Wood Formation under Severe Drought Invokes Adjustment of the Hormonal and Transcriptional Landscape in Poplar
Abstract
:1. Introduction
2. Results
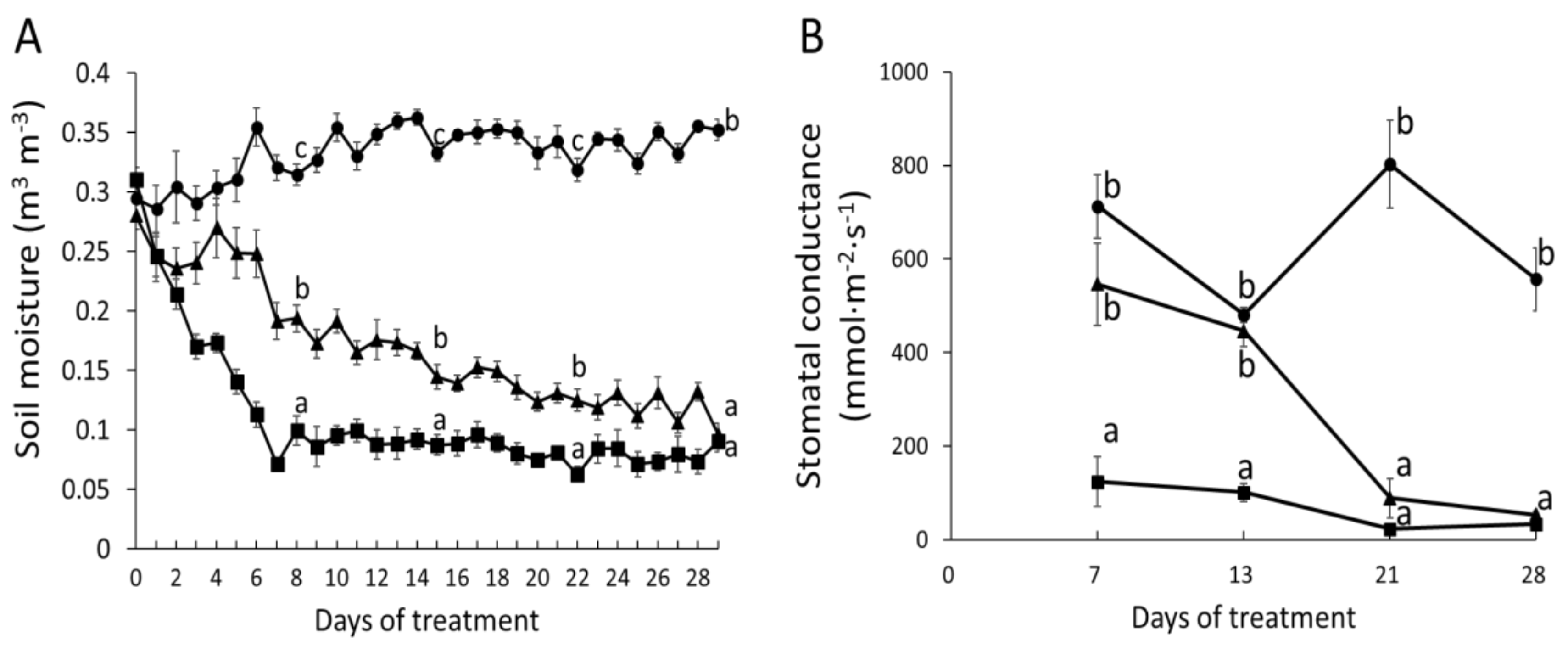
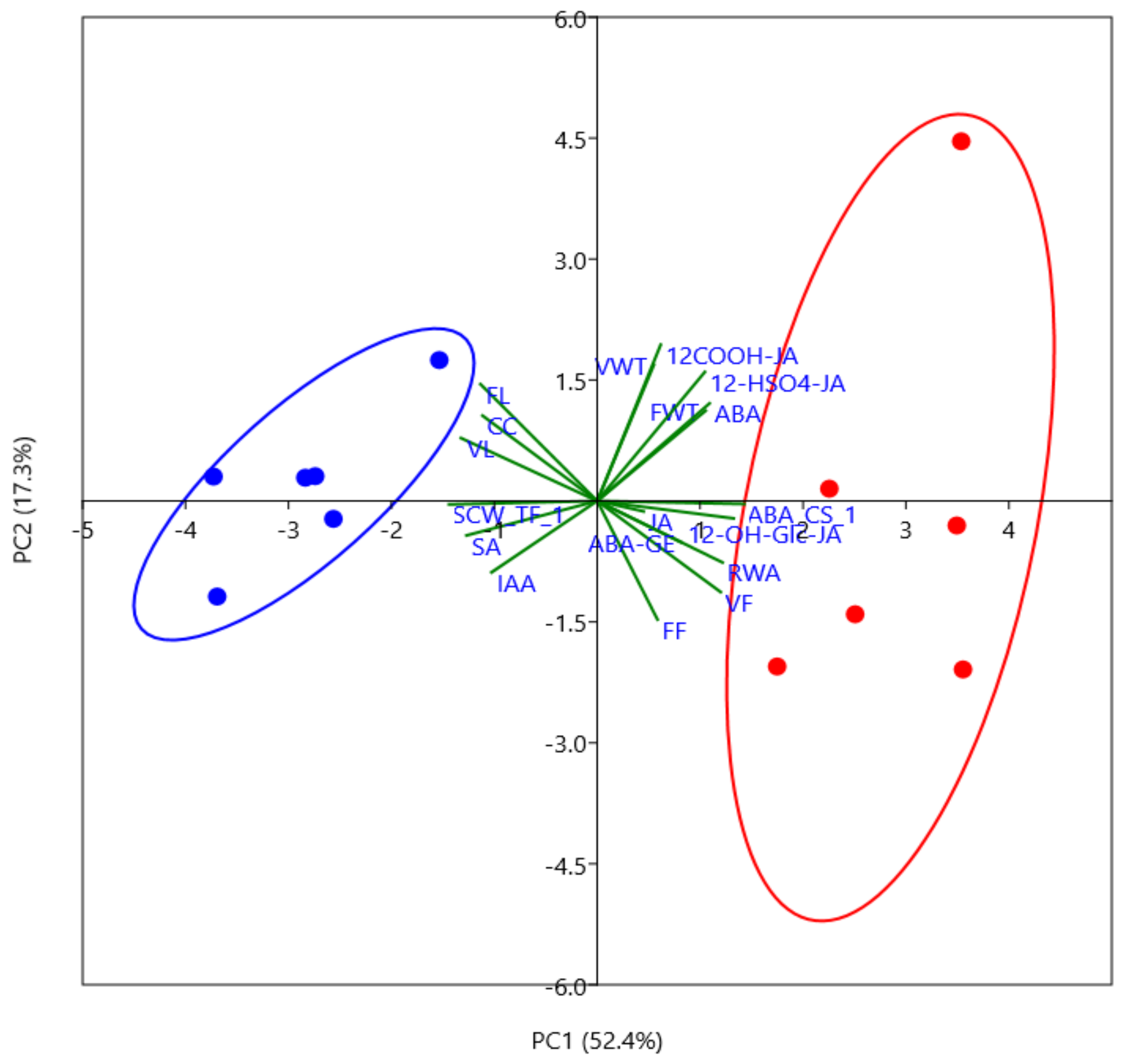
2.1. Phytohormone Profiling Detects Tissue-Specific Drought Responses Concurring with Physiological Stress Adaptation
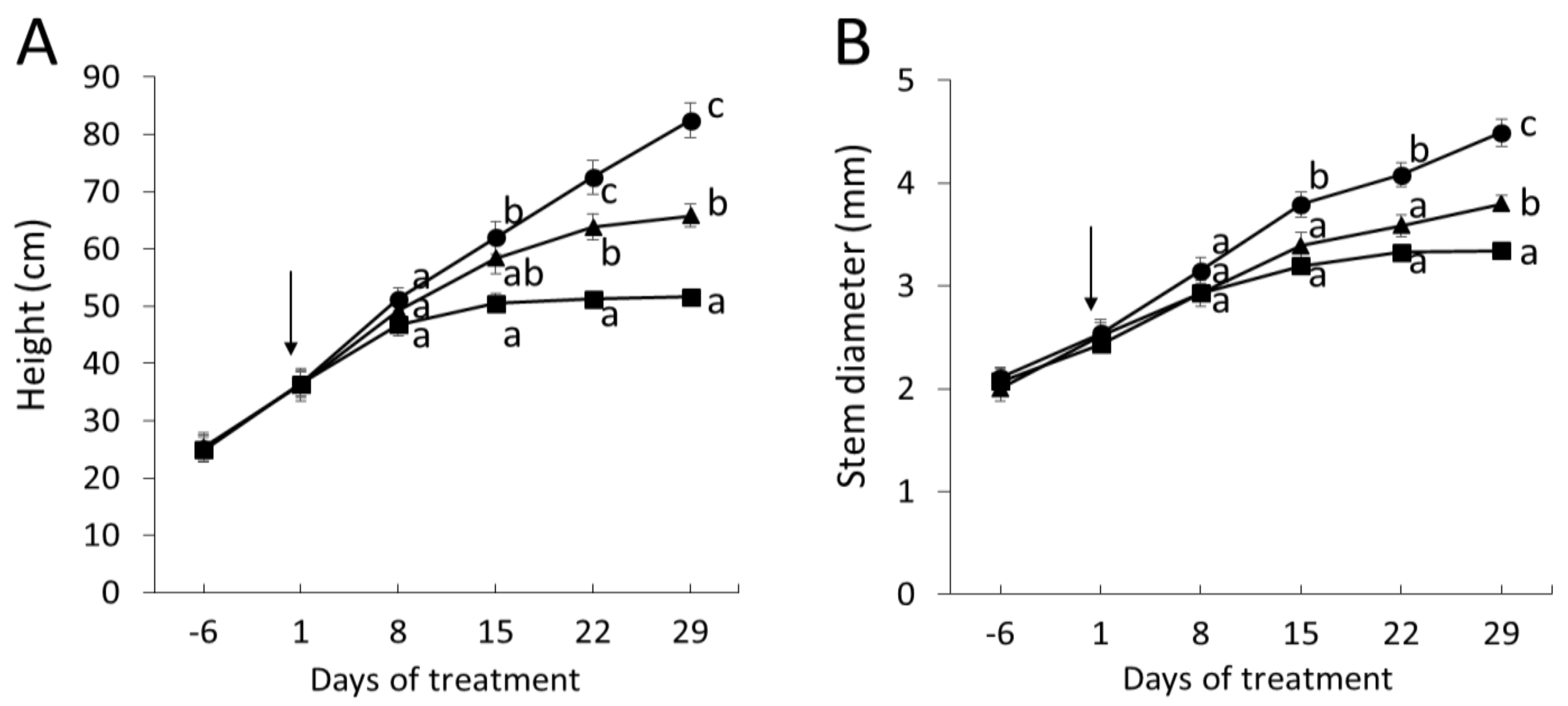
2.2. Wood Anatomical Characteristics Are Strongly Changed in Response to Severe Drought
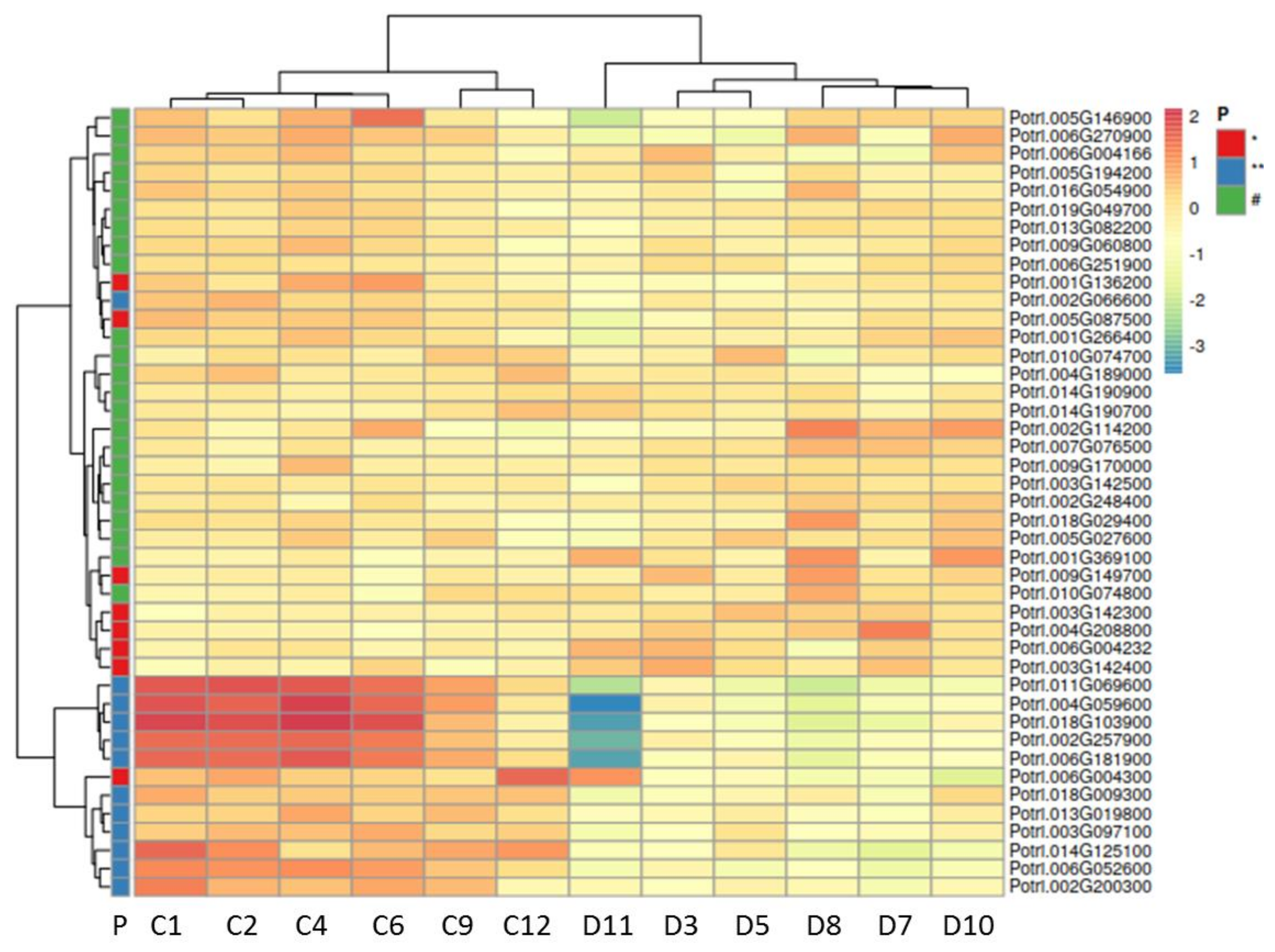
2.3. Drought Stress Reprograms the Wood Transcriptome
2.4. Transcriptional Regulation of ABA and Other Phytohormones in Xylem under Severe Drought
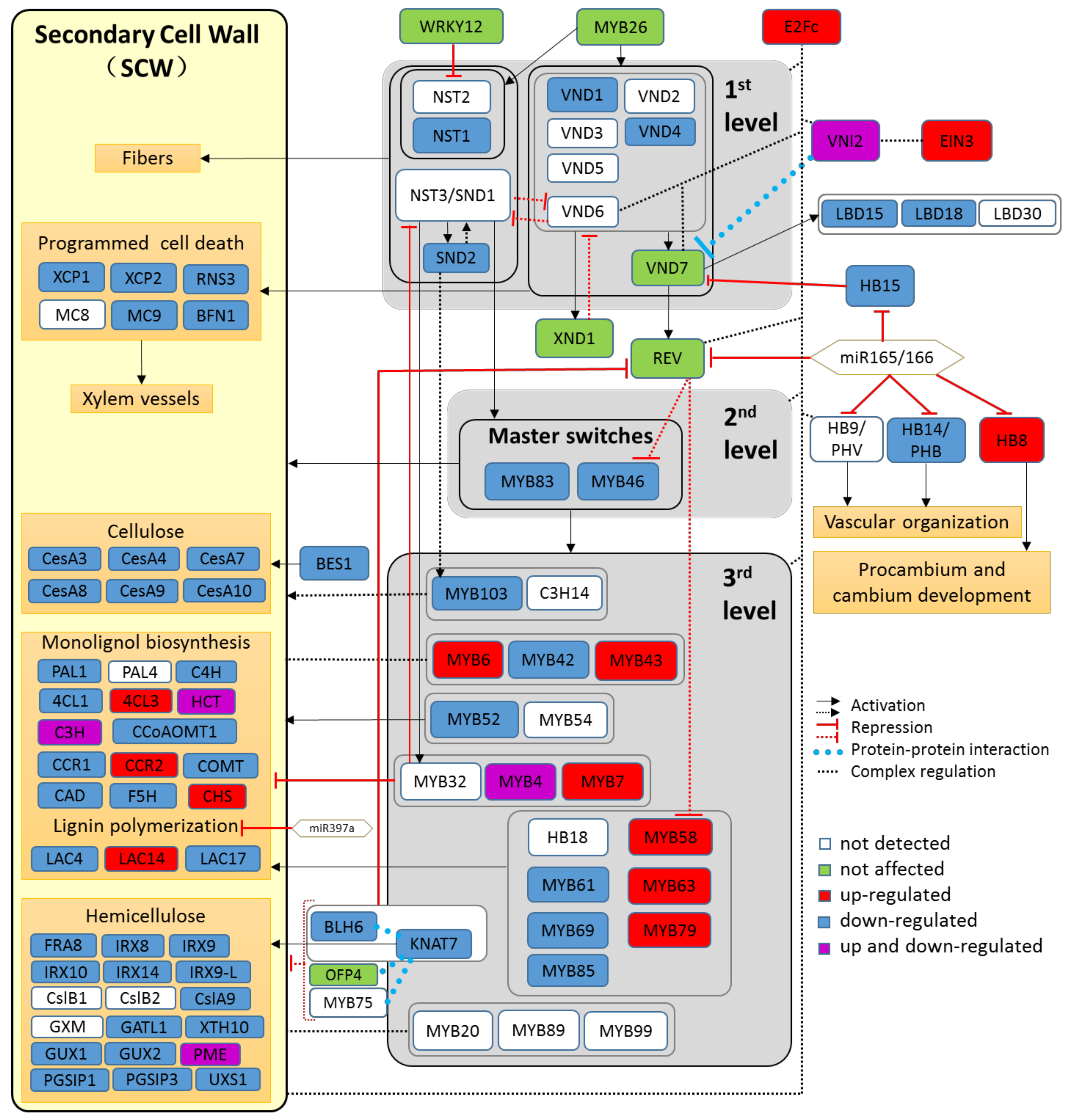
2.5. The Transcriptional Cascade Governing Secondary Cell Wall Formation Is Suppressed by Drought
2.6. Fiber Cell Wall Thickness Is Correlated with ABA Concentrations
3. Discussion
3.1. ABA Is Strongly Regulated in Drought-Stressed Wood
3.2. Drought Uncovers Antagonistic Effects on Wood Anatomy, Transcriptional Regulation of the SCW Cascade and ABA Core Signaling
4. Materials and Methods
4.1. Plant Materials and Drought Treatment
4.2. Sampling and Biomass
4.3. Wood Anatomical Analysis
4.4. Phytohormone Measurements
4.5. Statistical Analyses of Physiological Data
4.6. RNA Extraction and Sequencing
4.7. Bioinformatic Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, G.; Donnison, I.S.; Murphy-Bokern, D.; Morgante, M.; Bogeat-Triboulot, M.-B.; Bhalerao, R.; Hertzberg, M.; Polle, A.; Harfouche, A.; Alasia, F.; et al. Sustainable Bioenergy for Climate Mitigation: Developing Drought-Tolerant Trees and Grasses. Ann. Bot. 2019, 124, 513–520. [Google Scholar] [CrossRef] [Green Version]
- Bentsen, N.S.; Felby, C. Biomass for Energy in the European Union—A Review of Bioenergy Resource Assessments. Biotechnol. Biofuels 2012, 5, 25. [Google Scholar] [CrossRef] [Green Version]
- Polle, A.; Chen, S.L.; Eckert, C.; Harfouche, A. Engineering Drought Resistance in Forest Trees. Front. Plant Sci. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Harfouche, A.; Meilan, R.; Altman, A. Molecular and Physiological Responses to Abiotic Stress in Forest Trees and Their Relevance to Tree Improvement. Tree Physiol. 2014, 34, 1181–1198. [Google Scholar] [CrossRef] [Green Version]
- Passioura, J.B. Drought and Drought Tolerance. Plant Growth Regul. 1996, 20, 79–83. [Google Scholar] [CrossRef]
- Fischer, U.; Polle, A. Populus responses to abiotic stress. In Genetics and Genomics of Populus; Jansson, S., Bhalerao, R., Groover, A., Eds.; Springer: Berlin, Germany, 2010; pp. 225–247. [Google Scholar]
- Brunner, I.; Herzog, C.; Dawes, M.A.; Arend, M.; Sperisen, C. How Tree Roots Respond to Drought. Front. Plant Sci. 2015, 6, 547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agustí, J.; Blázquez, M.A. Plant Vascular Development: Mechanisms and Environmental Regulation. Cell. Mol. Life Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hertzberg, M.; Aspeborg, H.; Schrader, J.; Andersson, A.; Erlandsson, R.; Blomqvist, K.; Bhalerao, R.; Uhlen, M.; Teeri, T.T.; Lundeberg, J.; et al. A Transcriptional Roadmap to Wood Formation. Proc. Natl. Acad. Sci. USA 2001, 98, 14732–14737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. AspWood: High-Spatial-Resolution Transcriptome Profiles Reveal Uncharacterized Modularity of Wood Formation in Populus Tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hacke, U. Functional and Ecological Xylem Anatomy; Springer: Berlin/Heidelberg, Germany, 2015; ISBN 978-3-319-15783-2. [Google Scholar]
- Eckert, C.; Sharmin, S.; Kogel, A.; Yu, D.; Kins, L.; Strijkstra, G.-J.; Polle, A. What Makes the Wood? Exploring the Molecular Mechanisms of Xylem Acclimation in Hardwoods to an Ever-Changing Environment. Forests 2019, 10, 358. [Google Scholar] [CrossRef] [Green Version]
- Plomion, C.; Leprovost, G.; Stokes, A. Wood Formation in Trees. Plant Physiol. 2001, 127, 1513–1523. [Google Scholar] [CrossRef]
