LncRNA-CR11538 Decoys Dif/Dorsal to Reduce Antimicrobial Peptide Products for Restoring Drosophila Toll Immunity Homeostasis
Abstract
:1. Introduction
2. Results
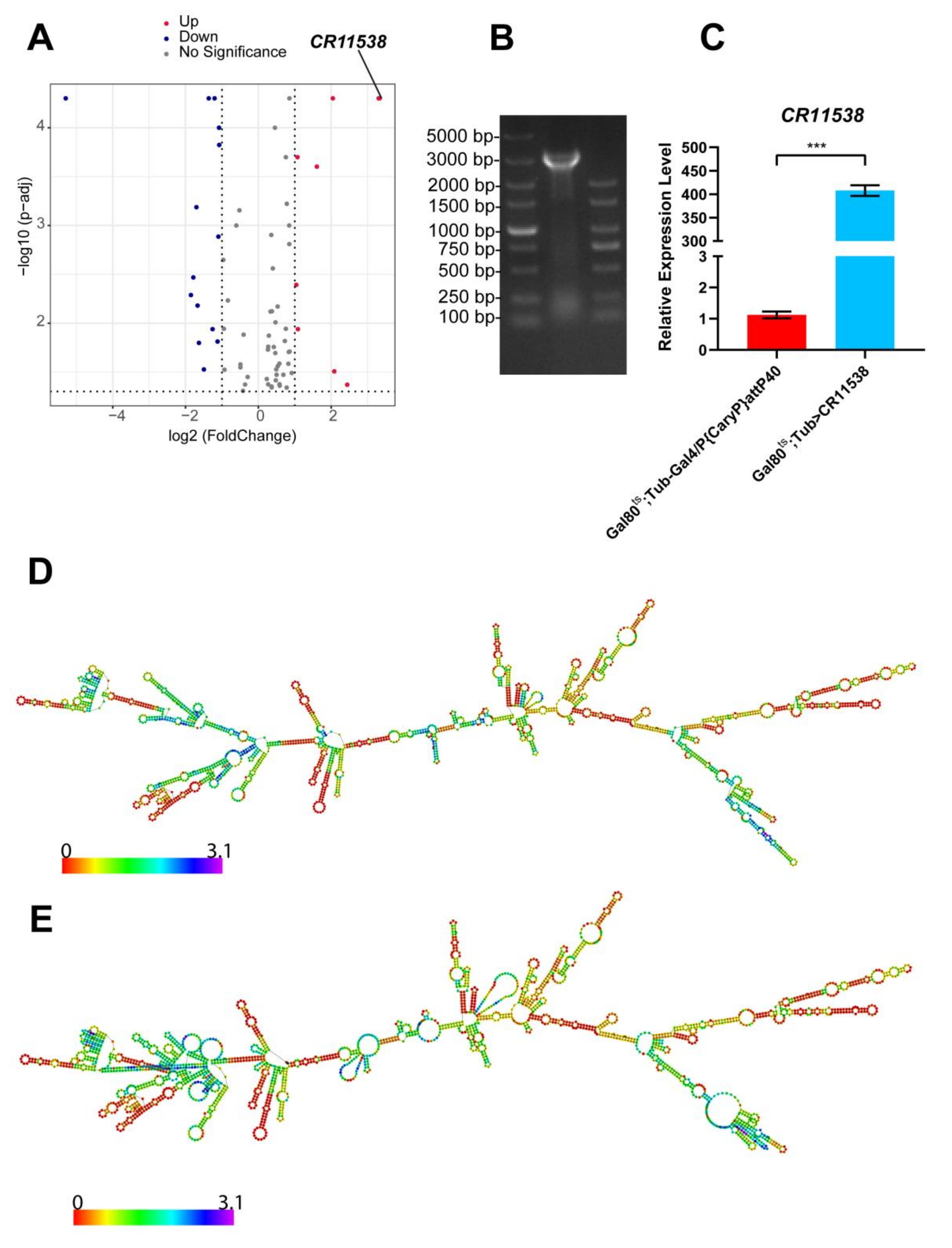
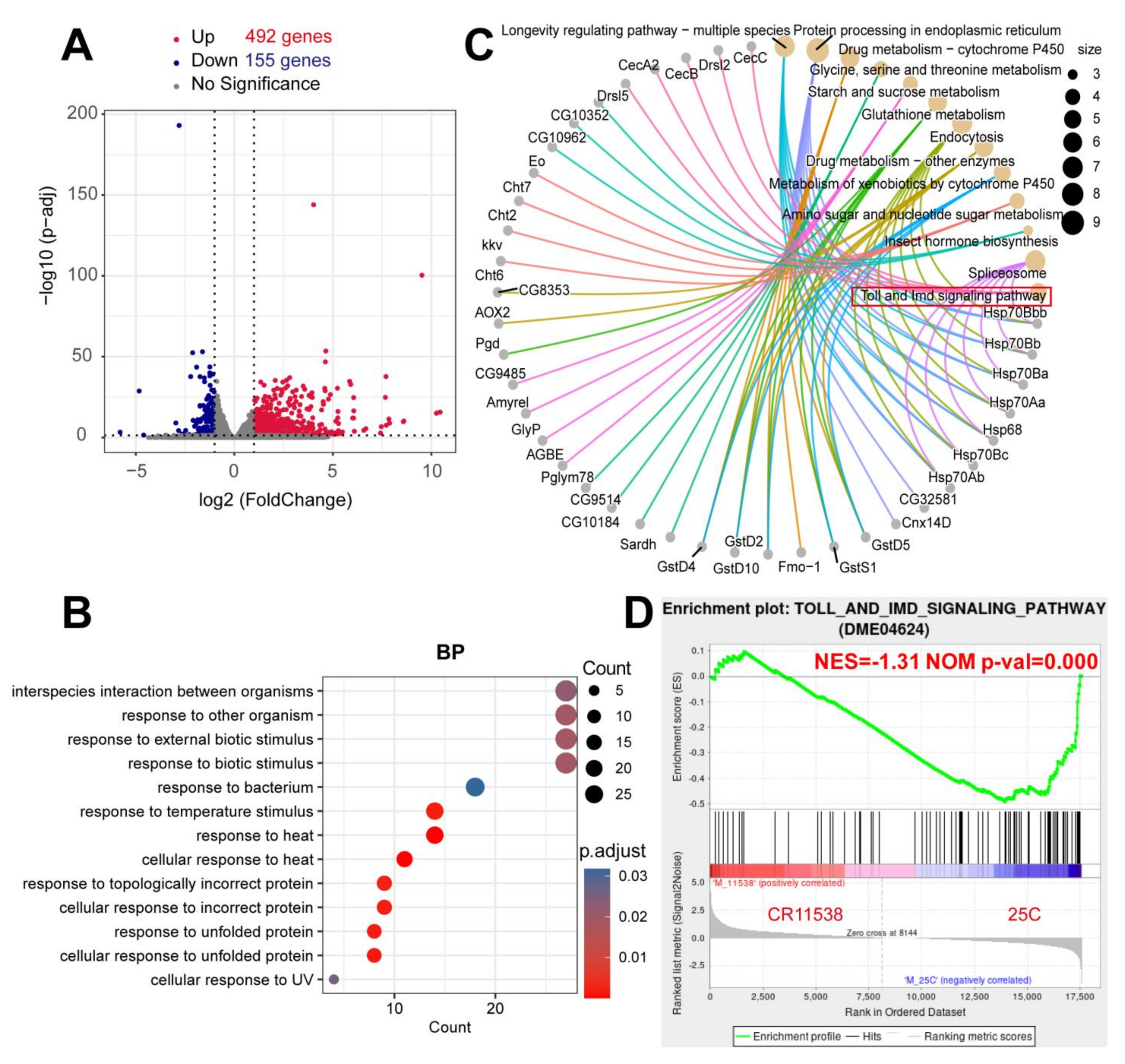
2.1. The Overexpression of lncRNA-CR11538 Reduces AMP Productions of Toll Pathway
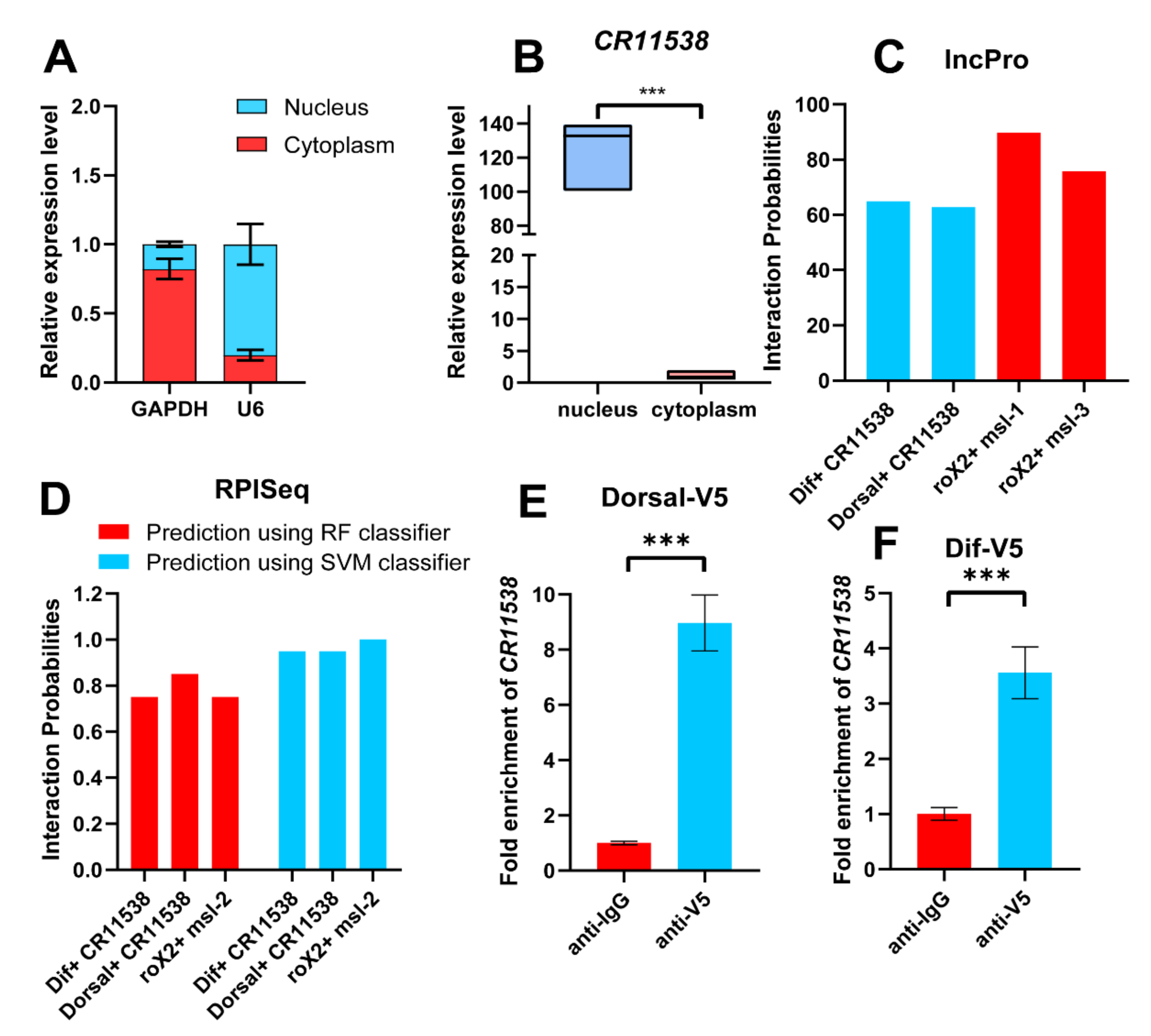
2.2. The Interaction of lncRNA-CR11538 with Dif/Dorsal
2.3. LncRNA-CR11538 Prevents Binding of Dif/Dorsal to AMPs Promoters
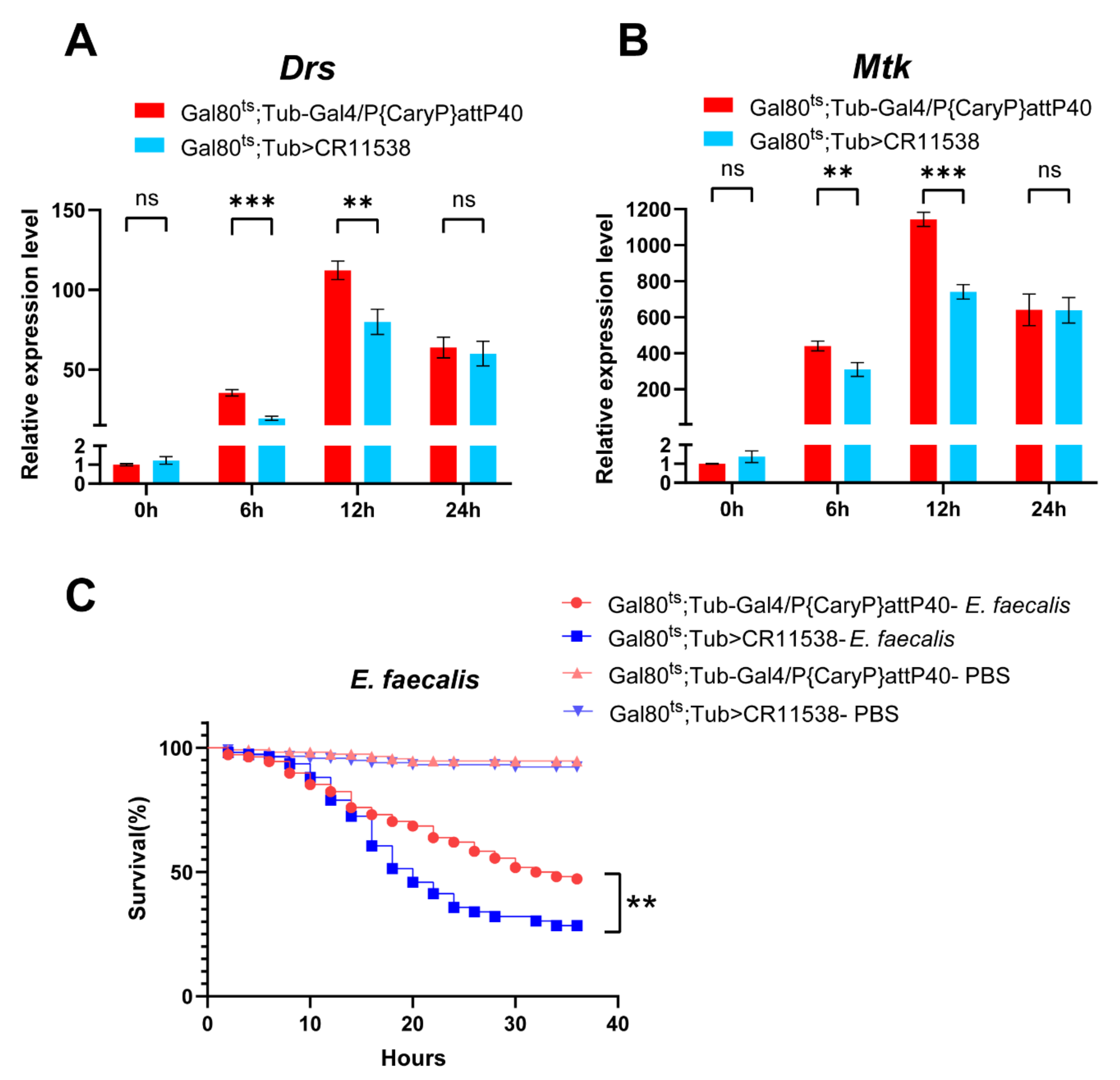
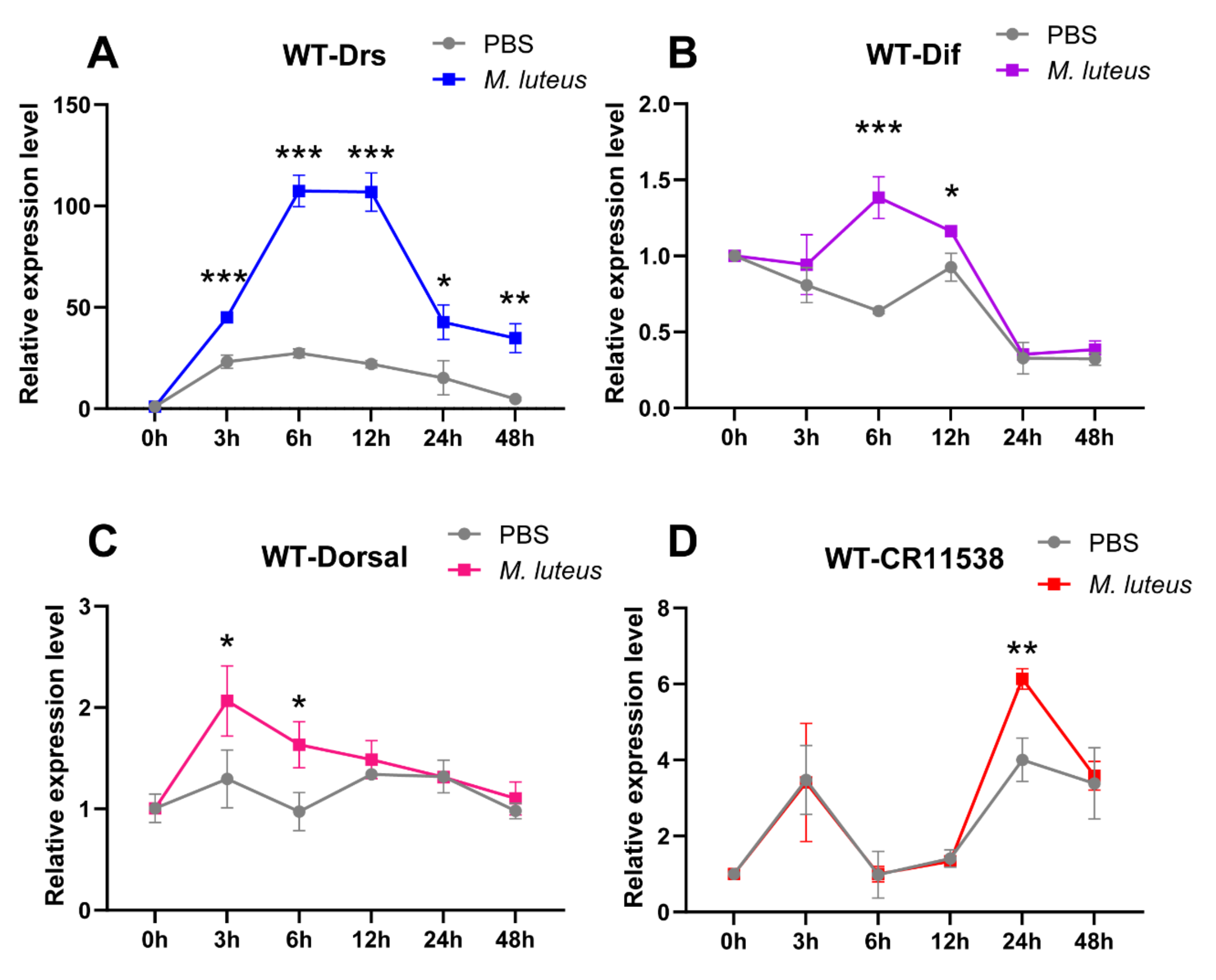
2.4. LncRNA-CR11538 Facilitates Homeostasis Recovery in the Wild-Type Flies after Stimulation
3. Discussion
4. Materials and Methods
4.1. Fly breeding, Strain and Model Construction
4.2. Sepsis and Survival Experiments
4.3. Genomic DNA Extraction and lncRNA Identification in Drosophila
4.4. RNA Extraction and RT-qPCR
4.5. RNA-Sequencing and Enrichment Analyses
4.6. Subcellular Fractionation
4.7. Cell culture and Transfection
4.8. Prediction of Interaction between RNA and Protein
4.9. RNA-Immunoprecipitation (RIP)
4.10. Transcription Factor (TF) Binding Site Prediction
4.11. Chromatin Immunoprecipitation (ChIP)
4.12. Dual Luciferase Reporter Assay
4.13. Data Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carpenter, J.A.; Obbard, D.J.; Maside, X.; Jiggins, F.M. The recent spread of a vertically transmitted virus through populations of Drosophila melanogaster. Mol. Ecol. 2007, 16, 3947–3954. [Google Scholar] [CrossRef] [PubMed]
- Galac, M.R.; Lazzaro, B.P. Comparative pathology of bacteria in the genus Providencia to a natural host, Drosophila melanogaster. Microbes Infect. 2011, 13, 673–683. [Google Scholar] [CrossRef] [Green Version]
