Urinary Extracellular Vesicles Are a Novel Tool to Monitor Allograft Function in Kidney Transplantation: A Systematic Review
Abstract
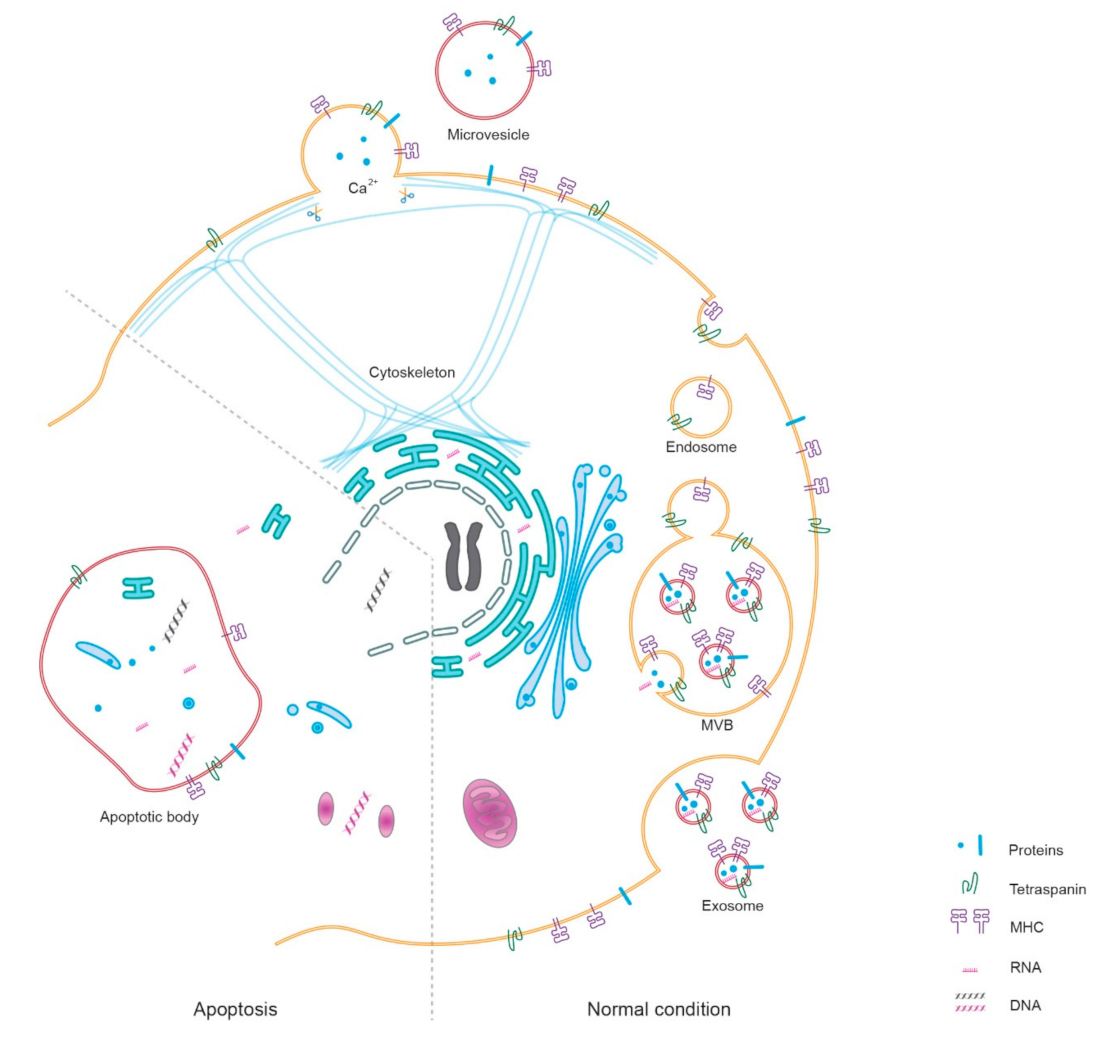
:1. Introduction
2. Materials and Methods
2.1. Data Sources and Searches
2.2. Study Selection and Eligibility Criteria
2.3. Data Extraction and Quality Assessment
3. Results
4. Discussion
4.1. Kidney Specific Markers
4.1.1. The Biogenesis of Kidney Specific Markers
4.1.2. The Diagnostic Potential of Kidney Specific Markers
4.2. Donor Specific Markers
4.2.1. HLA
4.2.2. Cell-Free DNA
4.3. Immune Response-Related Markers in uEV
4.3.1. Proteins
4.3.2. RNA
4.3.3. Viral Infection Related Markers
4.4. Challenges and Future Perspective
- many studies omit the necessity of normalizing urine dilution (Table 1);
- some studies merely detect the expression level of markers in the total uEV, rather than measure a particular “marker”-positive population of uEV;
- some clinical research is limited by small numbers of samples or a poor or non-existent description regarding the clinical properties of enrolled patients;
- the significant proteomic changes of uEV in some types of kidney dysfunction have been well studied, whereas the diagnostic accuracy (sensitivity, specificity, and AUC) of these changes is still unclear.
