Differential Contribution of Protein Factors and 70S Ribosome to Elongation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Kinetics of tRNA Interaction with the Ribosome Is Determined by the Properties of Elongation Factor EF-Tu
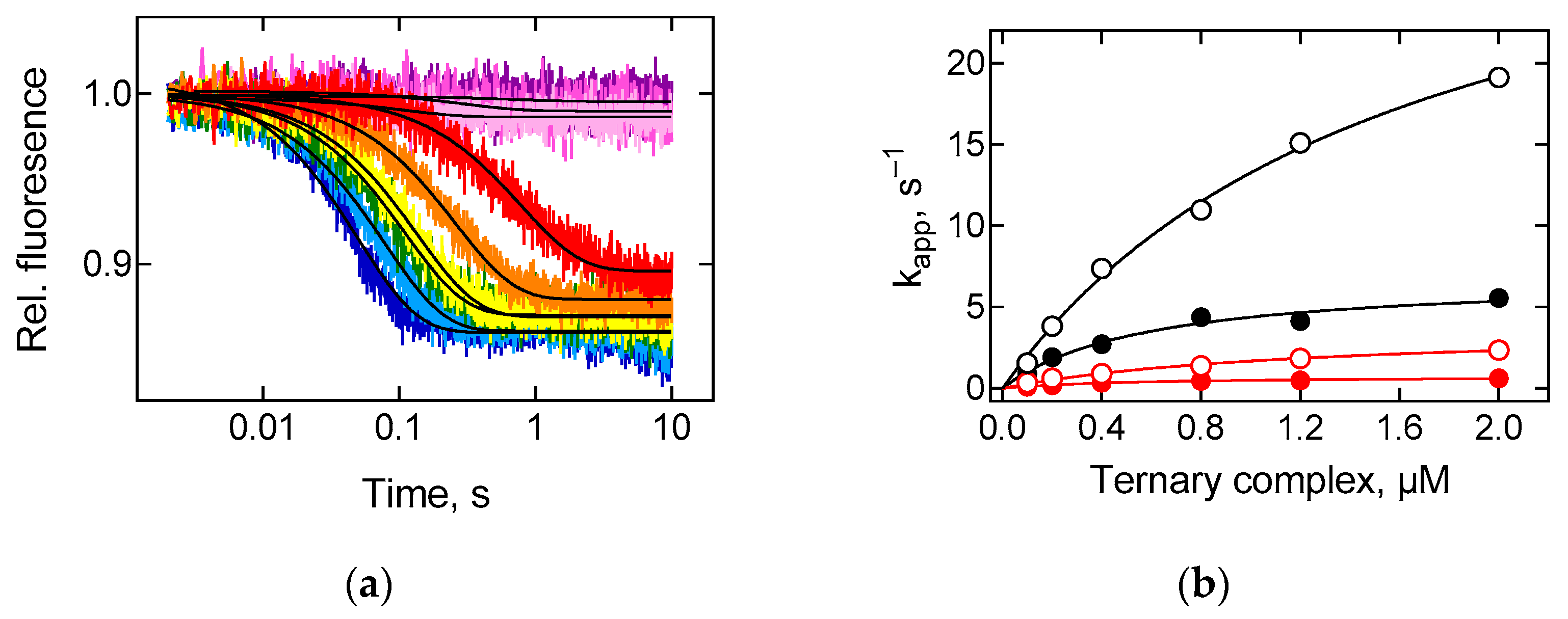
2.2. Tetracycline Interacts with the Ribosome and Kinetically Inhibits tRNA Binding to the A Site
2.3. Species Differences of Elongation Factor EF-G Do Not Influence Translocation Kinetics
2.4. Antibiotics Bind to the Small Ribosomal Subunit and Inhibit the Movement of the Elbow Region of tRNA
3. Materials and Methods
3.1. Materials
3.2. Protein Expression and Purification
3.3. Biochemical Assays
3.4. Rapid Kinetics
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| aa-tRNA | aminoacylated tRNA |
| EF-Tu | elongation factor Tu |
| A | aminoacyl |
| P | peptidyl |
| E | exit |
| Prf | proflavin |
| BPY | BODIPY FL |
References
- Williams, R.; Sharp, R. The taxonomy and the identification of Thermus. In Thermus Species; Williams, R., Sharp, R., Eds.; Plenum Press: New York, NY, USA, 1995; pp. 1–39. [Google Scholar]
- Henne, A.; Brüggemann, H.; Raasch, C.; Wiezer, A.; Hartsch, T.; Liesegang, H.; Johann, A.; Lienard, T.; Gohl, O.; Martinez-Arias, R.; et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004, 22, 547–553. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov (accessed on 17 May 2021).
