Tumor-Progressive Mechanisms Mediating miRNA–Protein Interaction
Abstract
1. Introduction
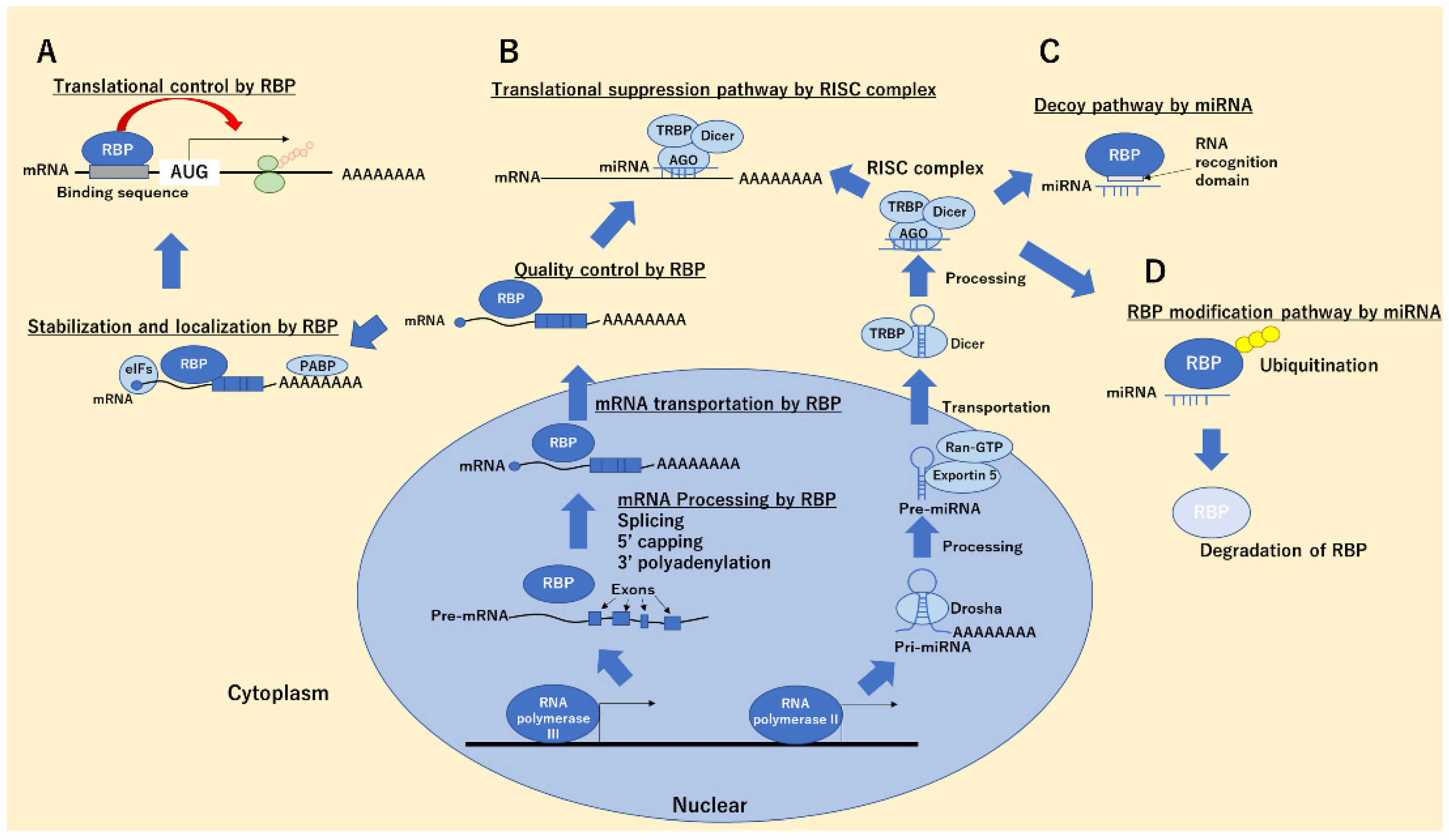
2. miRNA-Mediated Protein Regulation in Cancer Cells
2.1. RNA-Binding Proteins
2.2. RBPs in Cancer Progression and Suppression
2.3. miRNA–RBP Binding Functions in Cancer Cells
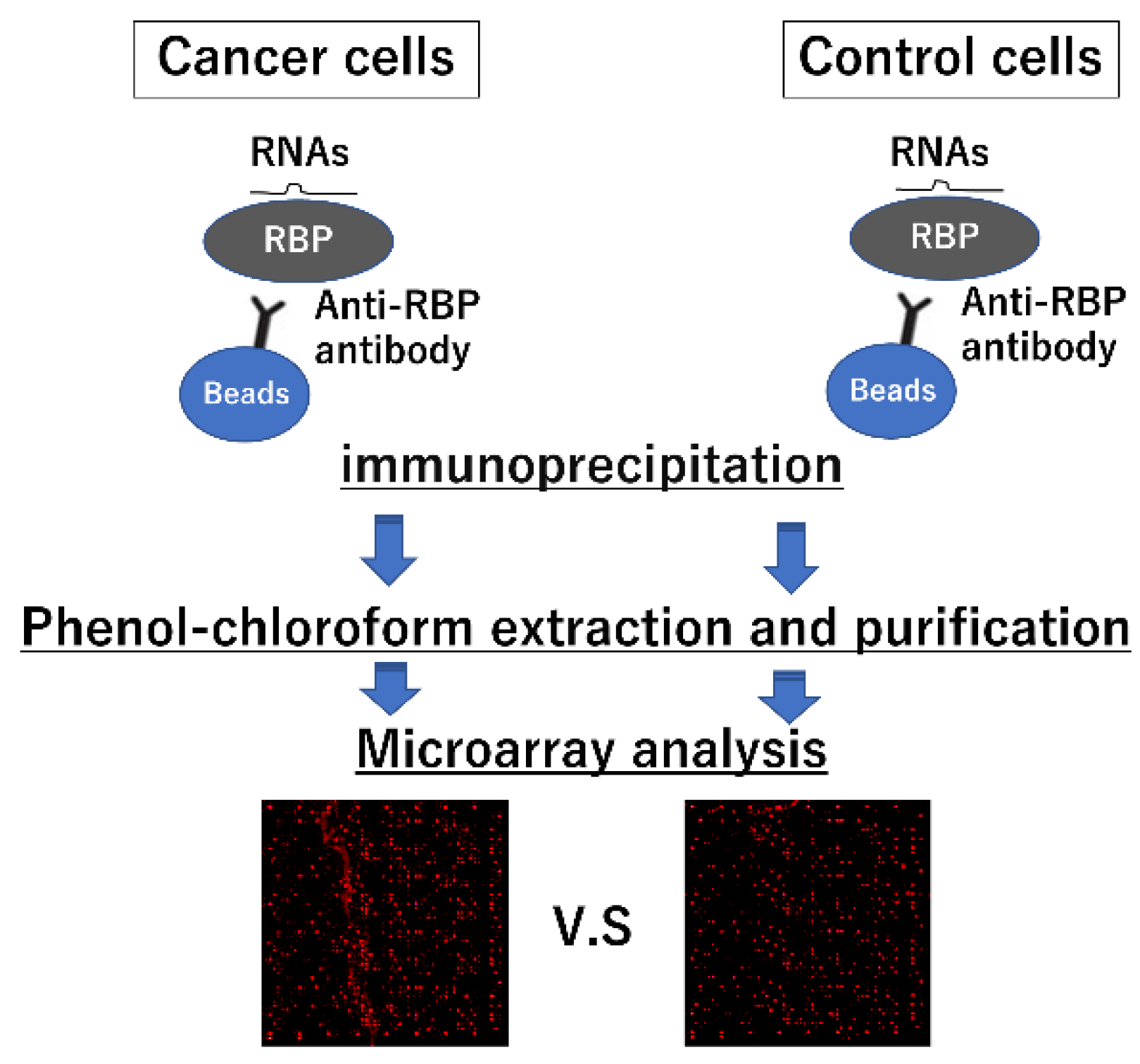
2.4. Strategies for Identifying Interactions between miRNAs and RBPs
3. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Calin, G.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- He, L.; He, X.; Lowe, S.W.; Hannon, G.J. microRNAs join the p53 Network—Another piece in the tumour-suppression puzzle. Nat. Rev. Cancer 2007, 7, 819–822. [Google Scholar] [CrossRef]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-N.; Zhi, Z.; Chen, L.-Y.; Zhou, Q.; Li, X.-M.; Gan, W.-J.; Chen, S.; Yang, M.; Liu, Y.; Shen, T.; et al. SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p. Cancer Lett. 2017, 409, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Chen, T.-H.; Huang, Y.-M.; Wei, P.-L.; Lin, J.-C. Involvement of microRNA in Solid Cancer: Role and Regulatory Mechanisms. Biomedicines 2021, 9, 343. [Google Scholar] [CrossRef]
- Lujambio, A.; Calin, G.; Villanueva, A.; Ropero, S.; Sanchez-Cespedes, M.; Blanco, D.; Montuenga, L.; Rossi, S.; Nicoloso, M.; Faller, W.; et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Zou, D.; Li, Z.; Luo, M.; Dong, L.; Wang, B.; Yin, H.; Ma, Y.; Liu, C.; Wang, F.; et al. MicroRNA-219-2-3p Functions as a Tumor Suppressor in Gastric Cancer and Is Regulated by DNA Methylation. PLoS ONE 2013, 8, e60369. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, J.; Song, Y.; Wang, G.; Gao, P.; Chen, X.; Gao, Z.; Xu, H. Clinical significance of promoter region hypermethylation of microRNA-148a in gastrointestinal cancers. Onco Targets Ther. 2014, 7, 853–863. [Google Scholar] [CrossRef]
- Li, Z.; Li, D.; Zhang, G.; Xiong, J.; Jie, Z.; Cheng, H.; Cao, Y.; Jiang, M.; Lin, L.; Le, Z.; et al. Methylation-associated silencing of MicroRNA-335 contributes tumor cell invasion and migration by interacting with RASA1 in gastric cancer. Am. J. Cancer Res. 2014, 4, 648–662. [Google Scholar]