- Arend, M.; Fromm, J. Seasonal Change in the Drought Response of Wood Cell Development in Poplar. Tree Physiol. 2007, 27, 985–992. [Google Scholar] [CrossRef] [Green Version]
- Bogeat-Triboulot, M.-B.; Brosche, M.; Renaut, J.; Jouve, L.; Le Thiec, D.; Fayyaz, P.; Vinocur, B.; Witters, E.; Laukens, K.; Teichmann, T.; et al. Gradual Soil Water Depletion Results in Reversible Changes of Gene Expression, Protein Profiles, Ecophysiology, and Growth Performance in Populus Euphratica, a Poplar Growing in Arid Regions. Plant Physiol. 2007, 143, 876–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wildhagen, H.; Paul, S.; Allwright, M.; Smith, H.K.; Malinowska, M.; Schnabel, S.K.; Paulo, M.J.; Cattonaro, F.; Vendramin, V.; Scalabrin, S.; et al. Genes and Gene Clusters Related to Genotype and Drought-Induced Variation in Saccharification Potential, Lignin Content and Wood Anatomical Traits in Populus Nigra. Tree Physiol. 2018, 38, 320–339. [Google Scholar] [CrossRef] [PubMed]
- Bhalerao, R.P.; Fischer, U. Environmental and Hormonal Control of Cambial Stem Cell Dynamics. J. Exp. Bot. 2017, 68, 79–87. [Google Scholar] [CrossRef]
- Didi, V.; Jackson, P.; Hejátko, J. Hormonal Regulation of Secondary Cell Wall Formation. J. Exp. Bot. 2015, 66, 5015–5027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, M.; Tuskan, G.A.; Muchero, W.; Chen, J.-G. Recent Advances in the Transcriptional Regulation of Secondary Cell Wall Biosynthesis in the Woody Plants. Front. Plant Sci. 2018, 9, 1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis Gene Regulatory Network for Secondary Cell Wall Synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turco, G.M.; Rodriguez-Medina, J.; Siebert, S.; Han, D.; Valderrama-Gómez, M.Á.; Vahldick, H.; Shulse, C.N.; Cole, B.J.; Juliano, C.E.; Dickel, D.E.; et al. Molecular Mechanisms Driving Switch Behavior in Xylem Cell Differentiation. Cell Rep. 2019, 28, 342–351.e4. [Google Scholar] [CrossRef] [Green Version]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription Switches for Protoxylem and Metaxylem Vessel Formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Demura, T.; Ye, Z.-H. SND1, a NAC Domain Transcription Factor, Is a Key Regulator of Secondary Wall Synthesis in Fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. Two NAC Domain Transcription Factors, SND1 and NST1, Function Redundantly in Regulation of Secondary Wall Synthesis in Fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC Transcription Factors, NST1 and NST3, Are Key Regulators of the Formation of Secondary Walls in Woody Tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC Domain Transcription Factor, Negatively Regulates Xylem Vessel Formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The Populus Class III HD ZIP Transcription Factor POPCORONA Affects Cell Differentiation during Secondary Growth of Woody Stems. PLoS ONE 2011, 6, e17458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Lee, C.; Ye, Z.-H. Functional Characterization of Poplar Wood-Associated NAC Domain Transcription Factors. Plant Physiol. 2010, 152, 1044–1055. [Google Scholar] [CrossRef] [Green Version]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX Gene KNAT7 Negatively Regulates Secondary Wall Formation in Arabidopsis and Is Functionally Conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, A.; Ahad, A.; Wang, S.; Mansfield, S.D.; Haughn, G.W.; Douglas, C.J.; Ellis, B.E. The Interacting MYB75 and KNAT7 Transcription Factors Modulate Secondary Cell Wall Deposition Both in Stems and Seed Coat in Arabidopsis. Planta 2013, 237, 1199–1211. [Google Scholar] [CrossRef]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional Repression by AtMYB4 Controls Production of UV-Protecting Sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.-H.; Kim, W.-C.; Han, K.-H. Ectopic Expression of MYB46 Identifies Transcriptional Regulatory Genes Involved in Secondary Wall Biosynthesis in Arabidopsis. Plant J. 2009, 60, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Ye, Z.-H. Evolutionary Conservation of the Transcriptional Network Regulating Secondary Cell Wall Biosynthesis. Trends Plant Sci. 2010, 15, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Junghans, U.; Langenfeld-Heyser, R.; Polle, A.; Teichmann, T. Effect of Auxin Transport Inhibitors and Ethylene on the Wood Anatomy of Poplar. Plant Biol. 2004, 6, 22–29. [Google Scholar] [PubMed]