- The normalization of uEV concentration/urine dilution is necessary in the research protocol. According to the newest guidelines published by the Urine Task Force of the International Society for Extracellular Vesicles, variable dilution of urine is a major challenge in uEV research [76]. Whether in healthy or unwell subjects, the concentration of uEV is highly influenced by water-loading [77]. For this, two methods are commonly recommended to realize normalization: (1) the relative excretion rate is based on the ratio between target-marker and other markers of uEV (e.g., numbers, the total yield of protein or RNA, tetraspanin, prostate-specific markers, etc.); (2) the absolute excretion rate is based on a long-term collection of urine or the ratio between target-marker and urinary creatinine [76].
- The measurement of markers in uEV mostly depends on isolation. This could cause bias, especially loss or selection of uEV due to diverse methods, e.g., ultracentrifugation, size exclusion chromatography, precipitation [78,79,80]. More efforts are still needed to compare the yield and purity of uEV between techniques. To avoid isolation bias, more sensitive techniques, such as imaging flow cytometry, might provide the possibility of independence of isolation, also facilitating the detection of a particular population of uEV [81], especially the donor-specific HLA-positive uEV [78]. Analysis of subpopulations of uEVs will reveal their specific contribution in clinical events by zooming in on uEV subpopulations; their role in rejection and other complications after transplantation will then become evident. This improves the value of uEV measurements like Western blot and immunoblot analysis.
- The data is based on small, single-center studies, which makes it difficult to draw conclusions. Future studies should take a multi-center approach which enables large patient numbers. This should take into account clinical properties including (1) gender, race, age, body size, HLA-mismatch, and common chronic diseases (e.g., hypertension, diabetes mellitus, and hepatitis virus infection) of donor and recipient; (2) the type and proportion of donors, e.g., living donor or deceased donor; (3) immunosuppressor-treated or -free, and the type of medicine administered. This information is of critical value for multivariate analysis, which will provide results defining the diagnostic accuracy of uEV measurement in kidney transplantation.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Axelrod, D.A.; Schnitzler, M.A.; Xiao, H.; Irish, W.; Tuttle-Newhall, E.; Chang, S.H.; Kasiske, B.L.; Alhamad, T.; Lentine, K.L. An Economic Assessment of Contemporary Kidney Transplant Practice. Am. J. Transplant. 2018, 18, 1168–1176. [Google Scholar] [CrossRef] [Green Version]
- Hart, A.; Smith, J.M.; Skeans, M.A.; Gustafson, S.K.; Wilk, A.R.; Castro, S.; Foutz, J.; Wainright, J.L.; Snyder, J.J.; Kasiske, B.L.; et al. OPTN/SRTR 2018 Annual Data Report: Kidney. Am. J. Transplant. 2020, 20, 20–130. [Google Scholar] [CrossRef]
- Kramer, A.; Boenink, R.; Noordzij, M.; Bosdriesz, J.R.; Stel, V.S.; Beltrán, P.; Ruiz, J.C.; Seyahi, N.; Comas Farnés, J.; Stendahl, M.; et al. The ERA-EDTA Registry Annual Report 2017: A Summary. Clin. Kidney J. 2020, 13, 693–709. [Google Scholar] [CrossRef]
- Gielis, E.M.; Beirnaert, C.; Dendooven, A.; Meysman, P.; Laukens, K.; De Schrijver, J.; Van Laecke, S.; Van Biesen, W.; Emonds, M.P.; De Winter, B.Y.; et al. Plasma Donor-Derived Cell-Free DNA Kinetics after Kidney Transplantation Using a Single Tube Multiplex PCR Assay. PLoS ONE 2018, 13, e0208207. [Google Scholar] [CrossRef]
- Goldberg, R.J.; Weng, F.L.; Kandula, P. Acute and Chronic Allograft Dysfunction in Kidney Transplant Recipients. Med. Clin. N. Am. 2016, 100, 487–503. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 2001–3078. [Google Scholar] [CrossRef] [Green Version]