- Grady, W.M.; Parkin, R.K.; Mitchell, P.; Lee, J.H.; Kim, Y.-H.; Tsuchiya, K.D.; Washington, M.K.; Paraskeva, C.; Willson, J.K.V.; Kaz, A.M.; et al. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene 2008, 27, 3880–3888. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Suzuki, H.; Sasaki, Y.; Maruyama, R.; Imai, K.; Shinomura, Y.; Tokino, T. Epigenetic Silencing of MicroRNA-34b/c and B-Cell Translocation Gene 4 Is Associated with CpG Island Methylation in Colorectal Cancer. Cancer Res. 2008, 68, 4123–4132. [Google Scholar] [CrossRef] [PubMed]
- Balaguer, F.; Link, A.; Lozano, J.J.; Cuatrecasas, M.; Nagasaka, T.; Boland, C.R.; Goel, A. Epigenetic Silencing of miR-137 Is an Early Event in Colorectal Carcinogenesis. Cancer Res. 2010, 70, 6609–6618. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-T.; Wang, J.-L.; Du, W.; Hong, J.; Zhao, S.-L.; Wang, Y.-C.; Xiong, H.; Chen, H.-M.; Fang, J.-Y. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis 2011, 32, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.; Rasheed, S.A.K.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2007, 27, 2128–2136. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Fujiya, M.; Konishi, H.; Sasajima, J.; Fujibayashi, S.; Hayashi, A.; Utsumi, T.; Sato, H.; Iwama, T.; Ijiri, M.; et al. An elevated expression of serum exosomal microRNA-191, − 21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 2018, 18, 116. [Google Scholar] [CrossRef]
- Li, Q.; Li, B.; Wei, S.; He, Z.; Huang, X.; Wang, L.; Xia, Y.; Xu, Z.; Li, Z.; Wang, W.; et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018, 9, 854. [Google Scholar] [CrossRef]
