Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association
Abstract
:1. Introduction
2. Results
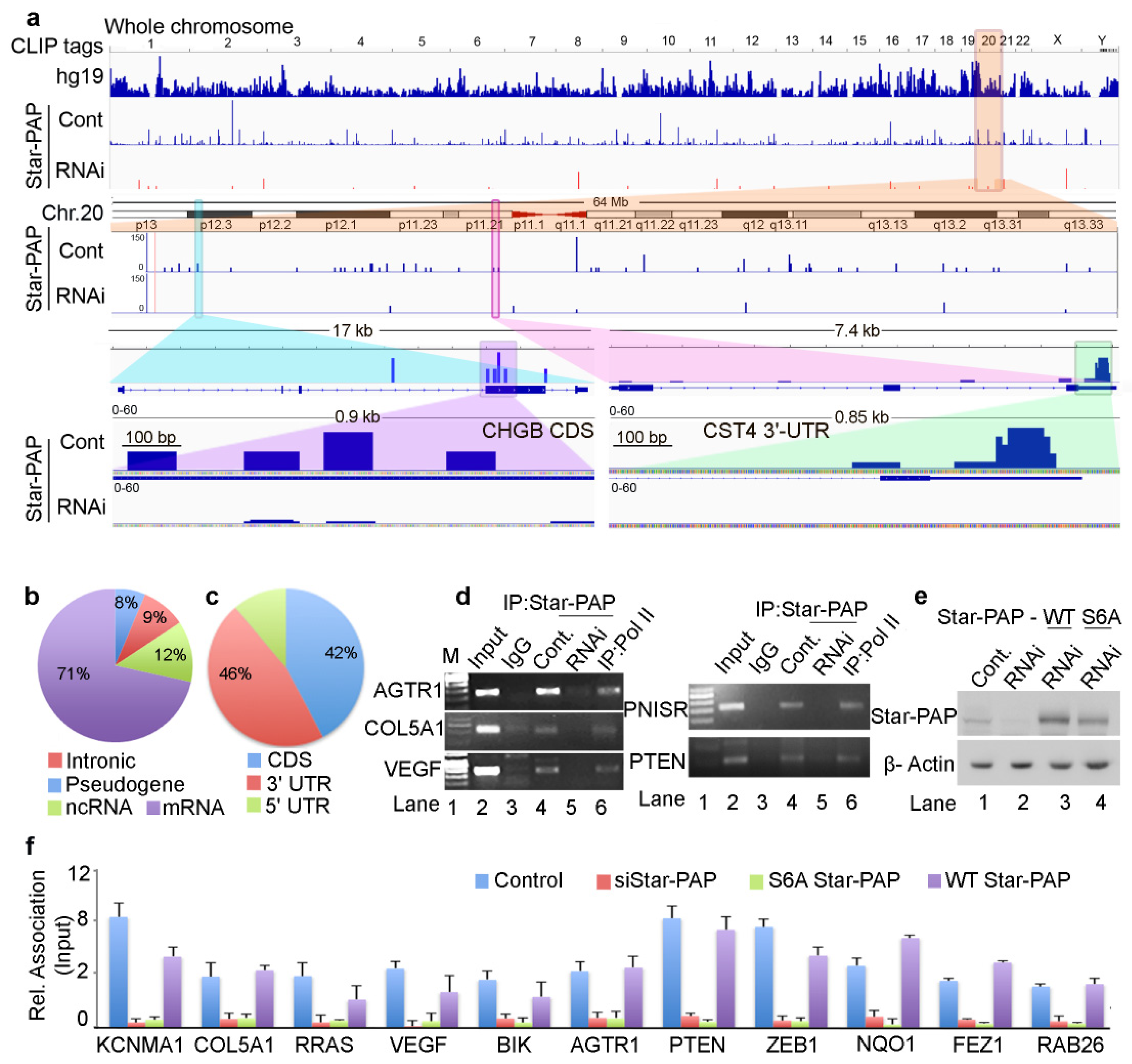
2.1. Genome-Wide Star-PAP RNA Binding Landscape Reveals Star-PAP Direct mRNA Targets
2.2. Star-PAP Associated mRNA Targets Show Wide Roles of Star-PAP in Human Diseases and Signaling Pathways
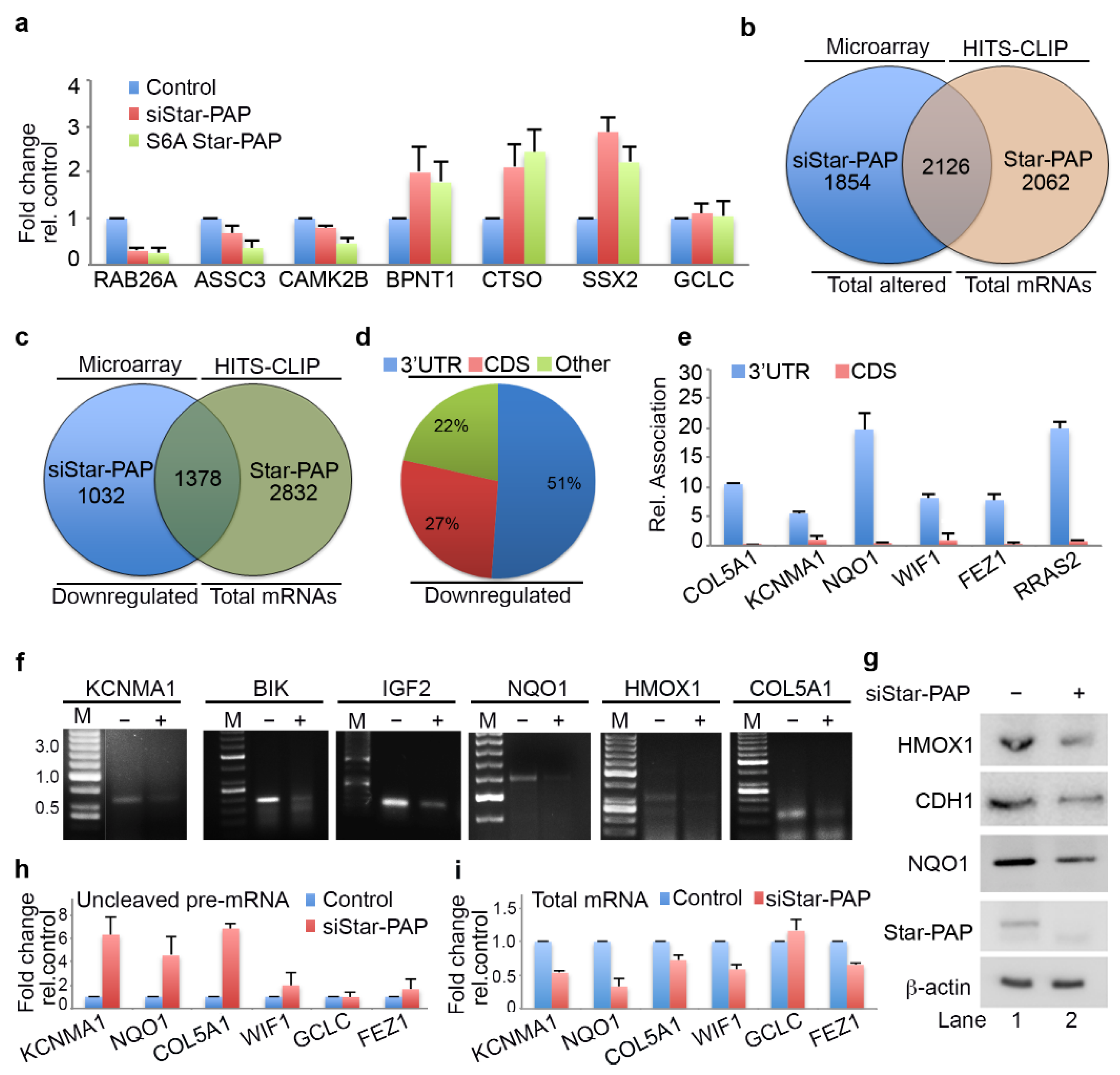
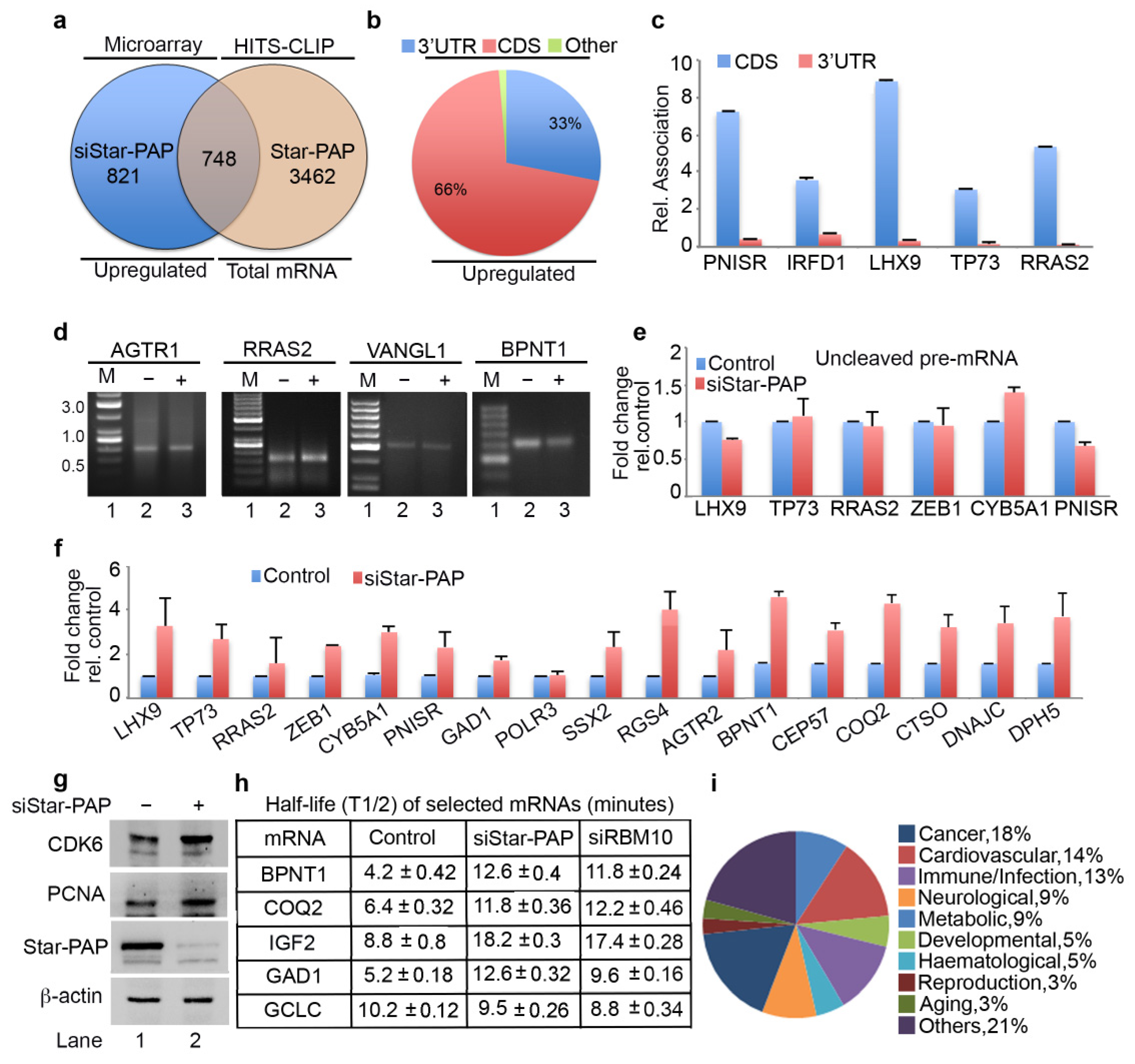
2.3. Star-PAP RNA Binding Can Both Down Regulate and Up-Regulate Target mRNA Expression
2.4. Star-PAP mRNA Binding Regulates Stability and Turnover Rate of Target mRNAs
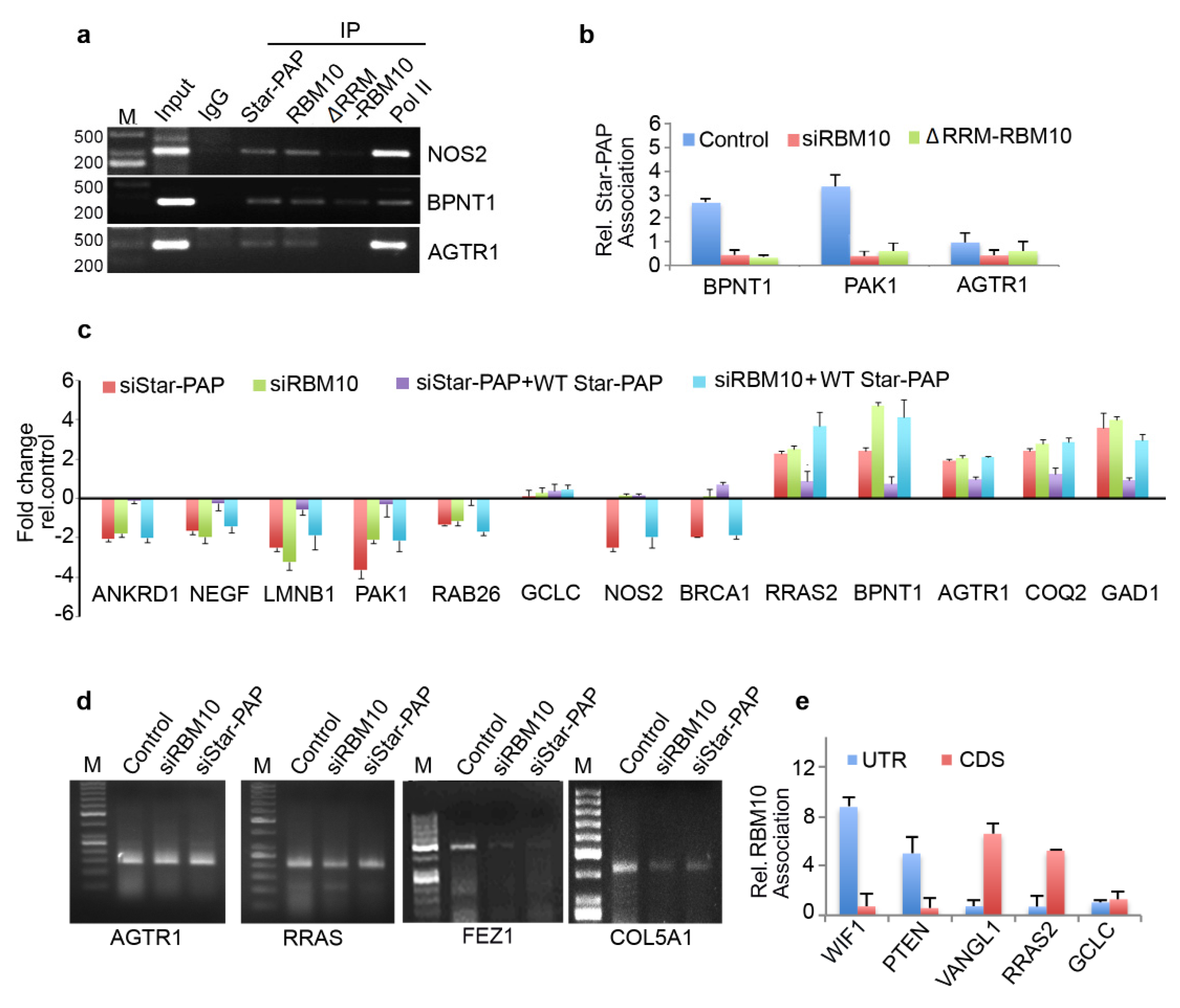
2.5. Transcriptome-Wide Star-PAP Binding Analysis after RBM10 Depletion Indicates Global Role of RBM10 in Star-PAP Target mRNA Association
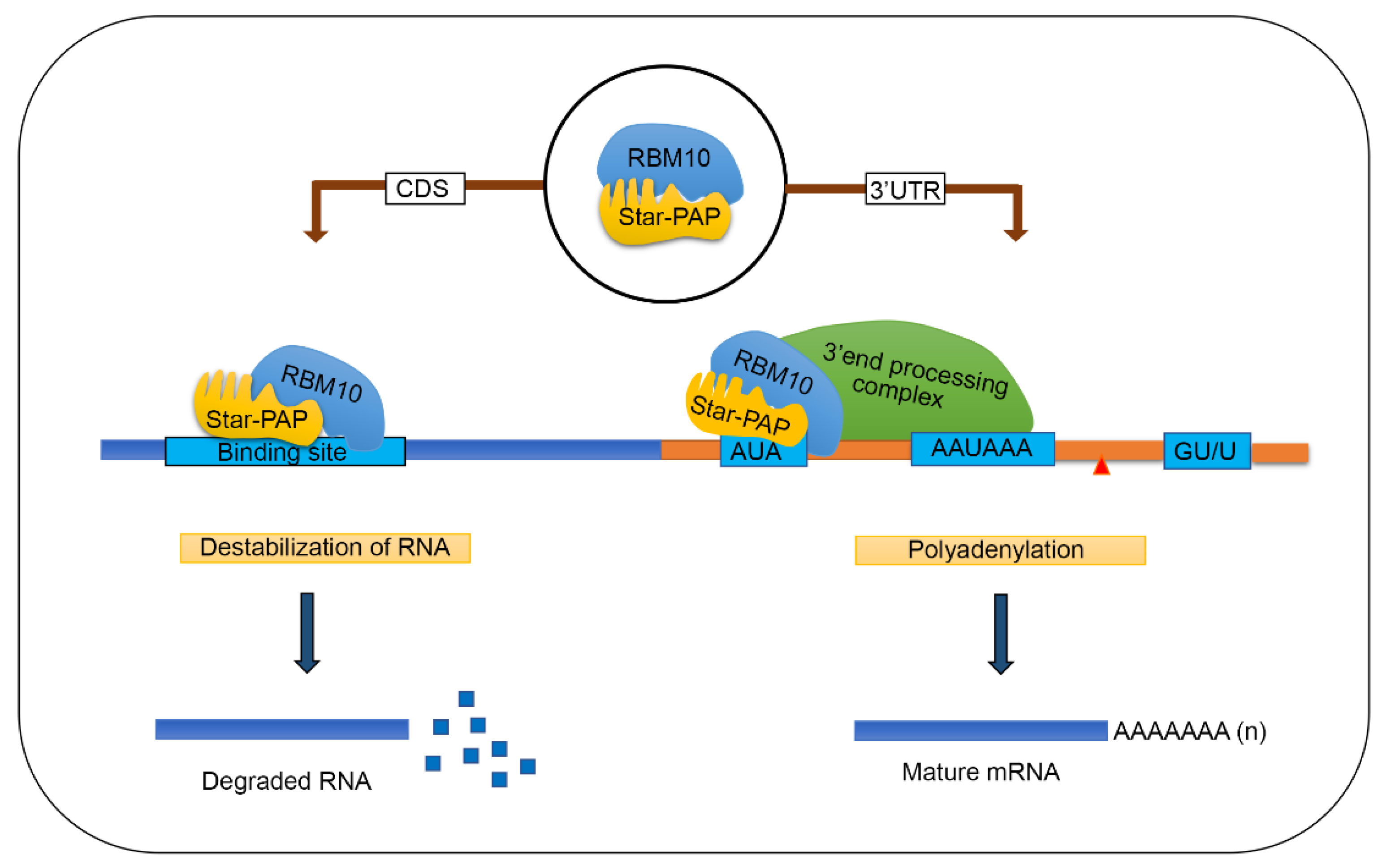
2.6. RBM10-RNA Association Regulates Star-PAP-Mediated mRNA Metabolism
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Transfections and Treatment
4.2. RNA Isolation
4.3. Quantitative Real-Time PCR (qRT-PCR)
4.4. Cleavage Assay
4.5. HITS-CLIP Sequencing and Analysis
4.6. RNA Immunoprecipitation (RIP)
4.7. Half-Life (T1/2) Measurement
4.8. 3′-RACE
4.9. Immunoblotting
4.10. Statistics
4.11. Primers and Antibodies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laishram, R.S. Poly(A) polymerase (PAP) diversity in gene expression–star-PAP vs canonical PAP. FEBS Lett. 2014, 588, 2185–2197. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 2004, 16, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Mandel, C.R.; Bai, Y.; Tong, L. Protein factors in pre-mRNA 3′-end processing. Cell Mol. Life Sci. 2008, 65, 1099–1122. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hyman, L.; Moore, C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999, 63, 405–445. [Google Scholar] [CrossRef]
