Na+-Coupled Respiration and Reshaping of Extracellular Polysaccharide Layer Counteract Monensin-Induced Cation Permeability in Prevotella bryantii B14
Abstract
:1. Introduction
2. Results
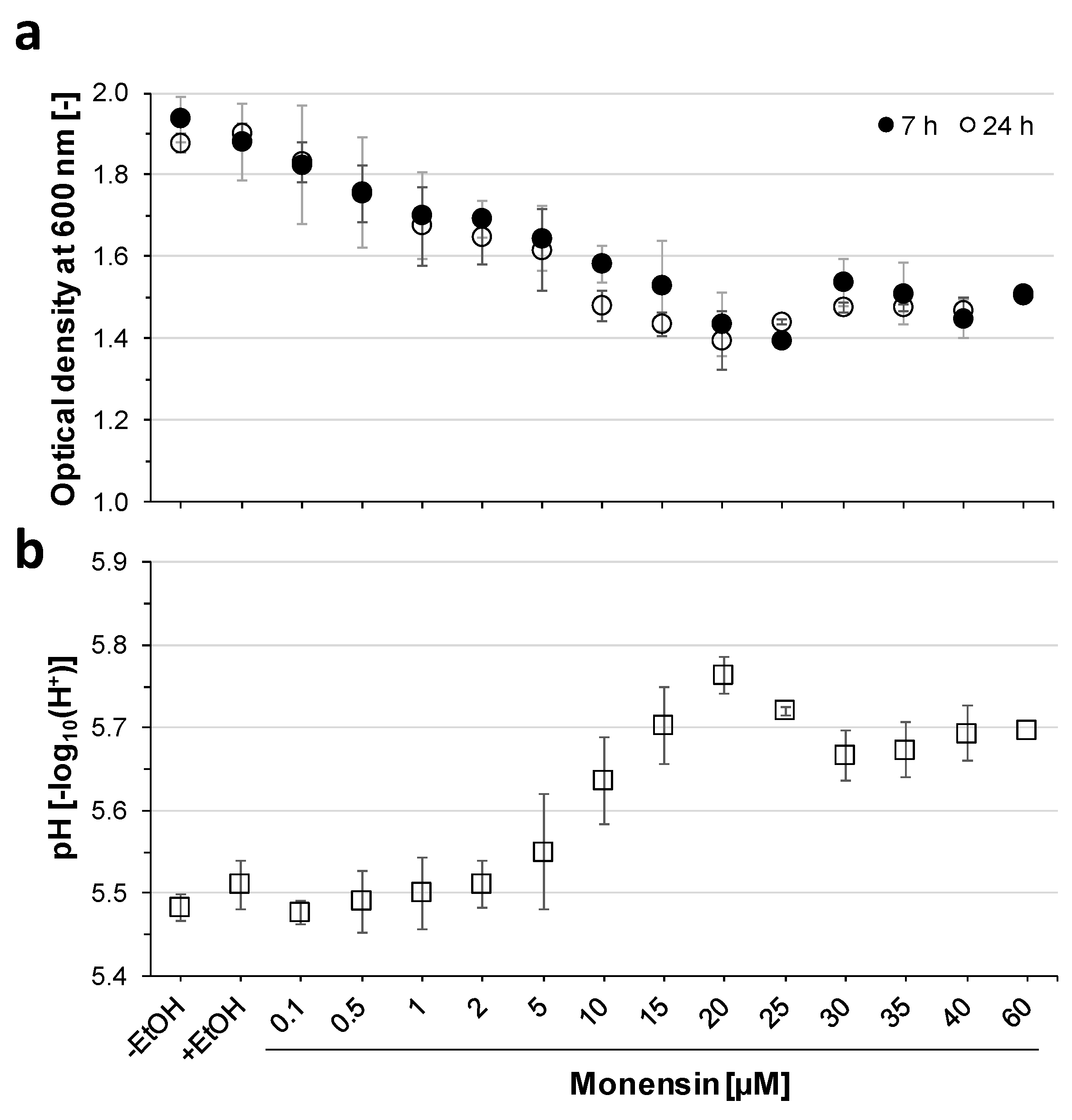
2.1. Inhibition of Prevotella Bryantii B14 Growth by Increasing Monensin Concentrations
2.2. Growth Adaptations towards Monensin over Time
