Alpha-1 Antitrypsin and Hepatocellular Carcinoma in Liver Cirrhosis: SERPINA1 MZ or MS Genotype Carriage Decreases the Risk
Abstract
:1. Introduction
2. Results
2.1. Genotype Frequency in Cases and Controls
2.2. Patients’ Demographic and Laboratory Data According to the SERPINA1 Genotype
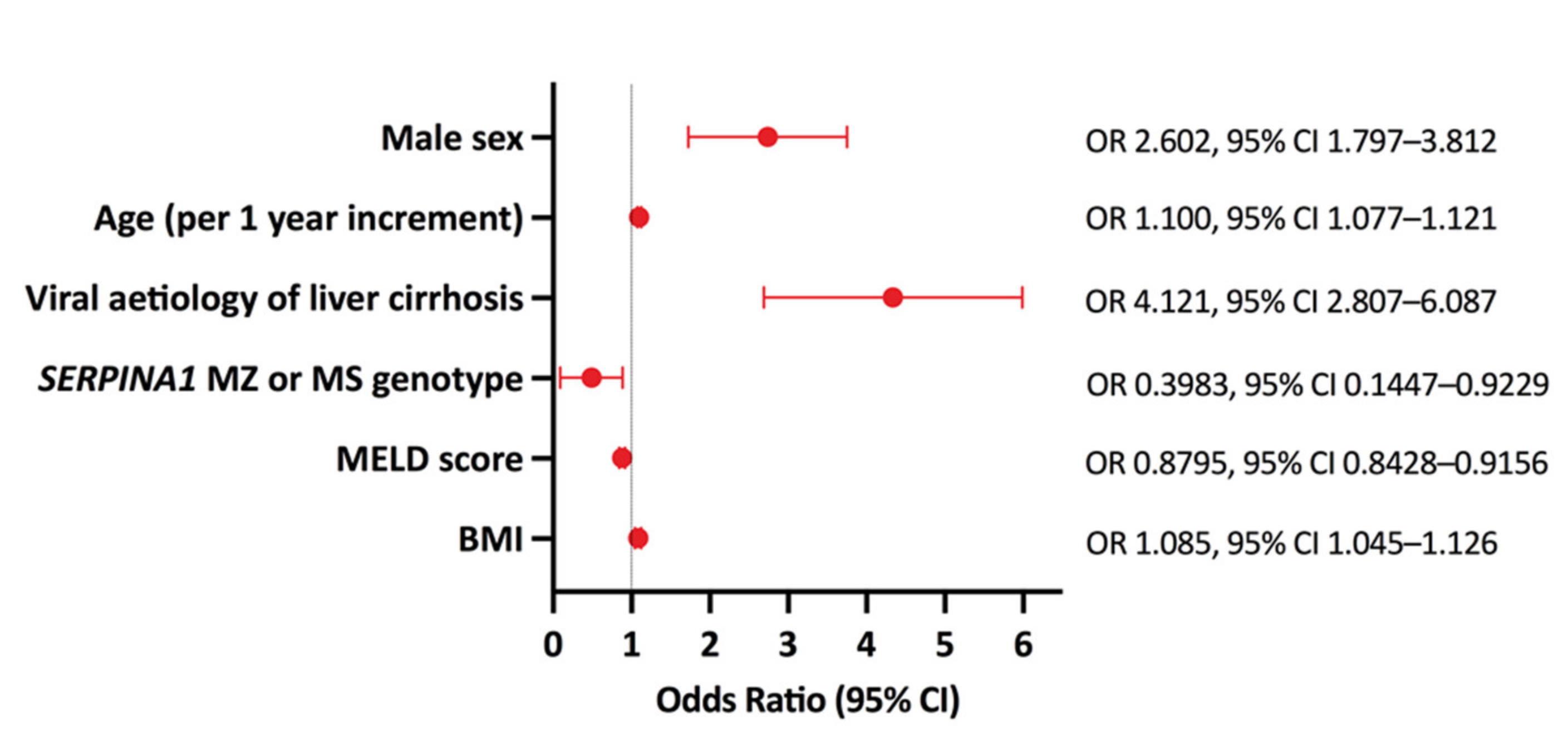
2.3. Multivariate Analysis of HCC Risk Factors
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Study Design and Eligibility of Patients
5.2. Genotyping
5.3. AAT and TNF-Alpha Serum Levels Assessment
5.4. Explanted Liver Tissue Histological Analysis
5.5. Statistical Analysis
5.6. Ethics Statement
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laurell, C.B.; Eriksson, S. [HYPO-ALPHA-1-ANTITRYPSINEMIA]. Verh. Dtsch. Ges. Inn. Med. 1964, 70, 537–539. [Google Scholar]
- Corley, M.; Solem, A.; Phillips, G.; Lackey, L.; Ziehr, B.; Vincent, H.A.; Mustoe, A.M.; Ramos, S.B.V.; Weeks, K.M.; Moorman, N.J.; et al. An RNA structure-mediated, posttranscriptional model of human α-1-antitrypsin expression. Proc. Natl. Acad. Sci. USA 2017, 114, E10244–E10253. [Google Scholar] [CrossRef] [Green Version]
- Lackey, L.; McArthur, E.; Laederach, A. Increased Transcript Complexity in Genes Associated with Chronic Obstructive Pulmonary Disease. PLoS ONE 2015, 10, e0140885. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Oton-Gonzalez, L.; Selvatici, R.; Rizzo, P.; Pavasini, R.; Campo, G.C.; Lanzillotti, C.; Mazziotta, C.; De Mattei, M.; Tognon, M.; et al. SERPINA1 Gene Promoter Is Differentially Methylated in Peripheral Blood Mononuclear Cells of Pregnant Women. Front. Cell Dev. Biol. 2020, 8, 550543. [Google Scholar] [CrossRef] [PubMed]
- Blanco, I.; Bueno, P.; Diego, I.; Pérez-Holanda, S.; Casas, F.; Esquinas, C.; Miravitlles, M. Alpha-1 antitrypsin Pi*Z gene frequency and Pi*ZZ genotype numbers worldwide: An update. Int. J. Chronic Obstr. Pulm. Dis. 2017, 12, 561–569. [Google Scholar] [CrossRef] [Green Version]
- Janciauskiene, S.; Eriksson, S.; Callea, F.; Mallya, M.; Zhou, A.; Seyama, K.; Hata, S.; Lomas, D.A. Differential detection of PAS-positive inclusions formed by the Z, Siiyama, and Mmalton variants of alpha1-antitrypsin. Hepatology 2004, 40, 1203–1210. [Google Scholar] [CrossRef]
- Le, A.; Ferrell, G.A.; Dishon, D.S.; Le, Q.Q.; Sifers, R.N. Soluble aggregates of the human PiZ alpha 1-antitrypsin variant are degraded within the endoplasmic reticulum by a mechanism sensitive to inhibitors of protein synthesis. J. Biol. Chem. 1992, 267, 1072–1080. [Google Scholar] [CrossRef]
