| 44.2 ± 2.6 | 49.5 ± 3.5 | 56.6 ± 3.7 | |
| 50.1 ± 3.3 | 46.7 ± 3.7 | 48.7 ± 4.2 | |
| 48.8 ± 4.1 | 42.2 ± 3.9 | 43.8 ± 2.9 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalinova, R.; Valchanova, M.; Dimitrov, I.; Turmanova, S.; Ugrinova, I.; Petrova, M.; Vlahova, Z.; Rangelov, S. Functional Polyglycidol-Based Block Copolymers for DNA Complexation. Int. J. Mol. Sci. 2021, 22, 9606. https://doi.org/10.3390/ijms22179606
Kalinova R, Valchanova M, Dimitrov I, Turmanova S, Ugrinova I, Petrova M, Vlahova Z, Rangelov S. Functional Polyglycidol-Based Block Copolymers for DNA Complexation. International Journal of Molecular Sciences. 2021; 22(17):9606. https://doi.org/10.3390/ijms22179606
Chicago/Turabian StyleKalinova, Radostina, Miroslava Valchanova, Ivaylo Dimitrov, Sevdalina Turmanova, Iva Ugrinova, Maria Petrova, Zlatina Vlahova, and Stanislav Rangelov. 2021. "Functional Polyglycidol-Based Block Copolymers for DNA Complexation" International Journal of Molecular Sciences 22, no. 17: 9606. https://doi.org/10.3390/ijms22179606
APA StyleKalinova, R., Valchanova, M., Dimitrov, I., Turmanova, S., Ugrinova, I., Petrova, M., Vlahova, Z., & Rangelov, S. (2021). Functional Polyglycidol-Based Block Copolymers for DNA Complexation. International Journal of Molecular Sciences, 22(17), 9606. https://doi.org/10.3390/ijms22179606