- Junghans, U.; Polle, A.; Düchting, P.; Weiler, E.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to High Salinity in Poplar Involves Changes in Xylem Anatomy and Auxin Physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef] [Green Version]
- Nieminen, K.; Immanen, J.; Laxell, M.; Kauppinen, L.; Tarkowski, P.; Dolezal, K.; Tähtiharju, S.; Elo, A.; Decourteix, M.; Ljung, K.; et al. Cytokinin Signaling Regulates Cambial Development in Poplar. Proc. Natl. Acad. Sci. USA 2008, 105, 20032–20037. [Google Scholar] [CrossRef] [Green Version]
- Dayan, J.; Schwarzkopf, M.; Avni, A.; Aloni, R. Enhancing Plant Growth and Fiber Production by Silencing GA 2-Oxidase. Plant Biotechnol. J. 2010, 8, 425–435. [Google Scholar] [CrossRef]
- Agusti, J.; Herold, S.; Schwarz, M.; Sanchez, P.; Ljung, K.; Dun, E.A.; Brewer, P.B.; Beveridge, C.A.; Sieberer, T.; Sehr, E.M.; et al. Strigolactone Signaling Is Required for Auxin-Dependent Stimulation of Secondary Growth in Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 20242–20247. [Google Scholar] [CrossRef] [Green Version]
- Aloni, R. Role of Hormones in Controlling Vascular Differentiation and the Mechanism of Lateral Root Initiation. Planta 2013, 238, 819–830. [Google Scholar] [CrossRef]
- Christmann, A.; Moes, D.; Himmelbach, A.; Yang, Y.; Tang, Y.; Grill, E. Integration of Abscisic Acid Signalling into Plant Responses. Plant Biol. 2006, 8, 314–325. [Google Scholar] [CrossRef]
- Dörffling, K.; Streich, J.; Kruse, W.; Muxfeldt, B. Abscisic Acid and the After-Effect of Water Stress on Stomatal Opening Potential. Z. Für Pflanzenphysiol. 1977, 81, 43–56. [Google Scholar] [CrossRef]
- Creelman, R.A.; Mason, H.S.; Bensen, R.J.; Boyer, J.S.; Mullet, J.E. Water Deficit and Abscisic Acid Cause Differential Inhibition of Shoot versus Root Growth in Soybean Seedlings: Analysis of Growth, Sugar Accumulation, and Gene Expression. Plant Physiol. 1990, 92, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Wang, S.; Hüttermann, A.; Altman, A. Xylem Abscisic Acid Accelerates Leaf Abscission by Modulating Polyamine and Ethylene Synthesis in Water-Stressed Intact Poplar. Trees 2002, 16, 16–22. [Google Scholar] [CrossRef]
- Tardieu, F.; Parent, B.; Simonneau, T. Control of Leaf Growth by Abscisic Acid: Hydraulic or Non-Hydraulic Processes? Plant Cell Environ. 2010, 33, 636–647. [Google Scholar] [CrossRef]
- Furukawa, J.; Abe, Y.; Mizuno, H.; Matsuki, K.; Sagawa, K.; Kojima, M.; Sakakibara, H.; Iwai, H.; Satoh, S. Seasonal Fluctuation of Organic and Inorganic Components in Xylem Sap of Populus Nigra. Plant Root 2011, 5, 56–62. [Google Scholar] [CrossRef] [Green Version]
- Kasper, K.; Abreu, I.N.; Feussner, K.; Zienkiewicz, K.; Herrfurth, C.; Ischebeck, T.; Janz, D.; Majcherczyk, A.; Schmitt, K.; Valerius, O.; et al. Multi-Omics Analysis of Xylem Sap Uncovers Dynamic Modulation of Poplar Defenses by Ammonium and Nitrate. bioRxiv 2021. [Google Scholar] [CrossRef]
- Yu, D.; Wildhagen, H.; Tylewicz, S.; Miskolczi, P.C.; Bhalerao, R.P.; Polle, A. Abscisic Acid Signalling Mediates Biomass Trade-off and Allocation in Poplar. New Phytol. 2019, 223, 1192–1203. [Google Scholar] [CrossRef]