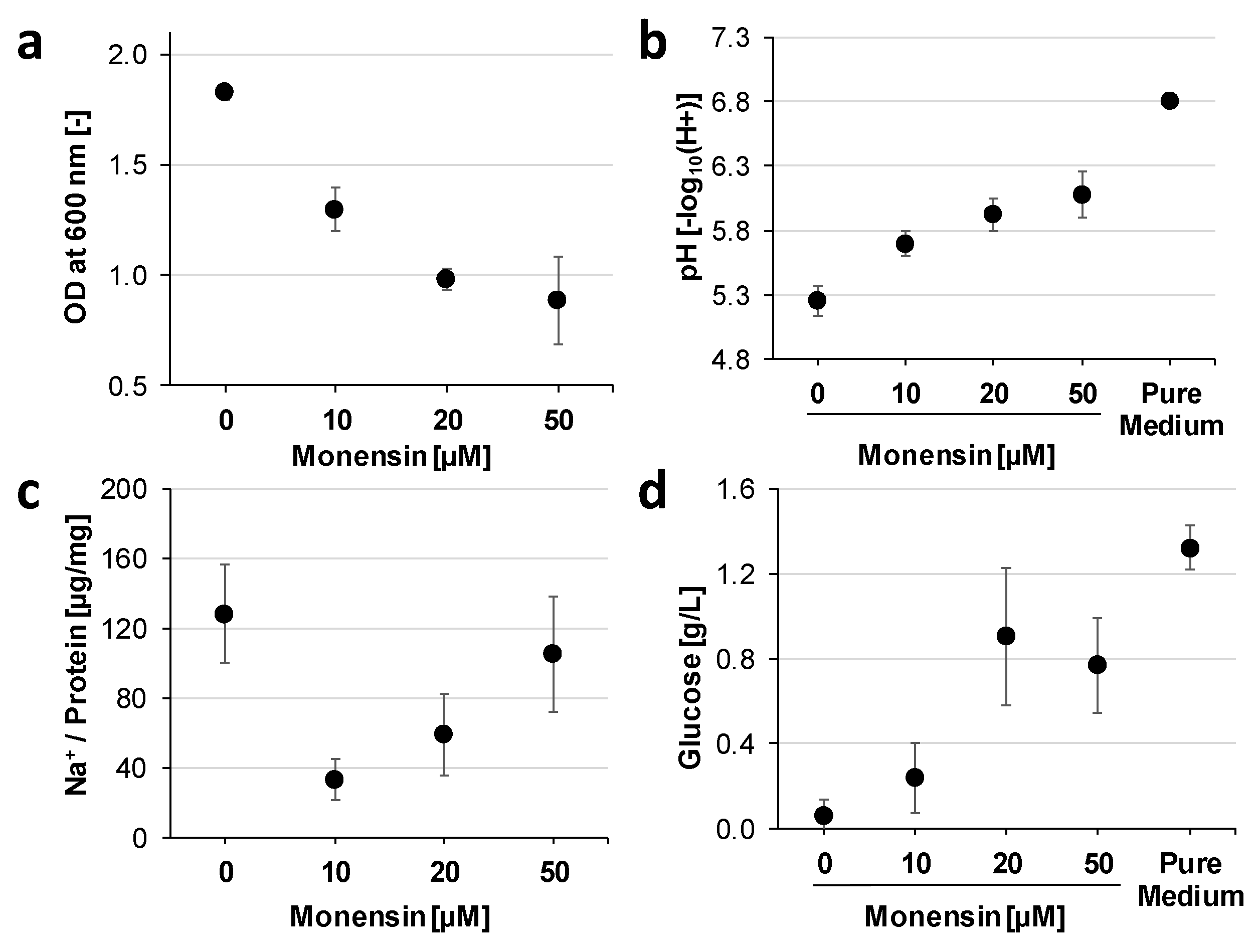
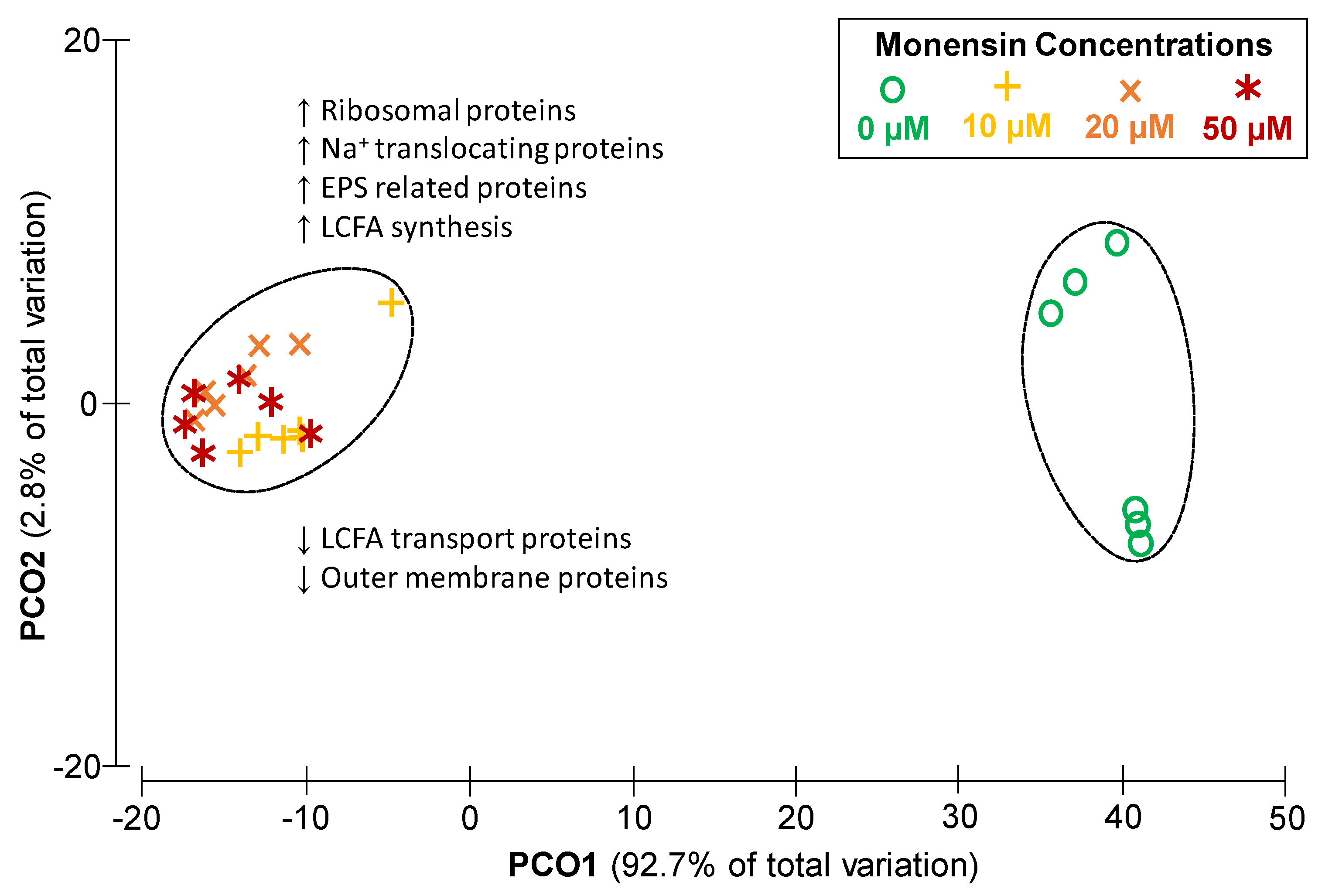
2.3. Dose Dependent Modulation of the Proteome of Prevotella bryantii B14 by Monensin
2.4. Protein Modification in Monensin Cultures over Time
2.5. Altered EPS Structure in Monensin Cultivations
3. Discussion
3.1. Degressive Dose–Response to Monensin
3.2. Cation Permeability Affects Proteome and Metabolism
3.3. Monensin Triggers Extracellular Polysaccharide Degradation by P. bryantii with a Concomitant Release of Glucose
- An increased amount of EPS degrading enzymes such as glucosidases or COG M class proteins;
- Elevated amounts of extracellular glucose;
- Zones with EPS depletions in P. bryantii cell clusters.