- Panayidou, S.; Ioannidou, E.; Apidianakis, Y. Human pathogenic bacteria, fungi, and viruses in Drosophila: Disease modeling, lessons, and shortcomings. Virulence 2014, 5, 253–269. [Google Scholar] [CrossRef] [Green Version]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, T.; Reichhart, J.M.; Hoffmann, J.A.; Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001, 414, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, D.; Liu, Y.; Xia, X.; Gong, W.; Qiu, Y.; Yang, J.; Zheng, Y.; Li, J.; Wang, Y.F.; et al. Drosophila Dicer-2 has an RNA interference-independent function that modulates Toll immune signaling. Sci. Adv. 2015, 1, e1500228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanoh, H.; Tong, L.L.; Kuraishi, T.; Suda, Y.; Momiuchi, Y.; Shishido, F.; Kurata, S. Genome-wide RNAi screening implicates the E3 ubiquitin ligase Sherpa in mediating innate immune signaling by Toll in Drosophila adults. Sci. Signal. 2015, 8, ra107. [Google Scholar] [CrossRef] [Green Version]
- Valanne, S.; Myllymaki, H.; Kallio, J.; Schmid, M.R.; Kleino, A.; Murumagi, A.; Airaksinen, L.; Kotipelto, T.; Kaustio, M.; Ulvila, J.; et al. Genome-wide RNA interference in Drosophila cells identifies G protein-coupled receptor kinase 2 as a conserved regulator of NF-kappaB signaling. J. Immunol. 2010, 184, 6188–6198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.R.; Chen, Z.J.; Kunes, S.; Chang, G.D.; Maniatis, T. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 8322–8327. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Liu, H.; Skolnik, E.Y.; Manley, J.L. Physical and functional interactions between Drosophila TRAF2 and Pelle kinase contribute to Dorsal activation. Proc. Natl. Acad. Sci. USA 2001, 98, 8596–8601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, P.; Huang, L.H.; Steward, R. Cactin, a conserved protein that interacts with the Drosophila IkappaB protein cactus and modulates its function. Mech. Dev. 2000, 94, 57–65. [Google Scholar] [CrossRef]