- Campbell, L.; Etchells, J.P.; Cooper, M.; Kumar, M.; Turner, S.R. An Essential Role for Abscisic Acid in the Regulation of Xylem Fibre Differentiation. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Yu, H.; Rao, X.; Li, L.; Dixon, R.A. Abscisic Acid Regulates Secondary Cell-Wall Formation and Lignin Deposition in Arabidopsis Thaliana through Phosphorylation of NST1. Proc. Natl. Acad. Sci. USA 2021, 118, e2010911118. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Augstein, F.; Mazumdar, S.; Nguyen, T.V.; Minina, E.A.; Melnyk, C.W.; Carlsbecker, A. Abscisic Acid Signaling Activates Distinct VND Transcription Factors to Promote Xylem Differentiation in Arabidopsis. Curr. Biol. 2021, 31, 3153–3161.e5. [Google Scholar] [CrossRef]
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.; Rieu, I.; Mariani, C.; van Dam, N.M. How Plants Handle Multiple Stresses: Hormonal Interactions Underlying Responses to Abiotic Stress and Insect Herbivory. Plant Mol. Biol. 2016, 91, 727–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuominen, H.; Sitbon, F.; Jacobsson, C.; Sandberg, G.; Olsson, O.; Sundberg, B. Altered Growth and Wood Characteristics in Transgenic Hybrid Aspen Expressing Agrobacterium Tumefaciens T-DNA Indoleacetic Acid-Biosynthetic Genes. Plant Physiol. 1995, 109, 1179–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uggla, C.; Moritz, T.; Sandberg, G.; Sundberg, B. Auxin as a Positional Signal in Pattern Formation in Plants. Proc. Natl. Acad. Sci. USA 1996, 93, 9282–9286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuominen, H.; Puech, L.; Fink, S.; Sundberg, B. A Radial Concentration Gradient of Indole-3-Acetic Acid Is Related to Secondary Xylem Development in Hybrid Aspen. Plant Physiol. 1997, 115, 577–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uggla, C.; Mellerowicz, E.J.; Sundberg, B. Indole-3-Acetic Acid Controls Cambial Growth in Scots Pine by Positional Signaling1. Plant Physiol. 1998, 117, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Fischer, U.; Kucukoglu, M.; Helariutta, Y.; Bhalerao, R.P. The Dynamics of Cambial Stem Cell Activity. Annu. Rev. Plant Biol. 2019, 70, 293–319. [Google Scholar] [CrossRef] [Green Version]
- Douglas, C.J. Populus as a Model Tree. In Comparative and Evolutionary Genomics of Angiosperm Trees; Groover, A., Cronk, Q., Eds.; Plant Genetics and Genomics: Crops and Models; Springer International Publishing: Cham, Switzerland, 2017; pp. 61–84. ISBN 978-3-319-49329-9. [Google Scholar]
- Tullus, A.; Rytter, L.; Tullus, T.; Weih, M.; Tullus, H. Short-Rotation Forestry with Hybrid Aspen (Populus Tremula L. × P. Tremuloides Michx.) in Northern Europe. Scand. J. For. Res. 2012, 27, 10–29. [Google Scholar] [CrossRef]
- Papacek, M.; Christmann, A.; Grill, E. Interaction Network of ABA Receptors in Grey Poplar. Plant J. 2017, 92, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In Vitro Reconstitution of an Abscisic Acid Signalling Pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF Transcription Factors Function Predominantly in Gene Expression Downstream of SnRK2 Kinases in Abscisic Acid Signalling in Response to Osmotic Stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Mano, Y.; Nemoto, K. The Pathway of Auxin Biosynthesis in Plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kombrink, E. Chemical and Genetic Exploration of Jasmonate Biosynthesis and Signaling Paths. Planta 2012, 236, 1351–1366. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic Acid Dynamics, Signaling, and Functions in Plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, S.; Li, L.; Sun, Y.-H.; Chiang, V.L. The Cellulose Synthase Gene Superfamily and Biochemical Functions of Xylem-Specific Cellulose Synthase-like Genes in Populus Trichocarpa. Plant Physiol. 2006, 142, 1233–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Zhang, D.; Hu, J.; Zhou, X.; Ye, X.; Reichel, K.L.; Stewart, N.R.; Syrenne, R.D.; Yang, X.; Gao, P.; et al. Comparative Genome Analysis of Lignin Biosynthesis Gene Families across the Plant Kingdom. BMC Bioinform. 2009, 10, S3. [Google Scholar] [CrossRef] [Green Version]