- Fra, A.; Cosmi, F.; Ordoñez, A.; Berardelli, R.; Perez, J.; Guadagno, N.A.; Corda, L.; Marciniak, S.; Lomas, D.A.; Miranda, E. Polymers of Z α1-antitrypsin are secreted in cell models of disease. Eur. Respir. J. 2016, 47, 1005–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Dickens, J.A.; DeMeo, D.L.; Miranda, E.; Perez, J.; Rashid, S.T.; Day, J.; Ordóñez, A.; Marciniak, S.J.; Haq, I.; et al. Circulating polymers in 1-antitrypsin deficiency. Eur. Respir. J. 2014, 43, 1501–1504. [Google Scholar] [CrossRef] [Green Version]
- Lomas, D.A.; Li-Evans, D.; Finch, J.T.; Carrell, R.W. The mechanism of Z α1-antitrypsin accumulation in the liver. Nature 1992, 357, 605–607. [Google Scholar] [CrossRef]
- Feldmann, G.; Martin, J.P.; Sesboue, R.; Ropartz, C.; Perelman, R.; Nathanson, M.; Seringe, P.; Benhamou, J.P. The ultrastructure of hepatocytes in alpha-1-antitrypsin deficiency with the genotype Pi. Gut 1975, 16, 796–799. [Google Scholar] [CrossRef]
- Silva, D.; Oliveira, M.J.; Guimarães, M.; Lima, R.; Gomes, S.; Seixas, S. Alpha-1-antitrypsin (SERPINA1) mutation spectrum: Three novel variants and haplotype characterization of rare deficiency alleles identified in Portugal. Respir. Med. 2016, 116, 8–18. [Google Scholar] [CrossRef] [Green Version]
- The 1000 Genomes Project Consortium; Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Tennessen, J.A.; Bigham, A.W.; O’Connor, T.D.; Fu, W.; Kenny, E.E.; Gravel, S.; McGee, S.; Do, R.; Liu, X.; Jun, G.; et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science 2012, 337, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, S. Pulmonary Emphysema and Alpha1-Antitrypsin Deficiency. Acta Medica Scand. 1964, 175, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.C.; Marek, G.; Liu, C.; Collinsworth, A.; Shuster, J.; Kurtz, T.; Nolte, J.; Brantly, M. Clinical and histologic features of adults with alpha-1 antitrypsin deficiency in a non-cirrhotic cohort. J. Hepatol. 2018, 69, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Hamesch, K.; Mandorfer, M.; Pereira, V.M.; Moeller, L.S.; Pons, M.; Dolman, G.E.; Reichert, M.C.; Schneider, C.V.; Woditsch, V.; Voss, J.; et al. Liver Fibrosis and Metabolic Alterations in Adults with alpha-1-antitrypsin Deficiency Caused by the Pi*ZZ Mutation. Gastroenterology 2019, 157, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef]