4. Materials and Methods
4.1. Cultivation
4.2. Growth in Presence of Monensin
4.3. Determination of Intracellular Sodium Content
4.4. Determination of D-Glucose and Glucose-6-Phosphate in Cell Cultures
4.5. Membrane Isolation and Enzyme Kinetics
4.6. Fluorescence Microscopy
4.7. Proteome Analysis Preparation
4.8. Tandem Mass Spectrometry
4.9. Proteome Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial growth promoters used in animal feed: Effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Perreten, V. Use of antimicrobials in food-producing animals in Switzerland and in the European Union (EU). Mitt. Lebensm. Hyg. 2003, 94, 155–163. [Google Scholar] [CrossRef]
- Castanon, J. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. J. 2007, 86, 2466–2471. [Google Scholar] [CrossRef] [PubMed]
- Kirchhelle, C. Pharming animals: A global history of antibiotics in food production (1935–2017). Palgrave Commun. 2018, 4, 96. [Google Scholar] [CrossRef] [Green Version]
- Newbold, C.; Wallace, R.; Walker, N.D. The effect of tetronasin and monensin on fermentation, microbial numbers and the development of ionophore-resistant bacteria in the rumen. J. Appl. Microbiol. 1993, 75, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Witzig, M.; Zeder, M.; Rodehutscord, M. Effect of the ionophore monensin and tannin extracts supplemented to grass silage on populations of ruminal cellulolytics and methanogens in vitro. Anaerobe 2018, 50, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.; Owens, F. Monensin supplementation and in vivo methane production by steers. Anim. Sci. J. 1981, 52, 628–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Commission Implementing Regulation (EU) 2020/994 of 9 July 2020 Concerning the Authorisation of Monensin and Nicarbazin (Monimax) as a Feed Additive for Turkeys for Fattening, Chickens for Fattening and Chickens Reared for Laying (Holder of Authorisation Huvepharma NV). Available online: https://eur-lex.europa.eu/eli/reg_impl/2020/994/oj (accessed on 10 July 2020).