- Shi, Y.; Di Giammartino, D.C.; Taylor, D.; Sarkeshik, A.; Rice, W.J.; Yates, J.R., III; Frank, J.; Manley, J.L. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 2009, 33, 365–376. [Google Scholar] [CrossRef]
- Wahle, E.; Keller, W. The biochemistry of polyadenylation. Trends Biochem. Sci. 1996, 21, 247–250. [Google Scholar] [CrossRef]
- Chan, S.L.; Huppertz, I.; Yao, C.; Weng, L.; Moresco, J.J.; Yates, J.R., III; Ule, J.; Manley, J.L.; Shi, Y. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 2014, 28, 2370–2380. [Google Scholar] [CrossRef]
- Clerici, M.; Faini, M.; Muckenfuss, L.M.; Aebersold, R.; Jinek, M. Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nat. Struct. Mol. Biol. 2018, 25, 135–138. [Google Scholar] [CrossRef]
- Schönemann, L.; Kühn, U.; Martin, G.; Schäfer, P.; Gruber, A.R.; Keller, W.; Zavolan, M.; Wahle, E. Reconstitution of CPSF active in polyadenylation: Recognition of the polyadenylation signal by WDR33. Genes Dev. 2014, 28, 2381–2393. [Google Scholar]
- Sun, Y.; Zhang, Y.; Hamilton, K.; Manley, J.L.; Shi, Y.; Walz, T.; Tong, L. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc. Natl. Acad. Sci. USA 2018, 115, E1419–E1428. [Google Scholar]
- Perez Canadillas, J.M.; Varani, G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003, 22, 2821–2830. [Google Scholar] [CrossRef] [PubMed]
- Takagaki, Y.; Manley, J.L. RNA recognition by the human polyadenylation factor CstF. Mol. Cell Biol. 1997, 17, 3907–3914. [Google Scholar] [CrossRef] [PubMed]
- Mandel, C.R.; Kaneko, S.; Zhang, H.; Gebauer, D.; Vethantham, V.; Manley, J.L.; Tong, L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 2006, 444, 953–956. [Google Scholar] [CrossRef]
- Ryan, K.; Calvo, O.; Manley, J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 2004, 10, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C.; Bonanno, J.B.; Sonenberg, N.; Burley, S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 1999, 98, 835–845. [Google Scholar] [CrossRef]