- Strnad, P.; Buch, S.; Hamesch, K.; Fischer, J.; Rosendahl, J.; Schmelz, R.; Brueckner, S.; Brosch, M.; Heimes, C.V.; Woditsch, V.; et al. Heterozygous carriage of the alpha1-antitrypsin Pi*Z variant increases the risk to develop liver cirrhosis. Gut 2018, 68, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Cacciottolo, T.M.; Gelson, W.T.; Maguire, G.; Davies, S.E.; Griffiths, W.J. Pi*Z heterozygous alpha-1 antitrypsin states accelerate parenchymal but not biliary cirrhosis. Eur. J. Gastroenterol. Hepatol. 2014, 26, 412–417. [Google Scholar] [CrossRef]
- Hurley, K.; Reeves, E.P.; Carroll, T.P.; McElvaney, N.G. Tumor necrosis factor-α driven inflammation in alpha-1 antitrypsin deficiency: A new model of pathogenesis and treatment. Expert Rev. Respir. Med. 2015, 10, 207–222. [Google Scholar] [CrossRef]
- Subramaniyam, D.; Virtala, R.; Pawłowski, K.; Clausen, I.G.; Warkentin, S.; Stevens, T.; Janciauskiene, S. TNF-α-induced self expression in human lung endothelial cells is inhibited by native and oxidized α1-antitrypsin. Int. J. Biochem. Cell Biol. 2008, 40, 258–271. [Google Scholar] [CrossRef]
- Hurley, K.; Lacey, N.; O’Dwyer, C.A.; Bergin, D.A.; McElvaney, O.J.; O’Brien, M.E.; McElvaney, O.F.; Reeves, E.P.; McElvaney, N.G. Alpha-1 Antitrypsin Augmentation Therapy Corrects Accelerated Neutrophil Apoptosis in Deficient Individuals. J. Immunol. 2014, 193, 3978–3991. [Google Scholar] [CrossRef] [Green Version]
- Eagan, T.M.L.; Ueland, T.; Wagner, P.D.; Hardie, J.A.; Mollnes, T.E.; Damas, J.K.; Aukrust, P.; Bakke, P.S. Systemic inflammatory markers in COPD: Results from the Bergen COPD Cohort Study. Eur. Respir. J. 2009, 35, 540–548. [Google Scholar] [CrossRef] [Green Version]
- Petrache, I.; Fijalkowska, I.; Medler, T.R.; Skirball, J.; Cruz, P.; Zhen, L.; Petrache, H.I.; Flotte, T.R.; Tuder, R.M. α-1 Antitrypsin Inhibits Caspase-3 Activity, Preventing Lung Endothelial Cell Apoptosis. Am. J. Pathol. 2006, 169, 1155–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockett, A.D.; Van Demark, M.; Gu, Y.; Schweitzer, K.S.; Sigua, N.; Kamocki, K.; Fijalkowska, I.; Garrison, J.; Fisher, A.J.; Serban, K.; et al. Effect of Cigarette Smoke Exposure and Structural Modifications on the α-1 Antitrypsin Interaction with Caspases. Mol. Med. 2012, 18, 445–454. [Google Scholar] [CrossRef]