- Găman, M.-A.; Cozma, M.-A.; Dobrică, E.-C.; Crețoiu, S.M.; Găman, A.M.; Diaconu, C.C. Liquid Biopsy and Potential Liquid Biopsy-Based Biomarkers in Philadelphia-Negative Classical Myeloproliferative Neoplasms: A Systematic Review. Life 2021, 11, 677. [Google Scholar] [CrossRef]
- Fekih, R.E.; Hurley, J.; Tadigotla, V.; Alghamdi, A.; Srivastava, A.; Coticchia, C.; Choi, J.; Allos, H.; Yatim, K.; Alhaddad, A.; et al. Discovery and Validation of a Urinary Exosome MRNA Signature for the Diagnosis of Human Kidney Transplant Rejection. J. Am. Soc. Nephrol. 2021, 32, 994–1004. [Google Scholar] [CrossRef]
- Santucci, L.; Bruschi, M.; Del Zotto, G.; Antonini, F.; Ghiggeri, G.M.; Panfoli, I.; Candiano, G. Biological Surface Properties in Extracellular Vesicles and Their Effect on Cargo Proteins. Sci. Rep. 2019, 9, 13048. [Google Scholar] [CrossRef] [Green Version]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, 30. [Google Scholar] [CrossRef]
- Svenningsen, P.; Sabaratnam, R.; Jensen, B.L. Urinary Extracellular Vesicles: Origin, Role as Intercellular Messengers and Biomarkers; Efficient Sorting and Potential Treatment Options. Acta Physiol. 2020, 228, e13346. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Poulsen, S.; Murali, S.; Grimm, P.R.; Su, X.-T.; Delpire, E.; Welling, P.; Ellison, D.; Fenton, R. Large-Scale Proteomic Assessment of Urinary Extracellular Vesicles Highlights Their Reliability in Reflecting Protein Changes in the Kidney. J. Am. Soc. Nephrol. 2021, 32, 2195–2209. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for Reporting Observational Studies. Lancet 2007, 370, 1453–1457. [Google Scholar] [CrossRef]
- Sonoda, H.; Yokota-Ikeda, N.; Oshikawa, S.; Kanno, Y.; Yoshinaga, K.; Uchida, K.; Ueda, Y.; Kimiya, K.; Uezono, S.; Ueda, A.; et al. Decreased Abundance of Urinary Exosomal Aquaporin-1 in Renal Ischemia-Reperfusion Injury. Am. J. Physiol.-Ren. Physiol. 2009, 297, F1006–F1016. [Google Scholar] [CrossRef] [Green Version]
- Pisitkun, T.; Gandolfo, M.T.; Das, S.; Knepper, M.A.; Bagnasco, S.M. Application of Systems Biology Principles to Protein Biomarker Discovery: Urinary Exosomal Proteome in Renal Transplantation. Proteom.-Clin. Appl. 2012, 6, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, S.; Suazo, C.; Boltansky, A.; Ursu, M.; Carvajal, D.; Innocenti, G.; Vukusich, A.; Hurtado, M.; Villanueva, S.; Carreño, J.E.; et al. Urinary Exosomes as a Source of Kidney Dysfunction Biomarker in Renal Transplantation. Transplant. Proc. 2013, 45, 3719–3723. [Google Scholar] [CrossRef] [PubMed]
- Dimuccio, V.; Ranghino, A.; Barbato, L.P.; Fop, F.; Biancone, L.; Camussi, G.; Bussolati, B. Urinary CD133+ Extracellular Vesicles Are Decreased in Kidney Transplanted Patients with Slow Graft Function and Vascular Damage. PLoS ONE 2014, 9, e104490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteva-Font, C.; Guillén-Gómez, E.; Diaz, J.M.; Guirado, L.; Facundo, C.; Ars, E.; Ballarin, J.A.; Fernández-Llama, P. Renal Sodium Transporters Are Increased in Urinary Exosomes of Cyclosporine-Treated Kidney Transplant Patients. Am. J. Nephrol. 2014, 39, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Ng, Y.W.; Lee, S.; Nicora, C.D.; Qian, W.J.; Smith, R.D.; Camp, D.G.; Sarwal, M.M. Perturbations in the Urinary Exosome in Transplant Rejection. Front. Med. 2015, 2, 57. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.H.; Lee, Y.H.; Seo, J.W.; Moon, H.; Kim, J.S.; Kim, Y.G.Y.H.; Jeong, K.H.; Moon, J.Y.; Lee, T.W.; Ihm, C.G.; et al. Urinary Exosomal Viral MicroRNA as a Marker of BK Virus Nephropathy in Kidney Transplant Recipients. PLoS ONE 2017, 12, e0190068. [Google Scholar] [CrossRef]
- Park, J.; Lin, H.-Y.Y.; Assaker, J.P.; Jeong, S.; Huang, C.-H.H.; Kurdi, A.; Lee, K.; Fraser, K.; Min, C.; Eskandari, S.; et al. Integrated Kidney Exosome Analysis for the Detection of Kidney Transplant Rejection. ACS Nano 2017, 11, 11041–11046. [Google Scholar] [CrossRef]