- Łowicki, D.; Huczyński, A. Structure and antimicrobial properties of monensin A and its derivatives: Summary of the achievements. BioMed Res. Int. 2013, 2013, 742149. [Google Scholar] [CrossRef] [PubMed]
- Saleh, O.; Bonitz, T.; Flinspach, K.; Kulik, A.; Burkard, N.; Mühlenweg, A.; Vente, A.; Polnick, S.; Lämmerhofer, M.; Gust, B. Activation of a silent phenazine biosynthetic gene cluster reveals a novel natural product and a new resistance mechanism against phenazines. Med. Chem. Commun. 2012, 3, 1009–1019. [Google Scholar] [CrossRef]
- Pospíšil, S. Resistance of Streptomyces cinnamonensis to butyrate and isobutyrate: Production and properties of a new anti-isobutyrate (AIB) factor. Microbiology 1991, 137, 2141–2146. [Google Scholar]
- Pospíšil, S.; Váňová, M.; Machurová, V. Activity of the AIB factor observed in prokaryotic and eukaryotic microorganisms. Folia Microbiol. 1993, 38, 147. [Google Scholar] [CrossRef]
- Chen, M.; Wolin, M. Effect of monensin and lasalocid-sodium on the growth of methanogenic and rumen saccharolytic bacteria. Appl. Environ. Microbiol. 1979, 38, 72–77. [Google Scholar] [CrossRef] [Green Version]
- Callaway, T.; Edrington, T.; Rychlik, J.; Genovese, K.; Poole, T.; Jung, Y.S.; Bischoff, K.; Anderson, R.; Nisbet, D.J. Ionophores: Their use as ruminant growth promotants and impact on food safety. Curr. Issues Intest. Microbiol. 2003, 4, 43–51. [Google Scholar] [PubMed]
- Morehead, M.; Dawson, K. Some growth and metabolic characteristics of monensin-sensitive and monensin-resistant strains of Prevotella (Bacteroides) ruminicola. Appl. Environ. Microbiol. 1992, 58, 1617–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newbold, C.; Wallace, R. Changes in the rumen bacterium, Bacteroides ruminicola, grown in the presence of the ionophore, tetronasin. Asian Australas. J. Anim. Sci. 1989, 2, 452–453. [Google Scholar] [CrossRef]
- Callaway, T.R.; Russell, J.B. Selection of a highly monensin-resistant Prevotella bryantii subpopulation with altered outer membrane characteristics. Appl. Environ. Microbiol. 1999, 65, 4753–4759. [Google Scholar] [CrossRef] [Green Version]
- Lewis, K.; Naroditskaya, V.; Ferrante, A.; Fokina, I. Bacterial resistance to uncouplers. J. Bioenerg. Biomembr. 1994, 26, 639–646. [Google Scholar] [CrossRef]
- Dunkley, E.; Clejan, S.; Krulwich, T. Mutants of Bacillus species isolated on the basis of protonophore resistance are deficient in fatty acid desaturase activity. J. Bacteriol. Res. 1991, 173, 7750–7755. [Google Scholar] [CrossRef] [Green Version]
- Rychlik, J.L.; Russell, J.B. The adaptation and resistance of Clostridium aminophilum F to the butyrivibriocin-like substance of Butyrivibrio fibrisolvens JL5 and monensin. FEMS Microbiol. Lett. 2002, 209, 93–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, J.M.; Van Kessel, J.A.S.; Russell, J.B. Binding of radiolabeled monensin and lasalocid to ruminal microorganisms and feed. J. Anim. Sci. 1994, 72, 1630–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callaway, T.R.; Russell, J.B. Variations in the ability of ruminal gram-negative Prevotella species to resist monensin. Curr. Microbiol. 2000, 40, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J.; Stevenson, D.M.; Mertens, D.R.; Thomas, E.E. Effect of monensin feeding and withdrawal on populations of individual bacterial species in the rumen of lactating dairy cows fed high-starch rations. Appl. Microbiol. Biotechnol. 2008, 80, 135–145. [Google Scholar] [CrossRef]
- Bisson-Filho, A.W.; Hsu, Y.-P.; Squyres, G.R.; Kuru, E.; Wu, F.; Jukes, C.; Sun, Y.; Dekker, C.; Holden, S.; VanNieuwenhze, M.S. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 2017, 355, 739–743. [Google Scholar] [CrossRef] [Green Version]
- Chow, J.; Russell, J. Effect of ionophores and pH on growth of Streptococcus bovis in batch and continuous culture. Appl. Environ. Microbiol. 1990, 56, 1588–1593. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.B. A proposed mechanism of monensin action in inhibiting ruminant bacterial growth: Effects on ion flux and protonmotive force. Anim. Sci. J. 1987, 64, 1519–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karpel, R.; Alon, T.; Glaser, G.; Schuldiner, S.; Padan, E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J. Biol. Chem. 1991, 266, 21753–21759. [Google Scholar] [CrossRef]