- Aldonytė, R.; Hutchinson, E.T.; Jin, B.; Brantly, M.; Block, E.; Patel, J.; Zhang, J. Endothelial Alpha-1-Antitrypsin Attenuates Cigarette Smoke Induced Apoptosis In Vitro. COPD J. Chronic Obstr. Pulm. Dis. 2008, 5, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Lomas, D.A. The Selective Advantage of α1-Antitrypsin Deficiency. Am. J. Respir. Crit. Care Med. 2006, 173, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Greulich, T.; Nell, C.; Hohmann, D.; Grebe, M.; Janciauskiene, S.; Koczulla, A.R.; Vogelmeier, C.F. The prevalence of diagnosed α1-antitrypsin deficiency and its comorbidities: Results from a large population-based database. Eur. Respir. J. 2016, 49, 1600154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkel, P.A.; Xie, G.; Monach, P.A.; Ji, X.; Ciavatta, D.J.; Byun, J.; Pinder, B.D.; Zhao, A.; Zhang, J.; Tadesse, Y.; et al. Identification of Functional and Expression Polymorphisms Associated with Risk for Antineutrophil Cytoplasmic Autoantibody–Associated Vasculitis. Arthritis Rheumatol. 2017, 69, 1054–1066. [Google Scholar] [CrossRef]
- Rahmattulla, C.; Mooyaart, A.; Van Hooven, D.; Schoones, J.W.; Bruijn, J.A.; Dekkers, O.; Bajema, I.M.; European Vasculitis Genetics Consortium. Genetic variants in ANCA-associated vasculitis: A meta-analysis. Ann. Rheum. Dis. 2015, 75, 1687–1692. [Google Scholar] [CrossRef]
- Callea, F. Natural history of hepatocellular carcinoma as viewed by the pathologist. Appl. Pathol. 1988, 6, 105–116. [Google Scholar]
- Giovannoni, I.; Callea, F.; Stefanelli, M.; Mariani, R.; Santorelli, F.M.; Francalanci, P. Alpha-1-antitrypsin deficiency: From genoma to liver disease. PiZ mouse as model for the development of liver pathology in human. Liver Int. 2014, 35, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Antoury, C. Alpha-1 antitrypsin deficiency and the risk of hepatocellular carcinoma in end-stage liver disease. World J. Hepatol. 2015, 7, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, B.; Mandorfer, M.; Viveiros, A.; Finkenstedt, A.; Ferenci, P.; Schneeberger, S.; Tilg, H.; Zoller, H. Heterozygosity for the alpha-1-antitrypsin Z allele in cirrhosis is associated with more advanced disease. Liver Transplant. 2018, 24, 744–751. [Google Scholar] [CrossRef]