- Tutakhel, O.A.Z.; Moes, A.D.; Valdez-Flores, M.A.; Kortenoeven, M.L.A.; Vrie, M.V.D.; Jelen, S.; Fenton, R.A.; Zietse, R.; Hoenderop, J.G.J.; Hoorn, E.J.; et al. NaCl Cotransporter Abundance in Urinary Vesicles Is Increased by Calcineurin Inhibitors and Predicts Thiazide Sensitivity. PLoS ONE 2017, 12, e0176220. [Google Scholar] [CrossRef] [Green Version]
- Hinrichs, G.R.; Michelsen, J.S.; Zachar, R.; Friis, U.G.; Svenningsen, P.; Birn, H.; Bistrup, C.; Jensen, B.L. Albuminuria in Kidney Transplant Recipients Is Associated with Increased Urinary Serine Proteases and Activation of the Epithelial Sodium Channel. Am. J. Physiol.-Ren. Physiol. 2018, 315, F151–F160. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Lee, C.H.; Kim, K.Y.; Jung, H.Y.; Choi, J.Y.; Cho, J.H.; Park, S.H.; Kim, Y.L.; Baek, M.C.; Park, J.B.; et al. Novel Urinary Exosomal Biomarkers of Acute T Cell-Mediated Rejection in Kidney Transplant Recipients: A Cross-Sectional Study. PLoS ONE 2018, 13, e0204204. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Planella, L.; Juega, J.; Taco, O.; Cañas, L.; Franquesa, M.; Lauzurica, R.; Borràs, F.E. Proteomic Characterization of Urinary Extracellular Vesicles from Kidney-Transplanted Patients Treated with Calcineurin Inhibitors. Int. J. Mol. Sci. 2020, 21, 7569. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.Y.; Lee, C.H.; Choi, J.Y.; Cho, J.H.; Park, S.H.; Kim, Y.L.; Moon, P.G.; Baek, M.C.; Berm Park, J.; Hoon Kim, Y.; et al. Potential Urinary Extracellular Vesicle Protein Biomarkers of Chronic Active Antibody-Mediated Rejection in Kidney Transplant Recipients. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1138, 121958. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Kamimura, D.; Jiang, J.J.; Higuchi, H.; Iwami, D.; Hotta, K.; Tanaka, Y.; Ota, M.; Higuchi, M.; Nishio, S.; et al. Increased Urinary Exosomal SYT17 Levels in Chronic Active Antibody-Mediated Rejection after Kidney Transplantation via the IL-6 Amplifier. Int. Immunol. 2020, 32, 653–662. [Google Scholar] [CrossRef]
- Liu, Y.; Li, S.; Rong, W.; Zeng, C.; Zhu, X.; Chen, Q.; Li, L.; Liu, Z.-H.; Zen, K. Podocyte-Released Migrasomes in Urine Serve as an Indicator for Early Podocyte Injury. Kidney Dis. 2020, 6, 422–433. [Google Scholar] [CrossRef]
- Medeiros, T.; Myette, R.L.; Almeida, J.R.; Silva, A.A.; Burger, D. Extracellular Vesicles: Cell-Derived Biomarkers of Glomerular and Tubular Injury. Cell. Physiol. Biochem. 2020, 54, 88–109. [Google Scholar] [CrossRef] [Green Version]
- Turco, A.E.; Lam, W.; Rule, A.D.; Denic, A.; Lieske, J.C.; Miller, V.M.; Larson, J.J.; Kremers, W.K.; Jayachandran, M. Specific Renal Parenchymal-Derived Urinary Extracellular Vesicles Identify Age-Associated Structural Changes in Living Donor Kidneys. J. Extracell. Vesicles 2016, 5, 29642. [Google Scholar] [CrossRef]
- Karpman, D.; Tontanahal, A. Extracellular Vesicles in Renal Inflammatory and Infectious Diseases. Sci. Total Environ. 2019, 171, 135907. [Google Scholar]
- Benichou, G.; Wang, M.; Ahrens, K.; Madsen, J.C. Extracellular Vesicles in Allograft Rejection and Tolerance. Cell. Immunol. 2020, 349, 104063. [Google Scholar] [CrossRef]
- Urbanelli, L.; Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Porcellati, S.; Emiliani, C. The Role of Extracellular Vesicles in Viral Infection and Transmission. Vaccines 2019, 7, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and Proteomic Profiling of Exosomes in Human Urine. Proc. Natl. Acad. Sci. USA. 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, F.; Rinschen, M.; Buchner, D.; Bohl, K.; Plagmann, I.; Bachurski, D.; Richard Späth, M.; Antczak, P.; Göbel, H.; Klein, C.; et al. The Proteomic Landscape of Small Urinary Extracellular Vesicles during Kidney Transplantation. J. Extracell. Vesicles 2020, 10, e12026. [Google Scholar] [CrossRef]