- Ortiz-Sánchez, P.; Orero, M.V.; López-Olañeta, M.M.; Larrasa-Alonso, J.; Sanchez-Cabo, F.; Martí-Gómez, C.; Camafeita, E.; Gómez-Salinero, J.M.; Hernández, L.R.; Nielsen, P.J.; et al. Loss of SRSF3 in Cardiomyocytes Leads to Decapping of Contraction-Related mRNAs and Severe Systolic Dysfunction. Circ. Res. 2019, 125, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Chen, X.; Cui, Y.; Li, W.; Dai, H.; Yue, Q.; Zhang, H.; Zheng, Y.; Guo, X.; Zhu, H. TULP2, a New RNA-Binding Protein, Is Required for Mouse Spermatid Differentiation and Male Fertility. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Tollenaere, M.; Tiedje, C.; Rasmussen, S.; Nielsen, J.C.; Vind, A.C.; Blasius, M.; Batth, T.S.; Mailand, N.; Olsen, J.; Gaestel, M.; et al. GIGYF1/2-Driven Cooperation between ZNF598 and TTP in Posttranscriptional Regulation of Inflammatory Signaling. Cell Rep. 2019, 26, 3511–3521.e4. [Google Scholar] [CrossRef]
- Ando, K.; Fujiya, M.; Konishi, H.; Ueno, N.; Inaba, Y.; Moriichi, K.; Ikuta, K.; Tanabe, H.; Ohtake, T.; Kohgo, Y. Heterogeneous Nuclear Ribonucleoprotein A1 Improves the Intestinal Injury by Regulating Apoptosis Through Trefoil Factor 2 in Mice with Anti-CD3–induced Enteritis. Inflamm. Bowel Dis. 2015, 21, 1541–1552. [Google Scholar] [CrossRef][Green Version]
- Guil, S.; Caceres, J. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007, 14, 591–596. [Google Scholar] [CrossRef]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Lee, Y.; Lee, J.-S. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12, 2699. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, H.; Yuan, T.; Zhou, G.; Li, X.; Wen, K. High hnRNP AB expression is associated with poor prognosis in patients with colorectal cancer. Oncol. Lett. 2019, 18, 6459–6468. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, B.; McKay, M.; Dundas, S.R.; Lawrie, L.C.; Telfer, C.; Murray, G.I. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br. J. Cancer 2006, 95, 921–927. [Google Scholar] [CrossRef]
- Takahashi, K.; Fujiya, M.; Konishi, H.; Murakami, Y.; Iwama, T.; Sasaki, T.; Kunogi, T.; Sakatani, A.; Ando, K.; Ueno, N.; et al. Heterogenous Nuclear Ribonucleoprotein H1 Promotes Colorectal Cancer Progression through the Stabilization of mRNA of Sphingosine-1-Phosphate Lyase 1. Int. J. Mol. Sci. 2020, 21, 4514. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Fu, H.; Zhang, J.; Lu, X.; Yu, F.; Jin, L.; Bai, L.; Huang, B.; Shen, L.; Feng, Y.; et al. RNA-Binding Protein Quaking, a Critical Regulator of Colon Epithelial Differentiation and a Suppressor of Colon Cancer. Gastroenterology 2010, 138, 231–240.e5. [Google Scholar] [CrossRef]
- Lee, H.H.; Son, Y.J.; Lee, W.H.; Park, Y.W.; Chae, S.W.; Cho, W.J.; Kim, Y.M.; Choi, H.-J.; Choi, D.H.; Jung, S.W.; et al. Tristetraprolin regulates expression of VEGF and tumorigenesis in human colon cancer. Int. J. Cancer 2009, 126, 1817–1827. [Google Scholar] [CrossRef] [PubMed]
- Konishi, H.; Kashima, S.; Goto, T.; Ando, K.; Sakatani, A.; Tanaka, H.; Ueno, N.; Moriichi, K.; Okumura, T.; Fujiya, M. The Identification of RNA-Binding Proteins Functionally Associated with Tumor Progression in Gastrointestinal Cancer. Cancers 2021, 13, 3165. [Google Scholar] [CrossRef]
- Konishi, H.; Fujiya, M.; Kashima, S.; Sakatani, A.; Dokoshi, T.; Ando, K.; Ueno, N.; Iwama, T.; Moriichi, K.; Tanaka, H.; et al. A tumor-specific modulation of heterogeneous ribonucleoprotein A0 promotes excessive mitosis and growth in colorectal cancer cells. Cell Death Dis. 2020, 11, 245. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Deng, R.; Zhao, X.; Chen, R.; Hou, G.; Zhang, H.; Wang, Y.; Xu, M.; Jiang, B.; Yu, J. SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis. Mol. Cancer 2017, 16, 157. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Li, S.; Guo, C.; Li, Q.; Zhang, X.; Li, M.; Mi, S. Heterogeneous Nuclear Ribonucleoprotein A1 Loads Batched Tumor-Promoting MicroRNAs Into Small Extracellular Vesicles with the Assist of Caveolin-1 in A549 Cells. Front. Cell Dev. Biol. 2021, 9, 687912. [Google Scholar] [CrossRef]