- Yamamoto-Hino, M.; Muraoka, M.; Kondo, S.; Ueda, R.; Okano, H.; Goto, S. Dynamic regulation of innate immune responses in Drosophila by Senju-mediated glycosylation. Proc. Natl. Acad. Sci. USA 2015, 112, 5809–5814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, S.; Sun, M.; Zheng, X.; Li, L.; Sun, L.; Chen, D.; Sun, Q. Cell-surface localization of Pellino antagonizes Toll-mediated innate immune signalling by controlling MyD88 turnover in Drosophila. Nat. Commun. 2014, 5, 3458. [Google Scholar] [CrossRef] [Green Version]
- Anjum, S.G.; Xu, W.; Nikkholgh, N.; Basu, S.; Nie, Y.; Thomas, M.; Satyamurti, M.; Budnik, B.A.; Ip, Y.T.; Veraksa, A. Regulation of Toll signaling and inflammation by beta-arrestin and the SUMO protease Ulp1. Genetics 2013, 195, 1307–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paddibhatla, I.; Lee, M.J.; Kalamarz, M.E.; Ferrarese, R.; Govind, S. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS Pathog. 2010, 6, e1001234. [Google Scholar] [CrossRef] [Green Version]
- Tipping, M.; Kim, Y.; Kyriakakis, P.; Tong, M.; Shvartsman, S.Y.; Veraksa, A. beta-arrestin Kurtz inhibits MAPK and Toll signalling in Drosophila development. EMBO J. 2010, 29, 3222–3235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belvin, M.P.; Jin, Y.; Anderson, K.V. Cactus protein degradation mediates Drosophila dorsal-ventral signaling. Genes Dev. 1995, 9, 783–793. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Yao, X.; Zhou, H.; Jin, P.; Ma, F. The Drosophila miR-959-962 Cluster Members Repress Toll Signaling to Regulate Antibacterial Defense during Bacterial Infection. Int. J. Mol. Sci. 2021, 22, 886. [Google Scholar] [CrossRef]