- Kuhn, U.; Gundel, M.; Knoth, A.; Kerwitz, Y.; Rudel, S.; Wahle, E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 2009, 284, 22803–22814. [Google Scholar] [CrossRef]
- Kuhn, U.; Wahle, E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 2004, 1678, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, W.; Laishram, R.S.; Hoque, M.; Ji, Z.; Tian, B.; Anderson, R.A. Distinct regulation of alternative polyadenylation and gene expression by nuclear poly(A) polymerases. Nucleic Acids Res. 2017, 45, 8930–8942. [Google Scholar] [CrossRef]
- Topalian, S.L.; Kaneko, S.; Gonzales, M.I.; Bond, G.L.; Ward, Y.; Manley, J.L. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol. Cell Biol. 2001, 21, 5614–5623. [Google Scholar] [CrossRef]
- Laishram, R.S.; Anderson, R.A. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. EMBO J. 2010, 29, 4132–4145. [Google Scholar] [CrossRef]
- Mellman, D.L.; Gonzales, M.L.; Song, C.; Barlow, C.A.; Wang, P.; Kendziorski, C.; Anderson, R.A. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 2008, 451, 1013–1017. [Google Scholar] [CrossRef]
- Gonzales, M.L.; Mellman, D.L.; Anderson, R.A. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J. Biol. Chem. 2008, 283, 12665–12673. [Google Scholar] [CrossRef]
- Sudheesh, A.; Mohan, N.; Francis, N.; Laishram, R.S.; Anderson, R.A. Star-PAP controlled alternative polyadenylation coupled poly (A) tail length regulates protein expression in hypertrophic heart. Nucleic Acids Res. 2019, 47, 10771–10787. [Google Scholar] [CrossRef] [PubMed]