- Schneider, C.V.; Hamesch, K.; Gross, A.; Mandorfer, M.; Moeller, L.S.; Pereira, V.; Pons, M.; Kuca, P.; Reichert, M.C.; Benini, F.; et al. Liver Phenotypes of European Adults Heterozygous or Homozygous for Pi*Z Variant of AAT (Pi*MZ vs Pi*ZZ genotype) and Noncarriers. Gastroenterology 2020, 159, 534–548. [Google Scholar] [CrossRef]
- Hakim, A.; Moll, M.; Qiao, D.; Liu, J.; Lasky-Su, J.A.; Silverman, E.K.; Vilarinho, S.; Jiang, Z.G.; Hobbs, B.D.; Cho, M.H. Heterozygosity of the Alpha-1-Antitrypsin Pi*Z Allele and Risk of Liver Disease. Hepatol. Commun. 2021, 5, 1348–1361. [Google Scholar] [CrossRef]
- Guillaud, O.; Jacquemin, E.; Couchonnal, E.; Vanlemmens, C.; Francoz, C.; Chouik, Y.; Conti, F.; Duvoux, C.; Hilleret, M.-N.; Kamar, N.; et al. Long term results of liver transplantation for alpha-1 antitrypsin deficiency. Dig. Liver Dis. 2020, 53, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Shahaf, G.; Moser, H.; Ozeri, E.; Mizrahi, M.; Abecassis, A.; Lewis, E.C. α-1-Antitrypsin Gene Delivery Reduces Inflammation, Increases T-Regulatory Cell Population Size and Prevents Islet Allograft Rejection. Mol. Med. 2011, 17, 1000–1011. [Google Scholar] [CrossRef]
- Guttman, O.; Baranovski, B.M.; Schuster, R.; Kaner, Z.; Freixo-Lima, G.S.; Bahar, N.; Kalay, N.; Mizrahi, M.I.; Brami, I.; Ochayon, D.E.; et al. Acute-phase protein α1-anti-trypsin: Diverting injurious innate and adaptive immune responses from non-authentic threats. Clin. Exp. Immunol. 2015, 179, 161–172. [Google Scholar] [CrossRef] [Green Version]
- Nita, I.M.; Serapinas, D.; Janciauskiene, S.M. α1-Antitrypsin regulates CD14 expression and soluble CD14 levels in human monocytes in vitro. Int. J. Biochem. Cell Biol. 2007, 39, 1165–1176. [Google Scholar] [CrossRef]
- Lewis, E.C.; Mizrahi, M.; Toledano, M.; DeFelice, N.; Wright, J.L.; Churg, A.; Shapiro, L.; Dinarello, C.A. 1-Antitrypsin monotherapy induces immune tolerance during islet allograft transplantation in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 16236–16241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergin, D.A.; Reeves, E.P.; Meleady, P.; Henry, M.; McElvaney, O.J.; Carroll, T.; Condron, C.; Chotirmall, S.H.; Clynes, M.; O’Neill, S.J.; et al. α-1 Antitrypsin regulates human neutrophil chemotaxis induced by soluble immune complexes and IL-8. J. Clin. Investig. 2010, 120, 4236–4250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergin, D.A.; Reeves, E.P.; Hurley, K.; Wolfe, R.; Jameel, R.; Fitzgerald, S.; McElvaney, N.G. The Circulating Proteinase Inhibitor α-1 Antitrypsin Regulates Neutrophil Degranulation and Autoimmunity. Sci. Transl. Med. 2014, 6, 217ra1. [Google Scholar] [CrossRef]