- Gao, Y.G.; Selmer, M.; Dunham, C.M.; Weixlbaumer, A.; Kelley, A.C.; Ramakrishnan, V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 2009, 326, 694–699. [Google Scholar] [CrossRef] [Green Version]
- Pulk, A.; Cate, J.H.D. Control of ribosomal subunit rotation by elongation factor G. Science 2013, 340, 1235970. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Gagnon, M.G.; Bulkley, D.; Steitz, T.A. Conformational changes of elongation factor g on the ribosome during tRNA translocation. Cell 2015, 160, 219–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, J.S.; Kavaliauskas, D.; Pfeil, S.H.; Blaise, M.; Cooperman, B.S.; Goldman, Y.E.; Thirup, S.S.; Knudsen, C.R. E. coli elongation factor Tu bound to a GTP analogue displays an open conformation equivalent to the GDP-bound form. Nucleic Acids Res. 2018, 46, 8641–8650. [Google Scholar] [CrossRef]
- Noble, C.G.; Song, H. Structural studies of elongation and release factors. Cell Mol. Life Sci. 2008, 65, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Andersen, G.R.; Nissen, P.; Nyborg, J. Elongation factors in protein biosynthesis. Trends Biochem. Sci. 2003, 28, 434–441. [Google Scholar] [CrossRef]
- Nock, S.; Grillenbeck, N.; Ahmadian, M.R.; Ribeiro, S.; Kreutzer, R.; Sprinzl, M. Properties of isolated domains of the elongation factor Tu from Thermus thermophilus HB8. Eur. J. Biochem. 1995, 234, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Cetin, R.; Anborgh, P.H.; Cool, R.H.; Parmeggiani, A. Functional role of the noncatalytic domains of elongation factor Tu in the interactions with ligands. Biochemistry 1998, 37, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Sanderová, H.; Hůlková, M.; Malon, P.; Kepková, M.; Jonák, J. Thermostability of multidomain proteins: Elongation factors EF-Tu from Escherichia coli and Bacillus stearothermophilus and their chimeric forms. Protein Sci. 2004, 13, 89–99. [Google Scholar] [CrossRef]
- Masullo, M.; Ianniciello, G.; Arcari, P.; Bocchini, V. Properties of truncated forms of the elongation factor 1alpha from the archaeon Sulfolobus solfataricus. Eur. J. Biochem. 1997, 243, 468–473. [Google Scholar] [CrossRef]
- Scarano, G.; Krab, I.M.; Bocchini, V.; Parmeggiani, A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: Its substitution by glutamine and alanine. FEBS Lett. 1995, 365, 214–218. [Google Scholar] [CrossRef] [Green Version]
- Daviter, T.; Wieden, H.J.; Rodnina, M.V. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J. Mol. Biol. 2003, 332, 689–699. [Google Scholar] [CrossRef] [Green Version]
- Zeidler, W.; Egle, C.; Ribeiro, S.; Wagner, A.; Katunin, V.; Kreutzer, R.; Rodnina, M.; Wintermeyer, W.; Sprinzl, M. Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur. J. Biochem. 1995, 229, 596–604. [Google Scholar] [CrossRef]
- Yikilmaz, E.; Chapman, S.J.; Schrader, J.M.; Uhlenbeck, O.C. The interface between Escherichia coli elongation factor Tu and aminoacyl-tRNA. Biochemistry 2014, 53, 5710–5720. [Google Scholar] [CrossRef] [Green Version]
- Zeidler, W.; Kreutzer, R.; Sprinzl, M. Participation of the overproduced elongation factor Tu from Thermus thermophilus in protein biosynthesis of Escherichia coli. FEBS Lett. 1993, 319, 185–188. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Asahara, H.; Gaucher, E.A.; Chong, S. Reconstitution of translation from Thermus thermophilus reveals a minimal set of components sufficient for protein synthesis at high temperatures and functional conservation of modern and ancient translation components. Nucleic Acids Res. 2012, 40, 7932–7945. [Google Scholar] [CrossRef] [Green Version]
- Raj, V.S.; Kaji, H.; Kaji, A. Interaction of RRF and EF-G from E. coli and T. thermophilus with ribosomes from both origins—Insight into the mechanism of the ribosome recycling step. RNA 2005, 11, 275–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.; Dahlberg, A.E. Testing the conservation of the translational machinery over evolution in diverse environments: Assaying Thermus thermophilus ribosomes and initiation factors in a coupled transcription-translation system from Escherichia coli. Nucleic Acids Res. 2004, 32, 5954–5961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodnina, M.V.; Fischer, N.; Maracci, C.; Stark, H. Ribosome dynamics during decoding. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160182. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Zhang, C.-M.M.; Kirillov, S.; Hou, Y.-M.M.; Cooperman, B.S. Perturbation of the tRNA tertiary core differentially affects specific steps of the elongation cycle. J. Biol. Chem. 2008, 283, 18431–18440. [Google Scholar] [CrossRef] [Green Version]