- Hussey, S.G.; Mizrachi, E.; Creux, N.M.; Myburg, A.A. Navigating the Transcriptional Roadmap Regulating Plant Secondary Cell Wall Deposition. Front. Plant Sci. 2013, 4, 325. [Google Scholar] [CrossRef] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Uggla, C.; Magel, E.; Moritz, T.; Sundberg, B. Function and Dynamics of Auxin and Carbohydrates during Earlywood/Latewood Transition in Scots Pine. Plant Physiol. 2001, 125, 2029–2039. [Google Scholar] [CrossRef] [Green Version]
- Abreu, I.N.; Johansson, A.I.; Sokołowska, K.; Niittylä, T.; Sundberg, B.; Hvidsten, T.R.; Street, N.R.; Moritz, T. A Metabolite Roadmap of the Wood-Forming Tissue in Populus Tremula. New Phytol. 2020, 228, 1559–1572. [Google Scholar] [CrossRef] [PubMed]
- Sorce, C.; Giovannelli, A.; Sebastiani, L.; Anfodillo, T. Hormonal Signals Involved in the Regulation of Cambial Activity, Xylogenesis and Vessel Patterning in Trees. Plant Cell Rep. 2013, 32, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.-W.; Zhou, Y.-T.; Mwange, K.-N.; Li, W.-F.; He, X.-Q.; Cui, K.-M. ABP1 Expression Regulated by IAA and ABA Is Associated with the Cambium Periodicity in Eucommia Ulmoides Oliv. J. Exp. Bot. 2006, 57, 3857–3867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aloni, R. Phytohormonal Mechanisms That Control Wood Quality Formation in Young and Mature Trees; University of Canterbury: Christchurch, New Zealand, 2008. [Google Scholar]
- Liu, J.; Sherif, S.M. Hormonal Orchestration of Bud Dormancy Cycle in Deciduous Woody Perennials. Front. Plant Sci. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Hartung, W.; Sauter, A.; Hose, E. Abscisic Acid in the Xylem: Where Does It Come from, Where Does It Go To? J. Exp. Bot. 2002, 53, 27–32. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Goodger, J.Q.D. Chemical Root to Shoot Signaling under Drought. Trends Plant Sci. 2008, 13, 281–287. [Google Scholar] [CrossRef]
- Merilo, E.; Jalakas, P.; Laanemets, K.; Mohammadi, O.; Hõrak, H.; Kollist, H.; Brosché, M. Abscisic Acid Transport and Homeostasis in the Context of Stomatal Regulation. Mol. Plant 2015, 8, 1321–1333. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.; Aoki, H.; Koiwai, H.; Kamiya, Y.; Nambara, E.; Koshiba, T. Comparative Studies on the Arabidopsis Aldehyde Oxidase (AAO) Gene Family Revealed a Major Role of AAO3 in ABA Biosynthesis in Seeds. Plant Cell Physiol. 2004, 45, 1694–1703. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Marion-Poll, A. Abscisic Acid Biosynthesis and Catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The Relationship of Drought-Related Gene Expression in Arabidopsis Thaliana to Hormonal and Environmental Factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef] [Green Version]
- Seiler, C.; Harshavardhan, V.T.; Rajesh, K.; Reddy, P.S.; Strickert, M.; Rolletschek, H.; Scholz, U.; Wobus, U.; Sreenivasulu, N. ABA Biosynthesis and Degradation Contributing to ABA Homeostasis during Barley Seed Development under Control and Terminal Drought-Stress Conditions. J. Exp. Bot. 2011, 62, 2615–2632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauter, A.; Dietz, K.-J.; Hartung, W. A Possible Stress Physiological Role of Abscisic Acid Conjugates in Root-to-Shoot Signalling. Plant Cell Environ. 2002, 25, 223–228. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of Glucosidase via Stress-Induced Polymerization Rapidly Increases Active Pools of Abscisic Acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.-Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A Vacuolar β-Glucosidase Homolog That Possesses Glucose-Conjugated Abscisic Acid Hydrolyzing Activity Plays an Important Role in Osmotic Stress Responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, E.-K.; Doucet, C.J.; Hou, B.; Jackson, R.G.; Abrams, S.R.; Bowles, D.J. Resolution of (+)-Abscisic Acid Using an Arabidopsis Glycosyltransferase. Tetrahedron Asymmetry 2005, 16, 143–147. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis Cytochrome P450 CYP707A Encodes ABA 8′-Hydroxylases: Key Enzymes in ABA Catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [Green Version]