- Ozeri, E.; Mizrahi, M.; Shahaf, G.; Lewis, E.C. α-1 Antitrypsin Promotes Semimature, IL-10–Producing and Readily Migrating Tolerogenic Dendritic Cells. J. Immunol. 2012, 189, 146–153. [Google Scholar] [CrossRef]
- O’Dwyer, C.A.; O’Brien, M.E.; Wormald, M.R.; White, M.; Banville, N.; Hurley, K.; McCarthy, C.; McElvaney, N.G.; Reeves, E.P. The BLT1 Inhibitory Function of α-1 Antitrypsin Augmentation Therapy Disrupts Leukotriene B4Neutrophil Signaling. J. Immunol. 2015, 195, 3628–3641. [Google Scholar] [CrossRef] [Green Version]
- Mizrahi, M.; Cal, P.; Rosenthal, M.; Ochayon, D.; Shahaf, G.; Kaner, Z.; Kachker, P.; Lewis, E.C. Human Alpha-1-Antitrypsin Modifies B Lymphocyte Responses During Allograft Transplantation. Immunology 2013, 140, 362–373. [Google Scholar] [CrossRef]
- Bergin, D.A.; Hurley, K.; McElvaney, N.G.; Reeves, E.P. Alpha-1 Antitrypsin: A Potent Anti-Inflammatory and Potential Novel Therapeutic Agent. Arch. Immunol. Ther. Exp. 2012, 60, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Petrache, I.; Fijalkowska, I.; Zhen, L.; Medler, T.R.; Brown, E.; Cruz, P.; Choe, K.-H.; Taraseviciene-Stewart, L.; Scerbavicius, R.; Shapiro, L.; et al. A Novel Antiapoptotic Role for α1-Antitrypsin in the Prevention of Pulmonary Emphysema. Am. J. Respir. Crit. Care Med. 2006, 173, 1222–1228. [Google Scholar] [CrossRef] [Green Version]
- Kalis, M.; Kumar, R.; Janciauskiene, S.; Salehi, A.; Cilio, C.M. α 1-antitrypsin enhances insulin secretion and prevents cytokine-mediated apoptosis in pancreatic β-cells. Islets 2010, 2, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Garten, A.; Grohmann, T.; Kluckova, K.; Lavery, G.; Kiess, W.; Penke, M. Sorafenib-Induced Apoptosis in Hepatocellular Carcinoma Is Reversed by SIRT1. Int. J. Mol. Sci. 2019, 20, 4048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Zhu, M.; Xu, Z.; Li, W.; Dong, X.; Chen, Y.; Lin, B.; Li, M. Ropivacaine promotes apoptosis of hepatocellular carcinoma cells through damaging mitochondria and activating caspase-3 activity. Biol. Res. 2019, 52, 36. [Google Scholar] [CrossRef] [PubMed]
- Cífková, R.; Škodová, Z.; Bruthans, J.; Adámková, V.; Jozífová, M.; Galovcová, M.; Wohlfahrt, P.; Krajčoviechová, A.; Poledne, R.; Stávek, P.; et al. Longitudinal trends in major cardiovascular risk factors in the Czech population between 1985 and 2007/Czech MONICA and Czech post-MONICA. Atherosclerosis 2010, 211, 676–681. [Google Scholar] [CrossRef]