- Li, R.; Huang, Y.; Zhang, Q.; Zhou, H.; Jin, P.; Ma, F. The miR-317 functions as a negative regulator of Toll immune response and influences Drosophila survival. Dev. Comp. Immunol. 2019, 95, 19–27. [Google Scholar] [CrossRef]
- Li, S.; Xu, J.; Sun, L.; Li, R.; Jin, P.; Ma, F. Drosophila miR-964 modulates Toll signaling pathway in response to bacterial infection. Dev. Comp. Immunol. 2017, 77, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Y.; Shen, L.; Jin, P.; Chen, L.; Ma, F. miR-958 inhibits Toll signaling and Drosomycin expression via direct targeting of Toll and Dif in Drosophila melanogaster. Am. J. Physiol. Cell Physiol. 2017, 312, C103–C110. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.J.; Hyun, S. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev. Comp. Immunol. 2014, 45, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.K.; Hyun, S. Conserved microRNA miR-8 in fat body regulates innate immune homeostasis in Drosophila. Dev. Comp. Immunol. 2012, 37, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Tian, Y.; Yuan, Y.; Fan, X.; Yang, M.; He, Z.; Yang, D. Insights into the Functions of LncRNAs in Drosophila. Int. J. Mol. Sci. 2019, 20, 4646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef] [Green Version]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Arai, S.; Song, X.; Reichart, D.; Du, K.; Pascual, G.; Tempst, P.; Rosenfeld, M.G.; Glass, C.K.; Kurokawa, R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008, 454, 126–130. [Google Scholar] [CrossRef]
- Pek, J.W.; Osman, I.; Tay, M.L.; Zheng, R.T. Stable intronic sequence RNAs have possible regulatory roles in Drosophila melanogaster. J. Cell Biol. 2015, 211, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Rios-Barrera, L.D.; Gutierrez-Perez, I.; Dominguez, M.; Riesgo-Escovar, J.R. acal is a long non-coding RNA in JNK signaling in epithelial shape changes during drosophila dorsal closure. PLoS Genet 2015, 11, e1004927. [Google Scholar] [CrossRef] [Green Version]