- Oshikawa-Hori, S.; Yokota-Ikeda, N.; Sonoda, H.; Sasaki, Y.; Ikeda, M. Reduced Urinary Release of AQP1- and AQP2-bearing Extracellular Vesicles in Patients with Advanced Chronic Kidney Disease. Physiol. Rep. 2021, 9, e15005. [Google Scholar] [CrossRef]
- Oshikawa-Hori, S.; Yokota-Ikeda, N.; Sonoda, H.; Ikeda, M. Urinary Extracellular Vesicular Release of Aquaporins in Patients with Renal Transplantation. BMC Nephrol. 2019, 20, 216. [Google Scholar] [CrossRef] [PubMed]
- Sagrinati, C.; Netti, G.S.; Mazzinghi, B.; Lazzeri, E.; Liotta, F.; Frosali, F.; Ronconi, E.; Meini, C.; Gacci, M.; Squecco, R.; et al. Isolation and Characterization of Multipotent Progenitor Cells from the Bowman’s Capsule of Adult Human Kidneys. J. Am. Soc. Nephrol. 2006, 17, 2443–2456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-Scale Proteomics and Phosphoproteomics of Urinary Exosomes. J. Am. Soc. Nephrol. 2009, 20, 363–379. [Google Scholar] [CrossRef] [Green Version]
- Prunotto, M.; Farina, A.; Lane, L.; Pernin, A.; Schifferli, J.; Hochstrasser, D.F.; Lescuyer, P.; Moll, S. Proteomic Analysis of Podocyte Exosome-Enriched Fraction from Normal Human Urine. J. Proteom. 2013, 82, 193–229. [Google Scholar] [CrossRef] [PubMed]
- Schütz, E.; Fischer, A.; Beck, J.; Harden, M.; Koch, M.; Wuensch, T.; Stockmann, M.; Nashan, B.; Kollmar, O.; Matthaei, J.; et al. Donor-Derived Cell-Free DNA Is a Novel Universal Biomarker for Allograft Rejection in Solid Organ Transplantation. Am. J. Transplant. 2019, 19, 108–112. [Google Scholar]
- Verhoeven, J.G.H.P.; Boer, K.; Van Schaik, R.H.N.; Manintveld, O.C.; Huibers, M.M.H.; Baan, C.C.; Hesselink, D.A. Liquid Biopsies to Monitor Solid Organ Transplant Function. Ther. Drug Monit. 2018, 40, 515–525. [Google Scholar] [CrossRef]
- Verhoeven, J.G.H.P.; Peeters, A.M.A.; Hesselink, D.A.; Boer, K. Pitfalls in the Detection of Donor-Derived Cell-Free DNA in Transplant Recipients. Clin. Chem. 2021, 67, 1030–1032. [Google Scholar] [CrossRef]
- Knight, S.R.; Thorne, A.; Lo Faro, M.L. Donor-Specific Cell-Free DNA as a Biomarker in Solid Organ Transplantation. A Systematic Review. Transplantation 2019, 103, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Cossarizza, A.; Chang, H.D.; Radbruch, A.; Akdis, M.; Andrä, I.; Annunziato, F.; Bacher, P.; Barnaba, V.; Battistini, L.; Bauer, W.M.; et al. Guidelines for the Use of Flow Cytometry and Cell Sorting in Immunological Studies. Eur. J. Immunol. 2017, 47, 1584–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma Cells Release Exosomes Carrying MtDNA. J. Neural Transm. 2010, 117, 1. [Google Scholar] [CrossRef]
- Kalluri, R.; Lebleu, V.S. Discovery of Double-Stranded Genomic DNA in Circulating Exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New Evidence That a Large Proportion of Human Blood Plasma Cell-Free DNA Is Localized in Exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [Green Version]
- Sigdel, T.K.; Vitalone, M.J.; Tran, T.Q.; Dai, H.; Hsieh, S.C.; Salvatierra, O.; Sarwal, M.M. A Rapid Noninvasive Assay for the Detection of Renal Transplant Injury. Transplantation 2013, 96, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Coemans, M.; Van Loon, E.; Lerut, E.; Gillard, P.; Sprangers, B.; Senev, A.; Emonds, M.P.; Van Keer, J.; Callemeyn, J.; Daniëls, L.; et al. Occurrence of Diabetic Nephropathy after Renal Transplantation despite Intensive Glycemic Control: An Observational Cohort Study. Diabetes Care 2019, 42, 625–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, K.; Karl, B.; Mathew, A.V.; Gangoiti, J.A.; Wassel, C.L.; Saito, R.; Pu, M.; Sharma, S.; You, Y.-H.; Wang, L.; et al. Metabolomics Reveals Signature of Mitochondrial Dysfunction in Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2013, 24, 1901–1912. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.Z.; Kwan, B.C.H.; Chow, K.M.; Cheng, P.M.S.; Luk, C.C.W.; Li, P.K.T.; Szeto, C.C. Urinary Mitochondrial DNA Level Is an Indicator of Intra-Renal Mitochondrial Depletion and Renal Scarring in Diabetic Nephropathy. Nephrol. Dial. Transplant. 