- Clarke, G.M.; Anderson, C.A.; Pettersson, F.H.; Cardon, L.R.; Morris, A.P.; Zondervan, K. Basic statistical analysis in genetic case-control studies. Nat. Protoc. 2011, 6, 121–133. [Google Scholar] [CrossRef] [Green Version]


| Genotype | Cases (Cirrhosis Group) (n = 1119) | Control Group (MONICA Study) (n = 3430) | p Value |
|---|---|---|---|
| MM | 1021 | 3240 | N.A. |
| MZ | 55 | 87 | <0.0001 OR 1.986; 95% CI 1.413–2.806 |
| MS | 32 | 101 | N.S. |
| SZ | 1 | 0 | N.A. |
| ZZ | 10 | 0 | <0.0001 OR Infinity; 95% CI 9.316–Infinity |
| SS | 0 | 2 | N.A. |
| SERPINA1 Genotype | Controls (n = 3430) | ALD Cirrhosis Subgroup (n = 360) | p Value | NASH Cirrhosis Subgroup (n = 146) | p Value | Pooled Subgroup of Other Cases * (n = 603) | p Value |
|---|---|---|---|---|---|---|---|
| MM | 3240 | 327 | N.A. | 125 | N.A. | 569 | N.A. |
| MZ | 87 | 24 | <0.001 OR 2.745 95% CI 1.744–4.325 | 16 | <0.001 OR 4.729 95% CI 2.733–8.298 | 15 | N.S. |
| MS | 101 | 9 | N.S. | 4 | N.S. | 19 | N.S. |
| SZ | 0 | 0 | N.A. | 0 | N.A. | 0 | N.A. |
| ZZ | 0 | 0 | N.A. | 0 | N.A. | 0 | N.A. |
| SS | 2 | 0 | N.A. | 0 | N.A. | 0 | N.A. |
| MM (%) (n = 1021) | MZ (%) (n = 55) | p Value MM vs. MZ | MS (%) (n = 32) | p Value MM vs. MS | ||
|---|---|---|---|---|---|---|
| Aetiology of liver cirrhosis | ALD | 327 (32%) | 24 (43.6%) | 0.002 | 9 (28.1%) | N.S. |
| NASH | 125 (12.2%) | 16 (29.1%) | 4 (12.5%) | |||
| VIR | 209 (20.5%) | 8 (14.6%) | 3 (9.4%) | |||
| AIH-CHOL | 302 (29.6%) | 6 (10.9%) | 12 (37.5%) | |||
| MET | 58 (5.7%) | 1 (1.8%) | 4 (12.5%) | |||
| HCC in liver cirrhosis | 243 (23.8%) | 5 (9.1%) | 0.0085 * | 1 (0.4%) | 0.0044 ** | |
| MM (n = 1021) | MZ (n = 55) | MS (n = 32) | ||||||
|---|---|---|---|---|---|---|---|---|
| Number of Analysed Subjects | Parameter | Number of Analysed Subjects | Parameter | p Value (MZ vs. MM) | Number of Analysed Subjects | Parameter | p Value (MS vs. MM) | |
| Age, years | 1021 | 56.7 (46.9–63.3) | 55 | 54.2 (48.4–62.2) | 0.279 | 32 | 48.2 (41.7–55.8) | 0.004 |
| BMI, kg/m2 | 1021 | 25.5 (22.4–28.7) | 55 | 27.5 (24.9–30.0) | 0.004 | 32 | 26.2 (21.3–29.7) | 0.894 |
| MELD score | 1008 | 14 (11–17) | 55 | 17 (14–20) | <0.001 | 32 | 17 (15–19) | 0.001 |
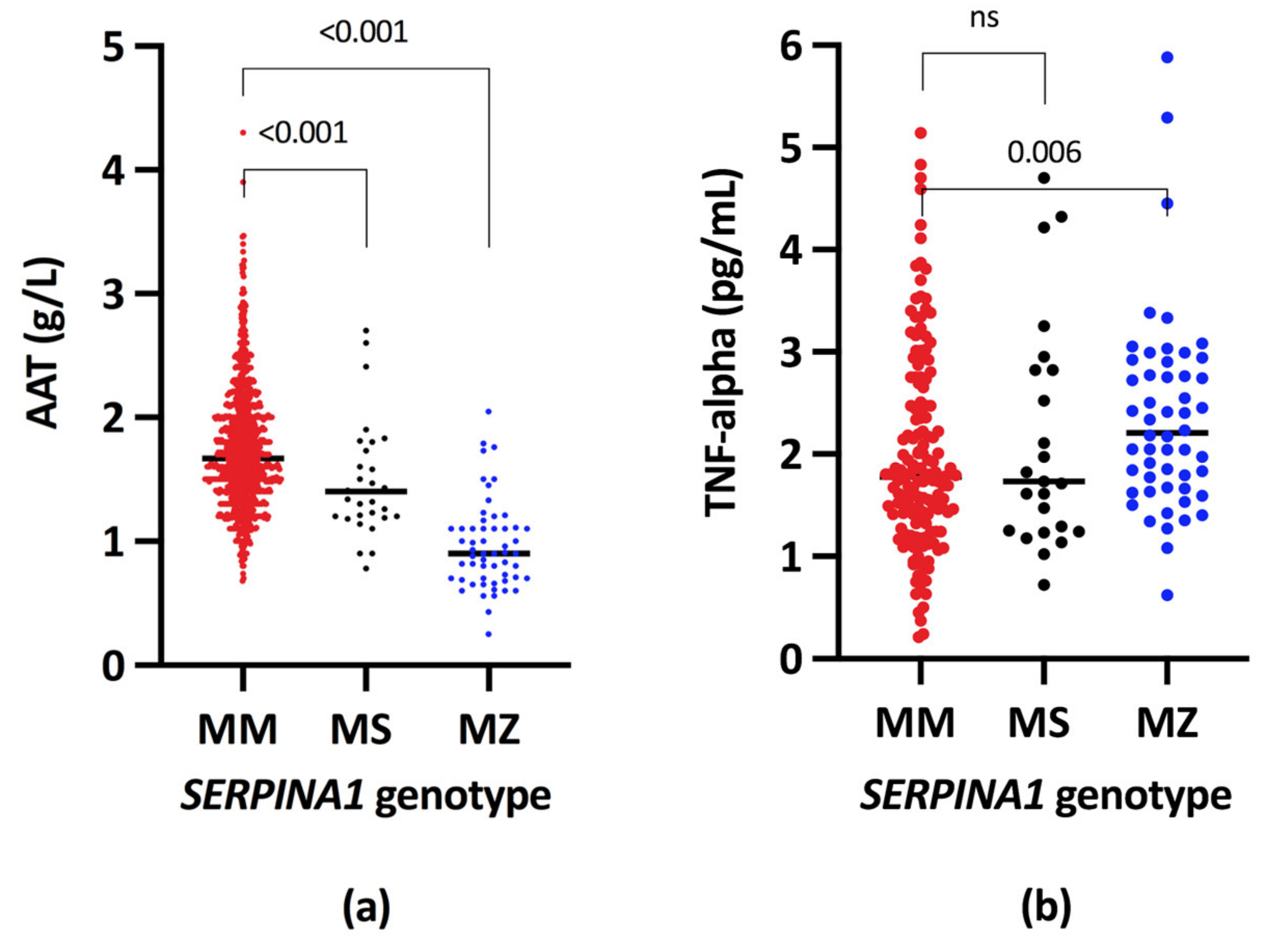