- Herzog, V.A.; Lempradl, A.; Trupke, J.; Okulski, H.; Altmutter, C.; Ruge, F.; Boidol, B.; Kubicek, S.; Schmauss, G.; Aumayr, K.; et al. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat. Genet. 2014, 46, 973–981. [Google Scholar] [CrossRef] [Green Version]
- Pease, B.; Borges, A.C.; Bender, W. Noncoding RNAs of the Ultrabithorax domain of the Drosophila bithorax complex. Genetics 2013, 195, 1253–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathak, R.U.; Mamillapalli, A.; Rangaraj, N.; Kumar, R.P.; Vasanthi, D.; Mishra, K.; Mishra, R.K. AAGAG repeat RNA is an essential component of nuclear matrix in Drosophila. RNA Biol. 2013, 10, 564–571. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.; Krueger, B.J.; Sedore, S.C.; Brogie, J.E.; Rogers, J.T.; Rajendra, T.K.; Saunders, A.; Matera, A.G.; Lis, J.T.; Uguen, P.; et al. The Drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res. 2012, 40, 5283–5297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Xiang, Y.; Liu, X.; Bai, B.; Chen, R.; Liu, L.; Li, M. Long noncoding RNA SMRG regulates Drosophila macrochaetes by antagonizing scute through E(spl)mbeta. RNA Biol. 2019, 16, 42–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardiman, K.E.; Brewster, R.; Khan, S.M.; Deo, M.; Bodmer, R. The bereft gene, a potential target of the neural selector gene cut, contributes to bristle morphogenesis. Genetics 2002, 161, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.K.; Sitnik, J.L.; Frei, Y.; Prince, E.; Gligorov, D.; Wolfner, M.F.; Karch, F. The lncRNA male-specific abdominal plays a critical role in Drosophila accessory gland development and male fertility. PLoS Genet. 2018, 14, e1007519. [Google Scholar] [CrossRef] [Green Version]