2018, 33, 784–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvia, S.L.; Musante, L.; Lannigan, J.; Gigliotti, J.C.; Le, T.H.; Erdbrugger, U. T-Cell Derived Extracellular Vesicles Are Elevated in Essential HTN. Am. J. Physiol.-Ren. Physiol. 2020, 319, F868–F875. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhong, X.; Ni, H.F.; Wang, C.; Tang, T.T.; Wang, L.T.; Song, K.Y.; Tang, R.N.; Liu, H.; Liu, B.C.; et al. Urinary Small Extracellular Vesicles Derived CCL21 MRNA as Biomarker Linked with Pathogenesis for Diabetic Nephropathy. J. Transl. Med. 2021, 19, 355. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sun, X.; Scicluna, B.J.; Coleman, B.M.; Hill, A.F. Characterization and Deep Sequencing Analysis of Exosomal and Non-Exosomal MiRNA in Human Urine. Kidney Int. 2014, 86, 433–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA Profiles in Subpopulations of Extracellular Vesicles: Apoptotic Bodies, Microvesicles and Exosomes. J. Extracell. Vesicles 2013, 2, 20677. [Google Scholar] [CrossRef]
- Yu, Y.; Bai, F.; Qin, N.; Liu, W.; Sun, Q.; Zhou, Y.; Yang, J. Non-Proximal Renal Tubule-Derived Urinary Exosomal MiR-200b as a Biomarker of Renal Fibrosis. Nephron 2018, 139, 269–282. [Google Scholar] [CrossRef]
- Franco-Acevedo, A.; Melo, Z.; Echavarria, R. Diagnostic, Prognostic, and Therapeutic Value of Non-Coding RNA Expression Profiles in Renal Transplantation. Diagnostics 2020, 10, 60. [Google Scholar] [CrossRef] [Green Version]
- Gildea, J.J.; Carlson, J.M.; Schoeffel, C.D.; Carey, R.M.; Felder, R.A. Urinary Exosome MiRNome Analysis and Its Applications to Salt Sensitivity of Blood Pressure. Clin. Biochem. 2013, 46, 1131–1134. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Ramos, S.I.; Bancu, I.; Carreras-Planella, L.; Monguió-Tortajada, M.; Cañas, L.; Juega, J.; Bonet, J.; Armengol, M.P.; Lauzurica, R.; Borràs, F.E. Molecular Profile of Urine Extracellular Vesicles from Normo-Functional Kidneys Reveal Minimal Differences between Living and Deceased Donors. BMC Nephrol. 2018, 19, 189. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.B.; Zheng, Y.; Jin, L.W.; Zhou, Z.H.; Li, Z.Y. Microvesicles Containing MicroRNA-21 Secreted by Proximal Tubular Epithelial Cells Are Involved in Renal Interstitial Fibrosis by Activating AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 707–714. [Google Scholar]
- Sonoda, H.; Lee, B.R.; Park, K.H.; Nihalani, D.; Yoon, J.H.; Ikeda, M.; Kwon, S.H. MiRNA Profiling of Urinary Exosomes to Assess the Progression of Acute Kidney Injury. Sci. Rep. 2019, 9, 4692. [Google Scholar] [CrossRef]
- Gniewkiewicz, M.S.; Paszkowska, I.; Gozdowska, J.; Czerwinska, K.; Sadowska-Jakubowicz, A.; Deborska-Materkowska, D.; Perkowska-Ptasinska, A.; Kosieradzki, M.; Durlik, M. Urinary MicroRNA-21-5p as Potential Biomarker of Interstitial Fibrosis and Tubular Atrophy (IFTA) in Kidney Transplant Recipients. Diagnostics 2020, 10, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalid, U.; Newbury, L.J.; Simpson, K.; Jenkins, R.H.; Bowen, T.; Bates, L.; Sheerin, N.S.; Chavez, R.; Fraser, D.J. A Urinary MicroRNA Panel That Is an Early Predictive Biomarker of Delayed Graft Function Following Kidney Transplantation. Sci. Rep. 2019, 9, 3584. [Google Scholar] [CrossRef]
- Zununi Vahed, S.; Omidi, Y.; Ardalan, M.; Samadi, N. Dysregulation of Urinary MiR-21 and MiR-200b Associated with Interstitial Fibrosis and Tubular Atrophy (IFTA) in Renal Transplant Recipients. Clin. Biochem. 2017, 50, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.M.; Volkmann, I.; Fiedler, J.; Schmidt, M.; Scheffner, I.; Haller, H.; Gwinner, W.; Thum, T. Urinary MiR-210 as a Mediator of Acute T-Cell Mediated Rejection in Renal Allograft Recipients. Am. J. Transplant. 2011, 11, 2221–2227. [Google Scholar] [CrossRef]
- Funahashi, Y. BK Virus-Associated Nephropathy after Renal Transplantation. Pathogens 2021, 10, 150. [Google Scholar] [CrossRef]