- Eiring, A.M.; Harb, J.G.; Neviani, P.; Garton, C.; Oaks, J.J.; Spizzo, R.; Liu, S.; Schwind, S.; Santhanam, R.; Hickey, C.J.; et al. miR-328 Functions as an RNA Decoy to Modulate hnRNP E2 Regulation of mRNA Translation in Leukemic Blasts. Cell 2010, 140, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Balkhi, M.Y.; Iwenofu, O.H.; Bakkar, N.; Ladner, K.J.; Chandler, D.S.; Houghton, P.J.; London, C.A.; Kraybill, W.; Perrotti, D.; Croce, C.M.; et al. miR-29 Acts as a Decoy in Sarcomas to Protect the Tumor Suppressor A20 mRNA from Degradation by HuR. Sci. Signal. 2013, 6, ra63. [Google Scholar] [CrossRef]
- Konishi, H.; Fujiya, M.; Ueno, N.; Moriichi, K.; Sasajima, J.; Ikuta, K.; Tanabe, H.; Tanaka, H.; Kohgo, Y. microRNA-26a and -584 inhibit the colorectal cancer progression through inhibition of the binding of hnRNP A1-CDK6 mRNA. Biochem. Biophys. Res. Commun. 2015, 467, 847–852. [Google Scholar] [CrossRef]
- Yao, P.; Wu, J.; Lindner, D.; Fox, P.L. Interplay between miR-574-3p and hnRNP L regulates VEGFA mRNA translation and tumorigenesis. Nucleic Acids Res. 2017, 45, 7950–7964. [Google Scholar] [CrossRef]
- Saul, M.J.; Baumann, I.; Bruno, A.; Emmerich, A.C.; Wellstein, J.; Ottinger, S.M.; Contursi, A.; Dovizio, M.; Donnini, S.; Tacconelli, S.; et al. miR-574-5p as RNA decoy for CUGBP1 stimulates human lung tumor growth by mPGES-1 induction. FASEB J. 2019, 33, 6933–6947. [Google Scholar] [CrossRef]
- Fujiya, M.; Konishi, H.; Kamel, M.K.M.; Ueno, N.T.; Inaba, Y.; Moriichi, K.; Tanabe, H.; Ikuta, K.; Ohtake, T.; Kohgo, Y. microRNA-18a induces apoptosis in colon cancer cells via the autophagolysosomal degradation of oncogenic heterogeneous nuclear ribonucleoprotein A1. Oncogene 2014, 33, 4847–4856. [Google Scholar] [CrossRef]
- Liu, T.-Y.; Chen, Y.-C.; Jong, Y.-J.; Tsai, H.-J.; Lee, C.-C.; Chang, Y.-S.; Chang, J.-G.; Chang, Y.-F. Muscle developmental defects in heterogeneous nuclear Ribonucleoprotein A1 knockout mice. Open Biol. 2017, 7, 160303. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef]
- Watanabe, S.; Hayashi, K.; Toh, K.; Kim, H.J.; Liu, X.; Chaya, H.; Fukushima, S.; Katsushima, K.; Kondo, Y.; Uchida, S.; et al. In vivo rendezvous of small nucleic acid drugs with charge-matched block catiomers to target cancers. Nat. Commun. 2019, 10, 1894. [Google Scholar] [CrossRef] [PubMed]