- Maksimova, E.; Vinogradova, D.; Osterman, I.; Kasatsky, P.; Nikonov, O.; Milón, P.; Dontsova, O.; Sergiev, P.; Paleskava, A.; Konevega, A. Multifaceted Mechanism of Amicoumacin A Inhibition of Bacterial Translation. Front. Microbiol. 2021, 12, 172. [Google Scholar] [CrossRef]
- Oshima, T.; Imahori, K. Physiochemical properties of deoxyribonucleic acid from an extreme thermophile. J. Biochem. 1974, 75, 179–183. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Jenner, L.; Starosta, A.L.; Terry, D.S.; Mikolajka, A.; Filonava, L.; Yusupov, M.; Blanchard, S.C.; Wilson, D.N.; Yusupova, G. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3812–3816. [Google Scholar] [CrossRef] [Green Version]
- Semenkov, Y.P.; Makarov, E.M.; Makhno, V.I.; Kirillov, S.V. Kinetic aspects of tetracycline action on the acceptor (A) site of Escherichia coli ribosomes. FEBS Lett. 1982, 144, 125–129. [Google Scholar] [CrossRef] [Green Version]
- Mittelstaet, J.; Konevega, A.L.; Rodnina, M.V. A kinetic safety gate controlling the delivery of unnatural amino acids to the ribosome. J. Am. Chem. Soc. 2013, 135, 17031–17038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodnina, M.V.; Fricke, R.; Kuhn, L.; Wintermeyer, W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 1995, 14, 2613–2619. [Google Scholar] [CrossRef] [Green Version]
- Holtkamp, W.; Cunha, C.E.; Peske, F.; Konevega, A.L.; Wintermeyer, W.; Rodnina, M.V. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014, 33, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Nekhai, S.A.; Parfenov, D.V.; Saminsky, E.M. tRNA regions which contact with the ribosomal poly(U)-programmed P-site. Biochim. Biophys. Acta 1994, 1218, 481–484. [Google Scholar] [CrossRef]
- Kuhn, C.-D. RNA versatility governs tRNA function: Why tRNA flexibility is essential beyond the translation cycle. Bioessays 2016, 38, 465–473. [Google Scholar] [CrossRef]
- Pan, D.; Kirillov, S.; Zhang, C.M.; Hou, Y.-M.M.; Cooperman, B.S. Rapid ribosomal translocation depends on the conserved 18–55 base pair in P-site transfer RNA. Nat. Struct. Mol. Biol. 2006, 13, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.V.; Blau, C.; Schröder, G.F.; Davydov, I.I.; Fischer, N.; Stark, H.; Rodnina, M.V.; Vaiana, A.C.; Grubmüller, H. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat. Struct. Mol. Biol. 2013, 20, 1390–1396. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Semenkov, Y.P.; Savelsbergh, A.; Katunin, V.I.; Peske, F.; Wilden, B.; Wintermeyer, W. Mechanism of tRNA translocation on the ribosome. Mol. Biol. 2001, 35, 559–568. [Google Scholar] [CrossRef]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef]
- Gromadski, K.B.; Rodnina, M.V. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 2004, 11, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Karimi, R.; Ehrenberg, M. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 1996, 15, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Karimi, R.; Ehrenberg, M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur. J. Biochem. 1994, 226, 355–360. [Google Scholar] [CrossRef]
- Ruusala, T.; Kurland, C.G. Streptomycin preferentially perturbs ribosomal proofreading. Mol. Gen. Genet. 1984, 198, 100–104. [Google Scholar] [CrossRef]
- Peske, F.; Savelsbergh, A.; Katunin, V.I.; Rodnina, M.V.; Wintermeyer, W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 2004, 343, 1183–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoji, S.; Walker, S.E.; Fredrick, K. Reverse Translocation of tRNA in the Ribosome. Mol. Cell 2006, 24, 931–942. [Google Scholar] [CrossRef] [Green Version]
- Stanley, R.E.; Blaha, G.; Grodzicki, R.L.; Strickler, M.D.; Steitz, T.A. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat. Struct. Mol. Biol. 2010, 17, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Arenz, S.; Wilson, D.N. Bacterial Protein Synthesis as a Target for Antibiotic Inhibition. Cold Spring Harb. Perspect. Med. 2016, 6, a025361. [Google Scholar] [CrossRef] [Green Version]