| Serum A1AT, g/L | 983 | 1.67 (1.40–2.00) | 55 | 0.90 (0.70–1.10) | <0.001 | 32 | 1.40 (1.20–1.67) | <0.001 |
| Serum TNFA, pg/L | 156 | 1.78 (1.32–2.49) | 53 | 2.18 (1.67–2.80) | 0.006 | 25 | 1.73 (1.25–2.82) | 0.742 |
| Serum CRP, mg/L | 689 | 8.3 (0–66.0) | 45 | 8.0 (0.4–69.4) | 0.740 | 23 | 9.0 (0.6–76.4) | 0.640 |
| Serum Creatinine, μmol/L | 1014 | 74 (61–90) | 55 | 78 (67–104) | 0.025 | 32 | 69 (61–86) | 0.632 |
| Serum Bilirubin, μmol/L | 1014 | 43 (25–86) | 55 | 63 (41–88) | 0.023 | 32 | 70 (35–92) | 0.032 |
| Prothrombin time, INR | 1009 | 1.30 (1.20–1.50) | 55 | 1,49 (1.33–1.70) | <0.001 | 32 | 1.49 (1.30–1.70) | 0.005 |
| Serum Albumin, g/L | 1014 | 29.5 (25.2–34.0) | 55 | 23.7 (21.6–27.9) | <0.001 | 32 | 30.0 (25.5–33.6) | 0.931 |
| Intrahepatic A1AT aggregates, N | 953 | 18 | 55 | 52 | <0.0001 | 29 | 1 | 0.4372 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabekova, Z.; Frankova, S.; Jirsa, M.; Neroldova, M.; Lunova, M.; Fabian, O.; Kveton, M.; Varys, D.; Chmelova, K.; Adamkova, V.; et al. Alpha-1 Antitrypsin and Hepatocellular Carcinoma in Liver Cirrhosis: SERPINA1 MZ or MS Genotype Carriage Decreases the Risk. Int. J. Mol. Sci. 2021, 22, 10560. https://doi.org/10.3390/ijms221910560
Rabekova Z, Frankova S, Jirsa M, Neroldova M, Lunova M, Fabian O, Kveton M, Varys D, Chmelova K, Adamkova V, et al. Alpha-1 Antitrypsin and Hepatocellular Carcinoma in Liver Cirrhosis: SERPINA1 MZ or MS Genotype Carriage Decreases the Risk. International Journal of Molecular Sciences. 2021; 22(19):10560. https://doi.org/10.3390/ijms221910560
Chicago/Turabian StyleRabekova, Zuzana, Sona Frankova, Milan Jirsa, Magdalena Neroldova, Mariia Lunova, Ondrej Fabian, Martin Kveton, David Varys, Klara Chmelova, Vera Adamkova, and et al. 2021. "Alpha-1 Antitrypsin and Hepatocellular Carcinoma in Liver Cirrhosis: SERPINA1 MZ or MS Genotype Carriage Decreases the Risk" International Journal of Molecular Sciences 22, no. 19: 10560. https://doi.org/10.3390/ijms221910560
APA StyleRabekova, Z., Frankova, S., Jirsa, M., Neroldova, M., Lunova, M., Fabian, O., Kveton, M., Varys, D., Chmelova, K., Adamkova, V., Hubacek, J. A., Spicak, J., Merta, D., & Sperl, J. (2021). Alpha-1 Antitrypsin and Hepatocellular Carcinoma in Liver Cirrhosis: SERPINA1 MZ or MS Genotype Carriage Decreases the Risk. International Journal of Molecular Sciences, 22(19), 10560. https://doi.org/10.3390/ijms221910560