- Mohan, N.; Kumar, V.; Kandala, D.T.; Kartha, C.C.; Laishram, R.S. A Splicing-Independent Function of RBM10 Controls Specific 3′ UTR Processing to Regulate Cardiac Hypertrophy. Cell Rep. 2018, 24, 3539–3553. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Laishram, R.S.; Ji, Z.; Barlow, C.A.; Tian, B.; Anderson, R.A. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIalpha and PKCdelta signaling. Mol. Cell 2012, 45, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Duan, A.; Kong, L.; An, T.; Zhou, H.; Yu, C.; Li, Y. Star-PAP regulates tumor protein D52 through modulating miR-449a/34a in breast cancer. Biol. Open 2019, 8, bio045914. [Google Scholar] [CrossRef]
- Yu, C.; Gong, Y.; Zhou, H.; Wang, M.; Kong, L.; Liu, J.; An, T.; Zhu, H.; Li, Y. Star-PAP, a poly (A) polymerase, functions as a tumor suppressor in an orthotopic human breast cancer model. Cell Death Dis. 2017, 8, e2582. [Google Scholar] [CrossRef]
- Yamashita, S.; Takagi, Y.; Nagaike, T.; Tomita, K. Crystal structures of U6 snRNA-specific terminal uridylyltransferase. Nat. Commun. 2017, 8, 15788. [Google Scholar] [CrossRef] [PubMed]
- Trippe, R.; Sandrock, B.; Benecke, B.J. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998, 26, 3119–3126. [Google Scholar] [CrossRef]
- Knouf, E.C.; Wyman, S.K.; Tewari, M. The human TUT1 nucleotidyl transferase as a global regulator of microRNA abundance. PLoS ONE 2013, 8, e69630. [Google Scholar] [CrossRef] [PubMed]
- Haas, G.; Cetin, S.; Messmer, M.; Chane-Woon-Ming, B.; Terenzi, O.; Chicher, J.; Kuhn, L.; Hammann, P.; Pfeffer, S. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 2016, 44, 2873–2887. [Google Scholar] [CrossRef]
- Hoque, M.; Ji, Z.; Zheng, D.; Luo, W.; Li, W.; You, B.; Park, J.Y.; Yehia, G.; Tian, B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods 2013, 10, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef]
- Divya, T.K.; Nimmy, M.; Vivekanand, A.; Sudheesh, A.P.; Reshmi, G.; Rakesh, S.L. CstF-64 and 3′-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding PAPalpha. Nucleic Acids Res. 2016, 44, 811–823. [Google Scholar]