- Huang, Y.; Zeng, G.; Randhawa, P.S. Detection of BKV Encoded Mature MicroRNAs in Kidney Transplant Patients: Clinical and Biologic Insights. J. Clin. Virol. 2019, 119, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Akalin, E.; Azzi, Y.; Bartash, R.; Seethamraju, H.; Parides, M.; Hemmige, V.; Ross, M.; Forest, S.; Goldstein, Y.D.; Ajaimy, M.; et al. Covid-19 and Kidney Transplantation. N. Engl. J. Med. 2020, 382, 2475–2477. [Google Scholar] [CrossRef]
- Banerjee, D.; Popoola, J.; Shah, S.; Ster, I.C.; Quan, V.; Phanish, M. COVID-19 Infection in Kidney Transplant Recipients. Kidney Int. 2020, 97, 1076–1082. [Google Scholar] [CrossRef]
- Braun, F.; Lütgehetmann, M.; Pfefferle, S.; Wong, M.N.; Carsten, A.; Lindenmeyer, M.T.; Nörz, D.; Heinrich, F.; Meißner, K.; Wichmann, D.; et al. SARS-CoV-2 Renal Tropism Associates with Acute Kidney Injury. Lancet 2020, 396, 597–598. [Google Scholar] [CrossRef]
- Wurtzer, S.; Waldman, P.; Ferrier-Rembert, A.; Frenois-Veyrat, G.; Mouchel, J.M.; Boni, M.; Maday, Y.; Marechal, V.; Moulin, L. Several Forms of SARS-CoV-2 RNA Can Be Detected in Wastewaters: Implication for Wastewater-Based Epidemiology and Risk Assessment. Water Res. 2021, 198, 117183. [Google Scholar] [CrossRef]
- Mishra, R.; Banerjea, A.C. SARS-CoV-2 Spike Targets USP33-IRF9 Axis via Exosomal MiR-148a to Activate Human Microglia. Front. Immunol. 2021, 12, 656700. [Google Scholar] [CrossRef] [PubMed]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Juan, M.; et al. Urinary Extracellular Vesicles: A Position Paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef] [PubMed]
- Blijdorp, C.J.; Tutakhel, O.A.Z.; Hartjes, T.A.; van den Bosch, T.P.P.; van Heugten, M.H.; Rigalli, J.P.; Willemsen, R.; Musterd-bhaggoe, U.M.; Barros, E.R.; Carles-fontana, R.; et al. Comparing Approaches to Normalize, Quantify, and Characterize Urinary Extracellular Vesicles. J. Am. Soc. Nephrol. 2021, 32, 1210–1226. [Google Scholar] [CrossRef] [PubMed]
- Mastoridis, S. Multiparametric Analysis of Circulating Exosomes and Other Small Extracellular Vesicles by Advanced Imaging Flow Cytometry. Front. Immunol. 2018, 9, 1583. [Google Scholar] [CrossRef] [Green Version]
- Freitas, D.; Balmaña, M.; Poças, J.; Campos, D.; Osório, H.; Konstantinidi, A.; Vakhrushev, S. Different Isolation Approaches Lead to Diverse Glycosylated Extracellular Vesicle Populations. J. Extracell. Vesicles 2019, 8, 1621131. [Google Scholar] [CrossRef] [Green Version]
- Liangsupree, T.; Multia, E.; Riekkola, M.L. Modern Isolation and Separation Techniques for Extracellular Vesicles. J. Chromatogr. A 2021, 1636, 461773. [Google Scholar] [CrossRef]
- Görgens, A.; Bremer, M.; Ferrer-Tur, R.; Murke, F.; Tertel, T.; Horn, P.A.; Thalmann, S.; Welsh, J.A.; Probst, C.; Guerin, C.; et al. Optimisation of Imaging Flow Cytometry for the Analysis of Single Extracellular Vesicles by Using Fluorescence-Tagged Vesicles as Biological Reference Material. J. Extracell. Vesicles 2019, 8, 1587567. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Authors | Year of Publication | Country of Origin | Outcomes and Population | Timing of uEV Measurement | Technique | Normalization | Marker in uEV | Diagnostic Information |
|---|---|---|---|---|---|---|---|---|
| Sonoda et al. [16] | 2009 | Japan | Early I/R after transplantation (n = 1) | Day 1 before and day 2, 6 after transplantation | Immunoblot analysis | No | AQP1 |
|
| Pisitkun et al. [17] | 2012 | USA | Nonspecific findings (n = 2); TI (n = 7); TCMR (n = 6); AMR (n = 3) | Biopsy after transplantation | Large-scale liquid chromatography-tandem mass spectrometry | No | Proteome | Compared to all KTR:
|
| Alvarez et al. [18] | 2013 | Chile | Non-DGF (n = 12); DGF (n = 3) | Day 1, 2, 3 after transplantation | Western blotting | No | NGAL |
|