- Umezu, T.; Takanashi, M.; Murakami, Y.; Ohno, S.-I.; Kanekura, K.; Sudo, K.; Nagamine, K.; Takeuchi, S.; Ochiya, T.; Kuroda, M. Acerola exosome-like nanovesicles to systemically deliver nucleic acid medicine via oral administration. Mol. Ther. Methods Clin. Dev. 2021, 21, 199–208. [Google Scholar] [CrossRef]
- Konishi, H.; Fujiya, M.; Kohgo, Y. Traffic Control of Bacteria-Derived Molecules: A New System of Host-Bacterial Crosstalk. Int. J. Cell Biol. 2013, 2013, 757148. [Google Scholar] [CrossRef] [PubMed][Green Version]

| miRNA | Type of Pathway | Target | Type of Cancer | Function | Reference |
|---|---|---|---|---|---|
| miR-328 | Decoy | hnRNP E2-CEBPα mRNA | Leukemic blasts | Differentiation | Eiring AM, Cell 2010 |
| Canonical | PIM1 mRNA | Decreased survival | |||
| miR-29 | Decoy | HuR-A20 mRNA | Sarcoma | Differentiation | Balkhi MY, Sci signal, 2013 |
| miR-26a, -584 | Decoy | hnRNP A1-CDK6 mRNA | Colorectal cancer | Cell growth suppression | Konishi H, Biochem Biophys Res Commun. 2015 |
| miR-574-3p | Decoy | hnRNP L-VEGFA mRNA | Myeloid cells | Inhibition of cell proliferation | Yao P, Nucleic Acids Research, 2017 |
| Canonical | EP300 mRNA | ||||
| miR-574-5p | Decoy | CUGBP1-mPGES-1 mRNA | Lung tumor | Cell growth promotion | Saul MJ, FASEB J, 2019 |
| miR-18a | Degradation | hnRNP A1 | Colorectal cancer | Apoptosis induction | Fujiya M, Oncogene, 2014 |
| miRs with Greater than 4-Fold Expression Compared to the Isotype Control IgG | |||
|---|---|---|---|
| Name | ID | Ratio (hnRNP A1/IgG) | LOG2ratio |
| hsa-miR-29a-3p | MIMAT0000086 | 11.49 | 3.52 |
| hsa-miR-26a-5p | MIMAT0000082 | 11.37 | 3.51 |
| hsa-miR-584-5p | MIMAT0003249 | 9.93 | 3.31 |
| hsa-miR-107 | MIMAT0000104 | 9.73 | 3.28 |
| hsa-miR-106b-5p | MIMAT0000680 | 8.99 | 3.17 |
| hsa-miR-1229-5p | MIMAT0022942 | 8.88 | 3.15 |
| hsa-miR-29b-3p | MIMAT0000100 | 8.07 | 3.01 |
| hsa-miR-194-5p | MIMAT0000460 | 8.07 | 3.01 |
| hsa-miR-142-3p | MIMAT0000434 | 7.97 | 2.99 |
| hsa-miR-18a-5p | MIMAT0000072 | 7.93 | 2.99 |
| hsa-let-7c-5p | MIMAT0000064 | 7.31 | 2.87 |