- Venturas, M.D.; Sperry, J.S.; Hacke, U.G. Plant Xylem Hydraulics: What We Understand, Current Research, and Future Challenges. J. Integr. Plant Biol. 2017, 59, 356–389. [Google Scholar] [CrossRef] [Green Version]
- Fichot, R.; Laurans, F.; Monclus, R.; Moreau, A.; Pilate, G.; Brignolas, F. Xylem Anatomy Correlates with Gas Exchange, Water-Use Efficiency and Growth Performance under Contrasting Water Regimes: Evidence from Populus Deltoides × Populus Nigra Hybrids. Tree Physiol. 2009, 29, 1537–1549. [Google Scholar] [CrossRef]
- Beniwal, R.S.; Langenfeld-Heyser, R.; Polle, A. Ectomycorrhiza and Hydrogel Protect Hybrid Poplar from Water Deficit and Unravel Plastic Responses of Xylem Anatomy. Environ. Exp. Bot. 2010, 69, 189–197. [Google Scholar] [CrossRef]
- Kao, Y.-Y.; Harding, S.A.; Tsai, C.-J. Differential Expression of Two Distinct Phenylalanine Ammonia-Lyase Genes in Condensed Tannin-Accumulating and Lignifying Cells of Quaking Aspen. Plant Physiol. 2002, 130, 796–807. [Google Scholar] [CrossRef] [Green Version]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic Acid Suppresses Jasmonic Acid Signaling Downstream of SCFCOI1-JAZ by Targeting GCC Promoter Motifs via Transcription Factor ORA59[C][W][OA]. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef] [Green Version]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How Salicylic Acid Takes Transcriptional Control over Jasmonic Acid Signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Sobczak, J.; Souza, U.; Mazzafera, P. Drought Stress and Changes in the Lignin Content and Composition in Eucalyptus. BMC Proc. 2011, 5, P103. [Google Scholar] [CrossRef] [Green Version]
- Janz, D.; Lautner, S.; Wildhagen, H.; Behnke, K.; Schnitzler, J.-P.; Rennenberg, H.; Fromm, J.; Polle, A. Salt Stress Induces the Formation of a Novel Type of ‘Pressure Wood’in Two Populus Species. New Phytol. 2012, 194, 129–141. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell Wall Remodeling under Abiotic Stress. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The Plant Cell Wall: A Complex and Dynamic Structure As Revealed by the Responses of Genes under Stress Conditions. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Weijde, T.; Huxley, L.M.; Hawkins, S.; Sembiring, E.H.; Farrar, K.; Dolstra, O.; Visser, R.G.F.; Trindade, L.M. Impact of Drought Stress on Growth and Quality of Miscanthus for Biofuel Production. GCB Bioenergy 2017, 9, 770–782. [Google Scholar] [CrossRef] [Green Version]
- Fichot, R.; Brignolas, F.; Cochard, H.; Ceulemans, R. Vulnerability to Drought-Induced Cavitation in Poplars: Synthesis and Future Opportunities. Plant Cell Environ. 2015, 38, 1233–1251. [Google Scholar] [CrossRef]
- Zhong, R.; Cui, D.; Ye, Z.-H. Secondary Cell Wall Biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollhöner, B.; Prestele, J.; Tuominen, H. Xylem Cell Death: Emerging Understanding of Regulation and Function. J. Exp. Bot. 2012, 63, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.-H.; Jeon, H.-W.; Kim, W.-C.; Kim, J.-Y.; Han, K.-H. The MYB46/MYB83-Mediated Transcriptional Regulatory Programme Is a Gatekeeper of Secondary Wall Biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef] [Green Version]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.R.; Pereira, A. Molecular and Physiological Analysis of Drought Stress in Arabidopsis Reveals Early Responses Leading to Acclimation in Plant Growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.; Dong, Y.; Liang, D.; Zhang, Z.; Ye, C.-Y.; Shuai, P.; Han, X.; Zhao, Y.; Yin, W.; Xia, X. Analysis of the Drought Stress-Responsive Transcriptome of Black Cottonwood (Populus Trichocarpa) Using Deep RNA Sequencing. Plant Mol. Biol. Rep. 2015, 33, 424–438. [Google Scholar] [CrossRef]
- Coleman, H.D.; Brunner, A.M.; Tsai, C.-J. Synergies and Entanglement in Secondary Cell Wall Development and Abiotic Stress Response in Trees. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.; Wheeler, J.; Heazlewood, J.; Li, S.F.; Parish, R.W. AtMYB32 Is Required for Normal Pollen Development in Arabidopsis Thaliana. Plant J. 2004, 40, 979–995. [Google Scholar] [CrossRef]