| Dimuccio et al. [19] | 2014 | Italy | Early graft function (n = 13); DGF (n = 12) | Hour 6, day 1, 7, 30 after transplantation | Cytofluorimetric analysis & Western blotting | Ratio of the expression level in CD133+ uEV and in total uEV | CD133 |
|
| Esteva-Font et al. [20] | 2014 | Spain | CsA-free (n = 8); CsA-treated (n = 39) | 1 year after transplantation | Immunoblotting | 24-h urine volume | NCC & NKCC2 |
|
| Sigdel et al. [21] | 2015 | USA | No rejection (n = 20); acute rejection (n = 10) | Biopsy after transplantation | Isobaric tags for relative and absolute quantitation & nanoLC-MS/MS | No | Proteome | DEFA5, CD5L, APOM, A2M, APOA2, PROS1, IGHM, FGA, and FGB were significantly increased in the acute rejection group. |
| Kim et al. [22] | 2017 | South Korea | Normal (n = 15); BKVN (n = 13); TCMR (n = 27); acute AMR (n = 9); CAMR (n = 16) | Biopsy after transplantation | Quantitative real-time polymerase chain reaction | MiR-16 | Viral microRNA |
|
| Park et al. [23] | 2017 | USA | No rejection (n = 22); acute rejection (n = 22) | Biopsy after transplantation | Integrated kidney exosome analysis | No | CD3 | Increased CD3 could diagnose acute rejection with AUC value of 0.911 and cut-off value of 0.298 μA (sensitivity 92.8%, specificity 87.5%) in discovery set, with AUC value of 0.837. (sensitivity 63.6%, specificity 100%) in validation set. |
| Tutakhel et al. [24] | 2017 | Netherlands | CNIs-free (n = 13); CsA-treated (n = 9); Tacrolimus-treated (n = 23) | At least 6 months after transplantation | Immunoblotting | Ratio of phosphorylated NCC and total NCC | NCC | The expression of total NCC or phosphorylated NCC (Thr60) in CNI-treated KTR was significantly higher. |
| Hinrichs et al. [25] | 2018 | Denmark | No albuminuria (n = 19); albuminuria (n = 18) | 1 year after transplantation | Western blotting | Urinary creatinine | γENaC | The expression of furin-cleaved γENaC and protease-cleaved γENaC (not full-length γENaC) was significantly increased in KTR with albuminuria. |
| Lim et al. [26] | 2018 | South Korea | Normal (n = 22); TCMR (n = 25) | Biopsy after transplantation | nanoLC-MS/MS & Western blotting | No | Proteome |
|
| Carreras-Planella et al. [27] | 2020 | Spain | Normal (n = 7); CNIs nephrotoxicity (n = 5); IFTA (n = 5) | Biopsy after transplantation | Mass spectrometry | Ezrin | Proteome | Compared to IFTA:
|
| Jung et al. [28] | 2020 | South Korea | Long-term graft survival (n = 57); CAMR (n = 26) | Biopsy after transplantation | Liquid chromatography–mass spectrometry | No | Proteome |
|
| Takada et al. [29] | 2020 | Japan | Normal (n = 20); IFTA (n = 19); CNIs nephrotoxicity (n = 17); CAMR (n = 22) | Biopsy after transplantation | Western blotting | CD9 | SYT17 |
|
| Fekih et al. [8] | 2021 | USA | No rejection (n = 133); acute AMR (n = 8); CAMR (n = 16); TCMR (n = 35); borderline TCMR (n = 23); BKVN (n = 5) | Biopsy after transplantation | Quantitative real-time polymerase chain reaction | No | Messenger RNA |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Boer, K.; Woud, W.W.; Udomkarnjananun, S.; Hesselink, D.A.; Baan, C.C. Urinary Extracellular Vesicles Are a Novel Tool to Monitor Allograft Function in Kidney Transplantation: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 10499. https://doi.org/10.3390/ijms221910499
Wu L, Boer K, Woud WW, Udomkarnjananun S, Hesselink DA, Baan CC. Urinary Extracellular Vesicles Are a Novel Tool to Monitor Allograft Function in Kidney Transplantation: A Systematic Review. International Journal of Molecular Sciences. 2021; 22(19):10499. https://doi.org/10.3390/ijms221910499
Chicago/Turabian StyleWu, Liang, Karin Boer, Wouter W. Woud, Suwasin Udomkarnjananun, Dennis A. Hesselink, and Carla C. Baan. 2021. "Urinary Extracellular Vesicles Are a Novel Tool to Monitor Allograft Function in Kidney Transplantation: A Systematic Review" International Journal of Molecular Sciences 22, no. 19: 10499. https://doi.org/10.3390/ijms221910499