- Jenny, A.; Hachet, O.; Zavorszky, P.; Cyrklaff, A.; Weston, M.D.; Johnston, D.S.; Erdelyi, M.; Ephrussi, A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 2006, 133, 2827–2833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muraoka, Y.; Nakamura, A.; Tanaka, R.; Suda, K.; Azuma, Y.; Kushimura, Y.; Lo Piccolo, L.; Yoshida, H.; Mizuta, I.; Tokuda, T.; et al. Genetic screening of the genes interacting with Drosophila FIG4 identified a novel link between CMT-causing gene and long noncoding RNAs. Exp. Neurol. 2018, 310, 1–13. [Google Scholar] [CrossRef]
- Lo Piccolo, L.; Jantrapirom, S.; Nagai, Y.; Yamaguchi, M. FUS toxicity is rescued by the modulation of lncRNA hsromega expression in Drosophila melanogaster. Sci. Rep. 2017, 7, 15660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valanne, S.; Salminen, T.S.; Jarvela-Stolting, M.; Vesala, L.; Ramet, M. Immune-inducible non-coding RNA molecule lincRNA-IBIN connects immunity and metabolism in Drosophila melanogaster. PLoS Pathog. 2019, 15, e1007504. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xu, W.; Gao, X.; Li, W.; Qi, S.; Guo, D.; Ajayi, O.E.; Ding, S.W.; Wu, Q. lncRNA Sensing of a Viral Suppressor of RNAi Activates Non-canonical Innate Immune Signaling in Drosophila. Cell Host. Microbe. 2020, 27, 115–128. [Google Scholar] [CrossRef]
- Zhou, H.; Ni, J.; Wu, S.; Ma, F.; Jin, P.; Li, S. lncRNA-CR46018 positively regulates the Drosophila Toll immune response by interacting with Dif/Dorsal. Dev. Comp. Immunol. 2021, 124, 104183. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Sun, L.; Li, R.; Li, L.; Xu, J.; Ma, F. Dynamic miRNA-mRNA regulations are essential for maintaining Drosophila immune homeostasis during Micrococcus luteus infection. Dev. Comp. Immunol. 2018, 81, 210–224. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef] [Green Version]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, 70–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.M.; Hoffmann, J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef] [Green Version]
- Vedelek, V.; Bodai, L.; Grezal, G.; Kovacs, B.; Boros, I.M.; Laurinyecz, B.; Sinka, R. Analysis of Drosophila melanogaster testis transcriptome. BMC Genom. 2018, 19, 697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.B.; Boley, N.; Eisman, R.; May, G.E.; Stoiber, M.H.; Duff, M.O.; Booth, B.W.; Wen, J.; Park, S.; Suzuki, A.M.; et al. Diversity and dynamics of the Drosophila transcriptome. Nature 2014, 512, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Xiang, S.; Zou, P.; Wu, J.; Zheng, F.; Tang, Q.; Zhou, J.; Hann, S.S. Crosstalk of NF-kappaB/P65 and LncRNA HOTAIR-Mediated Repression of MUC1 Expression Contribute to Synergistic Inhibition of Castration-Resistant Prostate Cancer by Polyphyllin 1-Enzalutamide Combination Treatment. Cell. Physiol. Biochem. 2018, 47, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Zhang, Z.; Liu, H.; Li, W.; Guo, X.; Zhang, Z.; Liu, Y.; Jia, L.; Li, Y.; Ren, Y.; et al. lincRNA-Cox2 regulates NLRP3 inflammasome and autophagy mediated neuroinflammation. Cell Death Differ. 2019, 26, 130–145. [Google Scholar] [CrossRef]
- Neyen, C.; Bretscher, A.J.; Binggeli, O.; Lemaitre, B. Methods to study Drosophila immunity. Methods 2014, 68, 116–128. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldwin, A.S., Jr. The NF-kappa B and I kappa B proteins: New discoveries and insights. Annu. Rev. Immunol. 1996, 14, 649–683. [Google Scholar] [CrossRef] [Green Version]