- Aravind, L.; Koonin, E.V. G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999, 24, 342–344. [Google Scholar] [CrossRef]
- Inoue, A.; Takahashi, K.P.; Kimura, M.; Watanabe, T.; Morisawa, S. Molecular cloning of a RNA binding protein, S1-1. Nucleic Acids Res. 1996, 24, 2990–2997. [Google Scholar] [CrossRef]
- Nguyen, C.D.; Mansfield, R.E.; Leung, W.; Vaz, P.M.; Loughlin, F.E.; Grant, R.P.; Mackay, J.P. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J. Mol. Biol. 2011, 407, 273–283. [Google Scholar] [CrossRef]
- Xiao, S.J.; Wang, L.Y.; Kimura, M.; Kojima, H.; Kunimoto, H.; Nishiumi, F.; Yamamoto, N.; Nishio, K.; Fujimoto, S.; Kato, T.; et al. S1-1/RBM10: Multiplicity and cooperativity of nuclear localisation domains. Biol. Cell 2013, 105, 162–174. [Google Scholar] [CrossRef]
- Gripp, K.W.; Hopkins, E.; Johnston, J.J.; Krause, C.; Dobyns, W.B.; Biesecker, L.G. Long-term survival in TARP syndrome and confirmation of RBM10 as the disease-causing gene. Am. J. Med. Genet. A 2011, 155, 2516–2520. [Google Scholar] [CrossRef]
- Imielinski, M.; Berger, A.H.; Hammerman, P.S.; Hernandez, B.; Pugh, T.J.; Hodis, E.; Cho, J.; Suh, J.; Capelletti, M.; Sivachenko, A.; et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012, 150, 1107–1120. [Google Scholar] [CrossRef]
- Sutherland, L.C.; Rintala-Maki, N.D.; White, R.D.; Morin, C.D. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? J. Cell Biochem. 2005, 94, 5–24. [Google Scholar] [CrossRef]
- Wang, Y.; Gogol-Doring, A.; Hu, H.; Frohler, S.; Ma, Y.; Jens, M.; Maaskola, J.; Murakawa, Y.; Quedenau, C.; Landthaler, M.; et al. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol. Med. 2013, 5, 1431–1442. [Google Scholar] [CrossRef]
- Inoue, A.; Yamamoto, N.; Kimura, M.; Nishio, K.; Yamane, H.; Nakajima, K. RBM10 regulates alternative splicing. FEBS Lett. 2014, 588, 942–947. [Google Scholar] [CrossRef]