- Fornalé, S.; Lopez, E.; Salazar-Henao, J.E.; Fernández-Nohales, P.; Rigau, J.; Caparros-Ruiz, D. AtMYB7, a New Player in the Regulation of UV-Sunscreens in Arabidopsis Thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.-H. MYB58 and MYB63 Are Transcriptional Activators of the Lignin Biosynthetic Pathway during Secondary Cell Wall Formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Li, E.; Porth, I.; Chen, J.-G.; Mansfield, S.D.; Douglas, C.J. Regulation of Secondary Cell Wall Biosynthesis by Poplar R2R3 MYB Transcription Factor PtrMYB152 in Arabidopsis. Sci. Rep. 2014, 4, 5054. [Google Scholar] [CrossRef] [Green Version]
- Geng, P.; Zhang, S.; Liu, J.; Zhao, C.; Wu, J.; Cao, Y.; Fu, C.; Han, X.; He, H.; Zhao, Q. MYB20, MYB42, MYB43, and MYB85 Regulate Phenylalanine and Lignin Biosynthesis during Secondary Cell Wall Formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef] [Green Version]
- Quan, M.; Liu, X.; Du, Q.; Xiao, L.; Lu, W.; Fang, Y.; Li, P.; Ji, L.; Zhang, D. Genome-Wide Association Studies Reveal the Coordinated Regulatory Networks Underlying Photosynthesis and Wood Formation in Populus. J. Exp. Bot. 2021, 72, 5372–5389. [Google Scholar] [CrossRef]
- Velasquez, S.M.; Guo, X.; Gallemi, M.; Aryal, B.; Venhuizen, P.; Barbez, E.; Dünser, K.A.; Darino, M.; Pĕnčík, A.; Novák, O.; et al. Xyloglucan Remodeling Defines Auxin-Dependent Differential Tissue Expansion in Plants. Int. J. Mol. Sci. 2021, 22, 9222. [Google Scholar] [CrossRef]
- Müller, A.; Volmer, K.; Mishra-Knyrim, M.; Polle, A. Growing Poplars for Research with and without Mycorrhizas. Front. Plant Sci. 2013, 4, 332. [Google Scholar] [CrossRef] [Green Version]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 2006, 15, 473–497. [Google Scholar] [CrossRef]
- Bogeat-Triboulot, M.B.; Buré, C.; Gerardin, T.; Chuste, P.A.; Le Thiec, D.; Hummel, I.; Durand, M.; Wildhagen, H.; Douthe, C.; Molins, A.; et al. Additive Effects of High Growth Rate and Low Transpiration Rate Drive Differences in Whole Plant Transpiration Efficiency among Black Poplar Genotypes. Environ. Exp. Bot. 2019, 166, 103784. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Herrfurth, C.; Feussner, I. Quantitative Jasmonate Profiling Using a High-Throughput UPLC-NanoESI-MS/MS Method. Methods Mol. Biol. 2020, 2085, 169–187. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Chang, S.; Puryear, J.; Cairney, J. A Simple and Efficient Method for Isolating RNA from Pine Trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus Trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Bauer, S.; Grossmann, S.; Vingron, M.; Robinson, P.N. Ontologizer 2.0—A Multifunctional Tool for GO Term Enrichment Analysis and Data Exploration. Bioinformatics 2008, 24, 1650–1651. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]



| Treatments | Well-Watered | Moderate Drought | Severe Drought | p-Value |
|---|---|---|---|---|
| Leaf number # | 15.38 (0.53) c | 11.50 (0.33) b | 7.50 (0.33) a | <0.001 |
| Leaf size (cm2 leaf−1) | 57.24 (2.28) b | 51.96 (2.35) b | 43.36 (2.40) a | 0.002 |
| SLA (cm2 g−1 dry mass) | 306.7 (7.6) a | 313.5 (12.7) a | 303.0 (11.0) a | 0.779 |
| Whole-plant leaf area (cm2 plant−1) | 1251.4 (55.4) c | 819.5 (20.4) b | 478.8 (32. 9) a | <0.001 |
| Biomass of leaves (g plant−1) | 4.18 (0.16) c | 2.77 (0.11) b | 2.11 (0.17) a | <0.001 |
| Biomass of stem (g plant−1) | 2.68 (0.23) c | 1.65 (0.13) b | 1.06 (0.07) a | <0.001 |
| Biomass of roots (g plant−1) | 1.86 (0.19) a | 1.45 (0.16) a | 1.40 (0.09) a | 0.099 |
| Whole-plant biomass (g plant−1) | 8.71 (0.53) b | 5.88 (0.38) a | 4.56 (0.28) a | <0.001 |
| Root-to-shoot ratio | 0.70 (0.05) a | 0.87 (0.06) a | 1.34 (0.10) b | <0.001 |
| Relative leaf water content (%) | 92.26 (2.77) b | 82.76 (2.99) ab | 74.45 (3.68) a | 0.003 |
| Phytohormone (nmol g−1 fw) | Tissue | Well-Watered | Moderate Drought | Severe Drought | p-Values | |