| hsa-miR-16-5p | MIMAT0000069 | 7.22 | 2.85 |
| hsa-miR-500a-3p | MIMAT0002871 | 6.96 | 2.8 |
| hsa-miR-200b-3p | MIMAT0000318 | 6.89 | 2.78 |
| hsa-miR-19a-3p | MIMAT0000073 | 6.55 | 2.71 |
| hsa-miR-222-3p | MIMAT0000279 | 6.4 | 2.68 |
| hsa-let-7b-5p | MIMAT0000063 | 5.98 | 2.58 |
| hsa-miR-23a-3p | MIMAT0000078 | 5.97 | 2.58 |
| hsa-let-7d-5p | MIMAT0000065 | 5.85 | 2.55 |
| hsa-miR-431-3p | MIMAT0004757 | 5.7 | 2.51 |
| hsa-miR-200c-3p | MIMAT0000617 | 5.62 | 2.49 |
| hsa-miR-23b-3p | MIMAT0000418 | 5.27 | 2.4 |
| hsa-miR-27b-3p | MIMAT0000419 | 5.23 | 2.39 |
| hsa-miR-19b-3p | MIMAT0000074 | 5.05 | 2.34 |
| hsa-miR-103a-3p | MIMAT0000101 | 4.95 | 2.31 |
| hsa-miR-1246 | MIMAT0005898 | 4.73 | 2.24 |
| hsa-let-7a-5p | MIMAT0000062 | 4.66 | 2.22 |
| hsa-miR-20a-5p | MIMAT0000075 | 4.5 | 2.17 |
| hsa-miR-27a-3p | MIMAT0000084 | 4.49 | 2.17 |
| hsa-miR-141-3p | MIMAT0000432 | 4.32 | 2.11 |
| hsa-miR-21-5p | MIMAT0000076 | 4.25 | 2.09 |
| hsa-miR-17-5p | MIMAT0000070 | 4.24 | 2.08 |
| hsa-miR-106a-5p | MIMAT0000103 | 4.15 | 2.05 |
| hsa-miR-20b-5p | MIMAT0001413 | 4.11 | 2.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konishi, H.; Sato, H.; Takahashi, K.; Fujiya, M. Tumor-Progressive Mechanisms Mediating miRNA–Protein Interaction. Int. J. Mol. Sci. 2021, 22, 12303. https://doi.org/10.3390/ijms222212303
Konishi H, Sato H, Takahashi K, Fujiya M. Tumor-Progressive Mechanisms Mediating miRNA–Protein Interaction. International Journal of Molecular Sciences. 2021; 22(22):12303. https://doi.org/10.3390/ijms222212303
Chicago/Turabian StyleKonishi, Hiroaki, Hiroki Sato, Kenji Takahashi, and Mikihiro Fujiya. 2021. "Tumor-Progressive Mechanisms Mediating miRNA–Protein Interaction" International Journal of Molecular Sciences 22, no. 22: 12303. https://doi.org/10.3390/ijms222212303
APA StyleKonishi, H., Sato, H., Takahashi, K., & Fujiya, M. (2021). Tumor-Progressive Mechanisms Mediating miRNA–Protein Interaction. International Journal of Molecular Sciences, 22(22), 12303. https://doi.org/10.3390/ijms222212303