- Murata, T.; Yamato, I.; Igarashi, K.; Kakinuma, Y. Intracellular Na+ regulates transcription of the ntp operon encoding a vacuolar-type Na+-translocating ATPase in Enterococcus hirae. J. Biol. Chem. 1996, 271, 23661–23666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mannarelli, B.M.; Ericsson, L.D.; Lee, D.; Stack, R.J. Taxonomic relationships among strains of the anaerobic bacterium Bacteroides ruminicola determined by DNA and extracellular polysaccharide analysis. Appl. Environ. Microbiol. 1991, 57, 2975–2980. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, T.; Yamane, K.; Furukawa, T.; Matsumoto-Mashimo, C.; Sugimori, C.; Nambu, T.; Obata, N.; Walker, C.B.; Leung, K.-P.; Fukushima, H. Comparison of the virulence of exopolysaccharide-producing Prevotella intermedia to exopolysaccharide non-producing periodontopathic organisms. BMC Infect. Dis. 2011, 11, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanin, S.L.; Sanin, F.D.; Bryers, J.D. Effect of starvation on the adhesive properties of xenobiotic degrading bacteria. Process Biochem. 2003, 38, 909–914. [Google Scholar] [CrossRef]
- Russell, J.B.; Houlihan, A.J. Ionophore resistance of ruminal bacteria and its potential impact on human health. FEMS Microbiol. Rev. 2003, 27, 65–74. [Google Scholar] [CrossRef]
- Yun, S.-H.; Choi, C.-W.; Park, S.-H.; Lee, J.C.; Leem, S.-H.; Choi, J.-S.; Kim, S.; Kim, S.I. Proteomic analysis of outer membrane proteins from Acinetobacter baumannii DU202 in tetracycline stress condition. J. Microbiol. 2008, 46, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Urfer, M.; Bogdanovic, J.; Monte, F.L.; Moehle, K.; Zerbe, K.; Omasits, U.; Ahrens, C.H.; Pessi, G.; Eberl, L.; Robinson, J.A. A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli. J. Biol. Chem. 2016, 291, 1921–1932. [Google Scholar] [CrossRef] [Green Version]
- King, L.B.; Pangburn, M.K.; McDaniel, L.S. Serine protease PKF of Acinetobacter baumannii results in serum resistance and suppression of biofilm formation. J. Infect. Dis. 2013, 207, 1128–1134. [Google Scholar] [CrossRef] [Green Version]
- Cabral, M.P.; Soares, N.C.; Aranda, J.; Parreira, J.R.; Rumbo, C.; Poza, M.; Valle, J.; Calamia, V.; Lasa, Í.; Bou, G. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 2011, 10, 3399–3417. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.H.; Jahn, A. Extraction of EPS. In Microbial Extracellular Polymeric Substances; Wingender, J., Neu, T.R., Flemming, H.C., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 49–72. [Google Scholar]
- Ding, Z.; Bourven, I.; Guibaud, G.; van Hullebusch, E.D.; Panico, A.; Pirozzi, F.; Esposito, G. Role of extracellular polymeric substances (EPS) production in bioaggregation: Application to wastewater treatment. Appl. Microbiol. Biotechnol. 2015, 99, 9883–9905. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, A.; Schleicher, L.; Deusch, S.; Gätgens, J.; Steuber, J.; Seifert, J. Short-chain fatty acids modulate metabolic pathways and membrane lipids in Prevotella bryantii B14. Proteomes 2020, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Vorburger, T.; Nedielkov, R.; Brosig, A.; Bok, E.; Schunke, E.; Steffen, W.; Mayer, S.; Götz, F.; Möller, H.M.; Steuber, J. Role of the Na+-translocating NADH: Quinone oxidoreductase in voltage generation and Na+ extrusion in Vibrio cholerae. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Terahara, N.; Sano, M.; Ito, M. A Bacillus flagellar motor that can use both Na+ and K+ as a coupling ion is converted by a single mutation to use only Na+. PLoS ONE 2012, 7, e46248. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- VDLUFA. Band VII: Umweltanalytik. In VDLUFA-Methodenbuch; Verband Deutscher Landwirtschaftlicher Untersuchungs-und Forschungsanstalten (VDLUFA) e. V.: Darmstadt, Germany, 2011; Available online: https://www.vdlufa.de/Methodenbuch/index.php?option=com_content&view=article&id=2&Itemid=113&lang=de (accessed on 11 January 2021).