|---|---|---|---|---|---|---|
| SA | Leaf | 4.81 (0.89) a | 5.09 (1.27) a | 3.88 (1.29) a | treat | 0.032 |
| Root | 0.74 (0.15) a | 4.64 (0.63) c | 2.49 (0.18) b | tissue | <0.001 | |
| Wood | 0.72 (0.05) b | 0.50 (0.08) a | 0.42 (0.08) a | treat:tissue | 0.020 | |
| IAA | Leaf | NA | NA | NA | treat | 0.310 |
| Root | NA | NA | NA | tissue | NA | |
| Wood | 0.44 (0.05) a | 0.39 (0.06) a | 0.32 (0.06) a | treat:tissue | NA | |
| JA | Leaf | 0.12 (0.05) a | 0.43 (0.31) a | 0.21 (0.10) a | treat | 0.206 |
| Root | 0.07 (0.02) a | 0.15 (0.08) a | 0.08 (0.01) a | tissue | 0.290 | |
| Wood | 0.07 (0.02) a | 0.21 (0.11) a | 0.20 (0.15) a | treat:tissue | 0.845 | |
| 12-HSO4-JA # | Leaf | 78.79 (57.87) a | 47.04 (34.56) a | 52.21 (33.24) a | treat | 0.826 |
| Root | 0.01 (0.005) a | 0.03 (0.02) a | 0.02 (0.01) a | tissue | 0.003 | |
| Wood | 0.005 (0.001) a | 0.006 (0.003) a | 0.008 (0.002) a | treat:tissue | 0.941 | |
| 12-OH-Glc-JA # | Leaf | 352.48 (252.74) a | 187.42 (128.69) a | 216.83 (150.81) a | treat | 0.759 |
| Root | 0.24 (0.10) a | 0.98 (0.60) a | 0.91 (0.35) a | tissue | 0.003 | |
| Wood | 0.13 (0.03) a | 0.26 (0.09) a | 0.74 (0.16) b | treat:tissue | 0.887 | |
| 12-COOH-JA # | Leaf | 138.86 (72.63) a | 114.28 (93.76) a | 94.89 (73.91) a | treat | 0.914 |
| Root | 0.66 (0.79) a | 0.32 (1.4) a | 0.52 (1.30) a | tissue | 0.002 | |
| Wood | 1.31 (0.30) a | 1.4 (0.61) a | 1.56 (0.36) a | treat:tissue | 0.983 | |
| ABA | Leaf | 0.37 (0.04) a | 3.95 (2.52) b | 4.94 (1.76) b | treat | 0.008 |
| Root | nd a | 0.31 (0.09) b | 0.38 (0.08) b | tissue | 0.013 | |
| Wood | 0.42 (0.05) a | 3.00 (1.89) ab | 6.09 (2.48) b | treat:tissue | 0.303 | |
| ABA-GE $ | Leaf | nd a | 4.34 (1.77) b | 7.28 (1.74) b | treat | 0.021 |
| Root | 0.15 (0.05) a | 1.04 (0.56) a | 0.61 (0.12) a | tissue | <0.001 | |
| Wood | 0.30 (0.09) a | 0.57 (0.08) ab | 0.68 (0.18) b | treat:tissue | 0.002 | |
| Anatomical Traits | Control | Drought | p-Value |
|---|---|---|---|
| Vessel frequency (number of vessels mm−2) | 240.20 (11.58) | 503.97 (46.74) | <0.001 |
| Vessel lumen size (μm2) | 749.19 (39.57) | 339.86 (37.95) | <0.001 |
| Fiber frequency (number of fibers mm−2) | 4162.0 (113.8) | 4865.0 (646.5) | 0.310 |
| Fiber lumen size (μm2) | 86.09 (3.35) | 56.36 (7.02) | 0.003 |
| Vessel cell wall thickness (µm) Double fiber cell wall thickness (μm) | 1.28 (0.09) 3.79 (0.13) | 1.45 (0.12) 4.22 (0.11) | 0.268 0.026 |
| Fraction of cell wall area (%) | 39.26 (1.70) | 48.40 (1.77) | <0.001 |
| Total cell wall area in mature xylem formed under treatments (mm2) | 2.35 (0.18) | 0.99 (0.10) | <0.001 |
| Number of cambial cell layers | 6.6 (0.5) | 3.8 (0.3) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, D.; Janz, D.; Zienkiewicz, K.; Herrfurth, C.; Feussner, I.; Chen, S.; Polle, A. Wood Formation under Severe Drought Invokes Adjustment of the Hormonal and Transcriptional Landscape in Poplar. Int. J. Mol. Sci. 2021, 22, 9899. https://doi.org/10.3390/ijms22189899
Yu D, Janz D, Zienkiewicz K, Herrfurth C, Feussner I, Chen S, Polle A. Wood Formation under Severe Drought Invokes Adjustment of the Hormonal and Transcriptional Landscape in Poplar. International Journal of Molecular Sciences. 2021; 22(18):9899. https://doi.org/10.3390/ijms22189899
Chicago/Turabian StyleYu, Dade, Dennis Janz, Krzysztof Zienkiewicz, Cornelia Herrfurth, Ivo Feussner, Shaoliang Chen, and Andrea Polle. 2021. "Wood Formation under Severe Drought Invokes Adjustment of the Hormonal and Transcriptional Landscape in Poplar" International Journal of Molecular Sciences 22, no. 18: 9899. https://doi.org/10.3390/ijms22189899
APA StyleYu, D., Janz, D., Zienkiewicz, K., Herrfurth, C., Feussner, I., Chen, S., & Polle, A. (2021). Wood Formation under Severe Drought Invokes Adjustment of the Hormonal and Transcriptional Landscape in Poplar. International Journal of Molecular Sciences, 22(18), 9899. https://doi.org/10.3390/ijms22189899