- Agafonov, D.E.; Deckert, J.; Wolf, E.; Odenwalder, P.; Bessonov, S.; Will, C.L.; Urlaub, H.; Luhrmann, R. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell Biol. 2011, 31, 2667–2682. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef]
- Makarov, E.M.; Owen, N.; Bottrill, A.; Makarova, O.V. Functional mammalian spliceosomal complex E contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucleic Acids Res. 2012, 40, 2639–2652. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002, 12, 1231–1245. [Google Scholar] [CrossRef] [PubMed]
- Sudheesh, A.P.; Laishram, R.S. Nuclear phosphatidyl-inositol-phosphate type I kinase alpha coupled Star-PAP polyadenylation regulates cell invasion. Mol. Cell Biol. 2017, 38, e00457-17. [Google Scholar]
- Mohan, N.; Sudheesh, A.P.; Francis, N.; Anderson, R.; Laishram, R.S. Phosphorylation regulates the Star-PAP-PIPKIalpha interaction and directs specificity toward mRNA targets. Nucleic Acids Res. 2015, 43, 7005–7020. [Google Scholar] [CrossRef]
- Laishram, R.S.; Barlow, C.A.; Anderson, R.A. CKI isoforms alpha and epsilon regulate Star-PAP target messages by controlling Star-PAP poly(A) polymerase activity and phosphoinositide stimulation. Nucleic Acids Res. 2011, 39, 7961–7973. [Google Scholar] [CrossRef]
- Trippe, R.; Guschina, E.; Hossbach, M.; Urlaub, H.; Luhrmann, R.; Benecke, B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 2006, 12, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Snoek, B.C.; Babion, I.; Koppers-Lalic, D.; Pegtel, D.M.; Steenbergen, R.D. Altered microRNA processing proteins in HPV-induced cancers. Curr. Opin. Virol. 2019, 39, 23–32. [Google Scholar] [CrossRef]
- Chen, C.Y.; Gherzi, R.; Ong, S.E.; Chan, E.L.; Raijmakers, R.; Pruijn, G.J.; Stoecklin, G.; Moroni, C.; Mann, M.; Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 2001, 107, 451–464. [Google Scholar] [CrossRef]