- Sacks, D.B. Carbohydrates. In Tietz Textbook of Clinical Chemistry and Molecular Diagnostics; Burtis, C.A., Ashwood, E.R., Bruns, D.E., Eds.; Elsevier: St. Louis, MO, USA, 2012; pp. 709–729. [Google Scholar]
- Deusch, S.; Bok, E.; Schleicher, L.; Seifert, J.; Steuber, J. Occurrence and function of the Na+-translocating NADH: Quinone oxidoreductase in Prevotella spp. Microorganisms 2019, 7, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juárez, O.; Athearn, K.; Gillespie, P.; Barquera, B. Acid residues in the transmembrane helices of the Na+-pumping NADH: Quinone oxidoreductase from Vibrio cholerae involved in sodium translocation. Biochemistry 2009, 48, 9516–9524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, V.; Ullmann, R.; Kröger, A. The direction of the proton exchange associated with the redox reactions of menaquinone during electron transport in Wolinella succinogenes. Biochim. Biophys. Acta Biomembr. 1994, 1184, 219–226. [Google Scholar] [CrossRef]
- Olsen, J.V.; de Godoy, L.M.F.; Li, G.; Macek, B.; Mortensen, P.; Pesch, R.; Makarov, A.; Lange, O.; Horning, S.; Mann, M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteom. 2005, 4, 2010–2021. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [Green Version]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Di Rienzo, J.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tablada, M.; Robledo, C. InfoStat, Software Estadístico, Manual del Usuario; Universidad Nacional de Córdoba: Cordoba, Argentina, 2008; 268p. [Google Scholar]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trautmann, A.; Schleicher, L.; Pfirrmann, J.; Boldt, C.; Steuber, J.; Seifert, J. Na+-Coupled Respiration and Reshaping of Extracellular Polysaccharide Layer Counteract Monensin-Induced Cation Permeability in Prevotella bryantii B14. Int. J. Mol. Sci. 2021, 22, 10202. https://doi.org/10.3390/ijms221910202
Trautmann A, Schleicher L, Pfirrmann J, Boldt C, Steuber J, Seifert J. Na+-Coupled Respiration and Reshaping of Extracellular Polysaccharide Layer Counteract Monensin-Induced Cation Permeability in Prevotella bryantii B14. International Journal of Molecular Sciences. 2021; 22(19):10202. https://doi.org/10.3390/ijms221910202
Chicago/Turabian StyleTrautmann, Andrej, Lena Schleicher, Jana Pfirrmann, Christin Boldt, Julia Steuber, and Jana Seifert. 2021. "Na+-Coupled Respiration and Reshaping of Extracellular Polysaccharide Layer Counteract Monensin-Induced Cation Permeability in Prevotella bryantii B14" International Journal of Molecular Sciences 22, no. 19: 10202. https://doi.org/10.3390/ijms221910202
APA StyleTrautmann, A., Schleicher, L., Pfirrmann, J., Boldt, C., Steuber, J., & Seifert, J. (2021). Na+-Coupled Respiration and Reshaping of Extracellular Polysaccharide Layer Counteract Monensin-Induced Cation Permeability in Prevotella bryantii B14. International Journal of Molecular Sciences, 22(19), 10202. https://doi.org/10.3390/ijms221910202