- Ermolenko, D.N.; Spiegel, P.C.; Majumdar, Z.K.; Hickerson, R.P.; Clegg, R.M.; Noller, H.F. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 2007, 14, 493–497. [Google Scholar] [CrossRef]
- Wintermeyer, W.; Savelsbergh, A.; Semenkov, Y.P.; Katunin, V.I.; Rodnina, M.V. Mechanism of elongation factor G function in tRNA translocation on the ribosome. Cold Spring Harb. Symp. Quant. Biol. 2001, 66, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, D.E.; Clemons, W.M.; Carter, A.P.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 2000, 103, 1143–1154. [Google Scholar] [CrossRef] [Green Version]
- Borovinskaya, M.A.; Shoji, S.; Holton, J.M.; Fredrick, K.; Cate, J.H.D. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem. Biol. 2007, 2, 545–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, D.; Kirillov, S.V.; Cooperman, B.S. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell 2007, 25, 519–529. [Google Scholar] [CrossRef] [Green Version]
- Rodnina, M.V.; Semenkov, Y.P.; Wintermeyer, W. Purification of fMET-tRNAfMET by Fast Protein Liquid Chromatography. Anal. Biochem. 1994, 219, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Wintermeyer, W.; Zachau, H.G. Fluorescent derivatives of yeast tRNAPhe. Eur. J. Biochem. 1979, 98, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Milon, P.; Konevega, A.L.; Peske, F.; Fabbretti, A.; Gualerzi, C.O.; Rodnina, M.V. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007, 430, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Rodnina, M.V.; Wintermeyer, W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl. Acad. Sci. USA 1995, 92, 1945–1949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calogero, R.A.; Pon, C.L.; Canonaco, M.A.; Gualerzi, C.O. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6427–6431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinogradova, D.; Zegarra, V.; Maksimova, E.; Nakamoto, J.; Kasatsky, P.; Paleskava, A.; Konevega, A.; Milón, P. How the initiating ribosome copes with ppGpp to translate mRNAs. PLoS Biol. 2020, 18, e3000593. [Google Scholar] [CrossRef] [Green Version]
- Fischer, N.; Neumann, P.; Konevega, A.; Bock, L.; Ficner, R.; Rodnina, M.; Stark, H. Structure of the E. coli ribosome-EF-Tu complex at. Nature 2015, 520, 567–570. [Google Scholar] [CrossRef] [Green Version]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (A complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Sampson, J.R.; Uhlenbeck, O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA 1988, 85, 1033–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelsbergh, A.; Matassova, N.B.; Rodnina, M.V.; Wintermeyer, W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 2000, 300, 951–961. [Google Scholar] [CrossRef]





| E. coli | T. thermophilus | |||
|---|---|---|---|---|
| T, °C | kelbow, s−1 | kcca, s−1 | kelbow, s−1 | kcca, s−1 |
| 15 | 2.1 | 4.9 | 1.9 | 2.3 |
| 20 | 5.2 | 7.7 | 5.7 | 4.7 |
| 25 | 8.7 | 11.9 | 8.9 | 6.9 |
| 30 | 15.7 | 18.2 | 15.5 | 8.9 |
| 37 | 31.7 | 26.1 | 31.5 | 19.0 |
| 42 | 52.2 | 39.2 | 55.8 | 28.7 |
| 50 | 83.0 | 66.1 | 111.0 | 62.1 |
| E. coli | T. thermophilus | |||||
|---|---|---|---|---|---|---|
| A/b | k1 elbow, s−1 | k2 elbow, s−1 | kcca, s−1 | k1 elbow, s−1 | k2 elbow, s−1 | kcca, s−1 |
| No a/b | 35 | 1 | 23 | 33 | 0.6 | 14 |
| Str | 8 | 1 | 15 | 10 | 0.6 | 7 |
| Vio | - | - | 47 | 0.004 | - | 22 |
| Hyg B | 0.02 | - | 33 | 0.05 | - | 18 |
| Spc | 53 | 0.07 | 44 | 33 | 0.03 | 27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paleskava, A.; Maksimova, E.M.; Vinogradova, D.S.; Kasatsky, P.S.; Kirillov, S.V.; Konevega, A.L. Differential Contribution of Protein Factors and 70S Ribosome to Elongation. Int. J. Mol. Sci. 2021, 22, 9614. https://doi.org/10.3390/ijms22179614
Paleskava A, Maksimova EM, Vinogradova DS, Kasatsky PS, Kirillov SV, Konevega AL. Differential Contribution of Protein Factors and 70S Ribosome to Elongation. International Journal of Molecular Sciences. 2021; 22(17):9614. https://doi.org/10.3390/ijms22179614
Chicago/Turabian StylePaleskava, Alena, Elena M. Maksimova, Daria S. Vinogradova, Pavel S. Kasatsky, Stanislav V. Kirillov, and Andrey L. Konevega. 2021. "Differential Contribution of Protein Factors and 70S Ribosome to Elongation" International Journal of Molecular Sciences 22, no. 17: 9614. https://doi.org/10.3390/ijms22179614