- Fabian, M.R.; Frank, F.; Rouya, C.; Siddiqui, N.; Lai, W.S.; Karetnikov, A.; Blackshear, P.J.; Nagar, B.; Sonenberg, N. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 2013, 20, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Gherzi, R.; Lee, K.Y.; Briata, P.; Wegmuller, D.; Moroni, C.; Karin, M.; Chen, C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 2004, 14, 571–583. [Google Scholar] [CrossRef]
- Loflin, P.; Chen, C.Y.; Shyu, A.B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999, 13, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Sanduja, S.; Blanco, F.F.; Dixon, D.A. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 2011, 2, 42–57. [Google Scholar] [CrossRef]
- Wu, X.; Chesoni, S.; Rondeau, G.; Tempesta, C.; Patel, R.; Charles, S.; Daginawala, N.; Zucconi, B.E.; Kishor, A.; Xu, G.; et al. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013, 41, 2644–2658. [Google Scholar] [CrossRef]
- Yoon, J.H.; Jo, M.H.; White, E.J.; De, S.; Hafner, M.; Zucconi, B.E.; Abdelmohsen, K.; Martindale, J.L.; Yang, X.; Wood, W.H., III; et al. AUF1 promotes let-7b loading on Argonaute 2. Genes Dev. 2015, 29, 1599–1604. [Google Scholar] [CrossRef]
- Min, K.W.; Jo, M.H.; Shin, S.; Davila, S.; Zealy, R.W.; Kang, S.I.; Lloyd, L.T.; Hohng, S.; Yoon, J.H. AUF1 facilitates microRNA-mediated gene silencing. Nucleic Acids Res. 2017, 45, 6064–6073. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef]
- Ule, J.; Jensen, K.; Mele, A.; Darnell, R.B. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods 2005, 37, 376–386. [Google Scholar] [CrossRef]
- Shah, A.; Qian, Y.; Weyn-Vanhentenryck, S.M.; Zhang, C. CLIP Tool Kit (CTK): A flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 2017, 33, 566–567. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Ma, W.; Noble, W.S.; Bailey, T.L. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat. Protoc. 2014, 9, 1428–1450. [Google Scholar] [CrossRef]