- Muppirala, U.K.; Honavar, V.G.; Dobbs, D. Predicting RNA-protein interactions using only sequence information. BMC Bioinform. 2011, 12, 489. [Google Scholar] [CrossRef] [Green Version]
- Busse, M.S.; Arnold, C.P.; Towb, P.; Katrivesis, J.; Wasserman, S.A. A kappaB sequence code for pathway-specific innate immune responses. EMBO J. 2007, 26, 3826–3835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Zhou, H.; Jia, C.; Jin, P.; Ma, F. Drosophila Myc restores immune homeostasis of Imd pathway via activating miR-277 to inhibit imd/Tab2. PLoS Genet. 2020, 16, e1008989. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Li, S.; Wu, S.; Jin, P.; Ma, F. LncRNA-CR11538 Decoys Dif/Dorsal to Reduce Antimicrobial Peptide Products for Restoring Drosophila Toll Immunity Homeostasis. Int. J. Mol. Sci. 2021, 22, 10117. https://doi.org/10.3390/ijms221810117
Zhou H, Li S, Wu S, Jin P, Ma F. LncRNA-CR11538 Decoys Dif/Dorsal to Reduce Antimicrobial Peptide Products for Restoring Drosophila Toll Immunity Homeostasis. International Journal of Molecular Sciences. 2021; 22(18):10117. https://doi.org/10.3390/ijms221810117
Chicago/Turabian StyleZhou, Hongjian, Shengjie Li, Shanshan Wu, Ping Jin, and Fei Ma. 2021. "LncRNA-CR11538 Decoys Dif/Dorsal to Reduce Antimicrobial Peptide Products for Restoring Drosophila Toll Immunity Homeostasis" International Journal of Molecular Sciences 22, no. 18: 10117. https://doi.org/10.3390/ijms221810117
APA StyleZhou, H., Li, S., Wu, S., Jin, P., & Ma, F. (2021). LncRNA-CR11538 Decoys Dif/Dorsal to Reduce Antimicrobial Peptide Products for Restoring Drosophila Toll Immunity Homeostasis. International Journal of Molecular Sciences, 22(18), 10117. https://doi.org/10.3390/ijms221810117