- Gilbert, C.; Kristjuhan, A.; Winkler, G.S.; Svejstrup, J.Q. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 2004, 14, 457–464. [Google Scholar] [CrossRef]
- Gilbert, C.; Svejstrup, J.Q. RNA immunoprecipitation for determining RNA-protein associations in vivo. Curr. Protoc. Mol. Biol. 2006, 75, 27.4.1–27.4.11. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Gilbert, C.; Svejstrup, J.Q. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb. Protoc. 2009, 2009, pdb-prot5234. [Google Scholar] [CrossRef]
- Krishnan, M.; Singh, A.B.; Smith, J.J.; Sharma, A.; Chen, X.; Eschrich, S.; Yeatman, T.J.; Beauchamp, R.D.; Dhawan, P. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene 2010, 29, 305–312. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshre, G.R.; Shaji, F.; Mohanan, N.K.; Mohan, N.; Ali, J.; Laishram, R.S. Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association. Int. J. Mol. Sci. 2021, 22, 9980. https://doi.org/10.3390/ijms22189980
Koshre GR, Shaji F, Mohanan NK, Mohan N, Ali J, Laishram RS. Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association. International Journal of Molecular Sciences. 2021; 22(18):9980. https://doi.org/10.3390/ijms22189980
Chicago/Turabian StyleKoshre, Ganesh R., Feba Shaji, Neeraja K. Mohanan, Nimmy Mohan, Jamshaid Ali, and Rakesh S. Laishram. 2021. "Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association" International Journal of Molecular Sciences 22, no. 18: 9980. https://doi.org/10.3390/ijms22189980
APA StyleKoshre, G. R., Shaji, F., Mohanan, N. K., Mohan, N., Ali, J., & Laishram, R. S. (2021). Star-PAP RNA Binding Landscape Reveals Novel Role of Star-PAP in mRNA Metabolism That Requires RBM10-RNA Association. International Journal of Molecular Sciences, 22(18), 9980. https://doi.org/10.3390/ijms22189980





