Abstract
Programmed cell death (PCD) is a highly regulated process that results in the orderly destruction of a cell. Many different forms of PCD may be distinguished, including apoptosis, PARthanatos, and cGMP-dependent cell death. Misregulation of PCD mechanisms may be the underlying cause of neurodegenerative diseases of the retina, including hereditary retinal degeneration (RD). RD relates to a group of diseases that affect photoreceptors and that are triggered by gene mutations that are often well known nowadays. Nevertheless, the cellular mechanisms of PCD triggered by disease-causing mutations are still poorly understood, and RD is mostly still untreatable. While investigations into the neurodegenerative mechanisms of RD have focused on apoptosis in the past two decades, recent evidence suggests a predominance of non-apoptotic processes as causative mechanisms. Research into these mechanisms carries the hope that the knowledge created can eventually be used to design targeted treatments to prevent photoreceptor loss. Hence, in this review, we summarize studies on PCD in RD, including on apoptosis, PARthanatos, and cGMP-dependent cell death. Then, we focus on a possible interplay between these mechanisms, covering cGMP-signaling targets, overactivation of poly(ADP-ribose)polymerase (PARP), energy depletion, Ca2+-permeable channels, and Ca2+-dependent proteases. Finally, an outlook is given into how specific features of cGMP-signaling and PARthanatos may be targeted by therapeutic interventions.
1. Introduction
In multicellular organisms, tight regulation and control of cell death are essential for development, tissue homeostasis, and survival [1]. Different cell types, tissues, and varying physiological conditions have resulted in the evolution of a multitude of cell death mechanisms for which collectively the term “programmed cell death” (PCD) has been introduced [2]. PCD mechanisms, such as apoptosis, usually involve a genetically controlled, program-driven activation of biochemical processes and cellular machinery that result in systematic cellular self-destruction [3].
Hereditary retinal degeneration (RD) is a genetically diverse group of diseases that typically result in progressive photoreceptor cell death, severe visual handicap, and blindness [4]. The most common disease within the RD group is retinitis pigmentosa (RP) [5], in which patients initially experience night blindness and gradual constriction of the visual field until complete blindness sets in [6,7]. The disease typically displays a two-step progression where primary loss of rod photoreceptors is followed by secondary degeneration of cone photoreceptors [6,7]. Approximately one in four thousand people are affected by RP [5,6]. Overall, RD-type blinding diseases are considered to be untreatable [8]. Key questions in the field of RD research concern the cellular mechanisms that bring about photoreceptor loss and whether these may be targeted by therapeutic interventions to prevent or delay the progression of RD.
While over the last 2–3 decades, research has overwhelmingly focused on apoptosis as a causative mechanism for RD, recent evidence indicates that apoptosis only plays a minor role in photoreceptor degeneration [9,10,11,12]. Instead, the importance of non-apoptotic mechanisms, for instance, triggered by exceedingly high levels of cyclic-guanosine-mono-phosphate (cGMP), is increasingly recognized [13]. Hence, for the purposes of this review, we will mention apoptosis only briefly to then focus on non-apoptotic cell death mechanisms. This will include cGMP-dependent cell death [13] and PARthanatos [14] as well as enzymes involved in either or both degenerative pathways. Finally, an outlook is given into how future therapeutic approaches may use an improved mechanistic understanding of PARthanatos and cGMP-signaling.
2. Photoreceptor Physiology and Phototransduction
Photoreceptors mediate the conversion of a photon of light into an electrochemical message that can be interpreted by second-order neurons, a process referred to as phototransduction. A feature of vertebrate phototransduction is their separation into rod and cone photoreceptors, adapted, respectively, for night and day vision. While rods and cones utilize distinct isoforms of the protein components of the transduction cascade, in both types of photoreceptors, cGMP plays a key role [15]. In darkness, cGMP concentration is controlled by the dynamic equilibrium of its synthesis and hydrolysis governed by the basal activities of retinal membrane guanylyl cyclases (GCs) and phosphodiesterase (PDE6), respectively [16,17]. Cyclic nucleotide-gated ion channels (CNGCs), belonging to the superfamily of pore-loop cation channels, are ion channels that are located in the outer segment plasma membrane of photoreceptors and that are activated by the binding of cGMP or cAMP [18,19]. Interestingly, mutations in genes encoding for CNGC subunits can cause retinal degeneration [18,20], attesting to their importance for photoreceptor physiology. A high level of free cGMP in darkness maintains ∼3% of CNGCs in the open state, allowing for an influx of Na+ and Ca2+ into the outer segment [16,17]. While Ca2+ is extruded from the outer segment by the Na+/Ca2+-K+ (NCKX) exchanger, Na+ is pumped out of the inner segment by the adenosine-5′-triphosphate (ATP) -driven Na+/K+ exchanger (NKX). This continuous flow of Na+ ions forms the circulating dark current (Figure 1) [21].
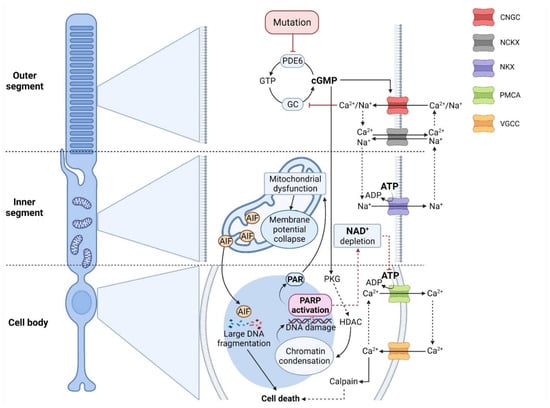
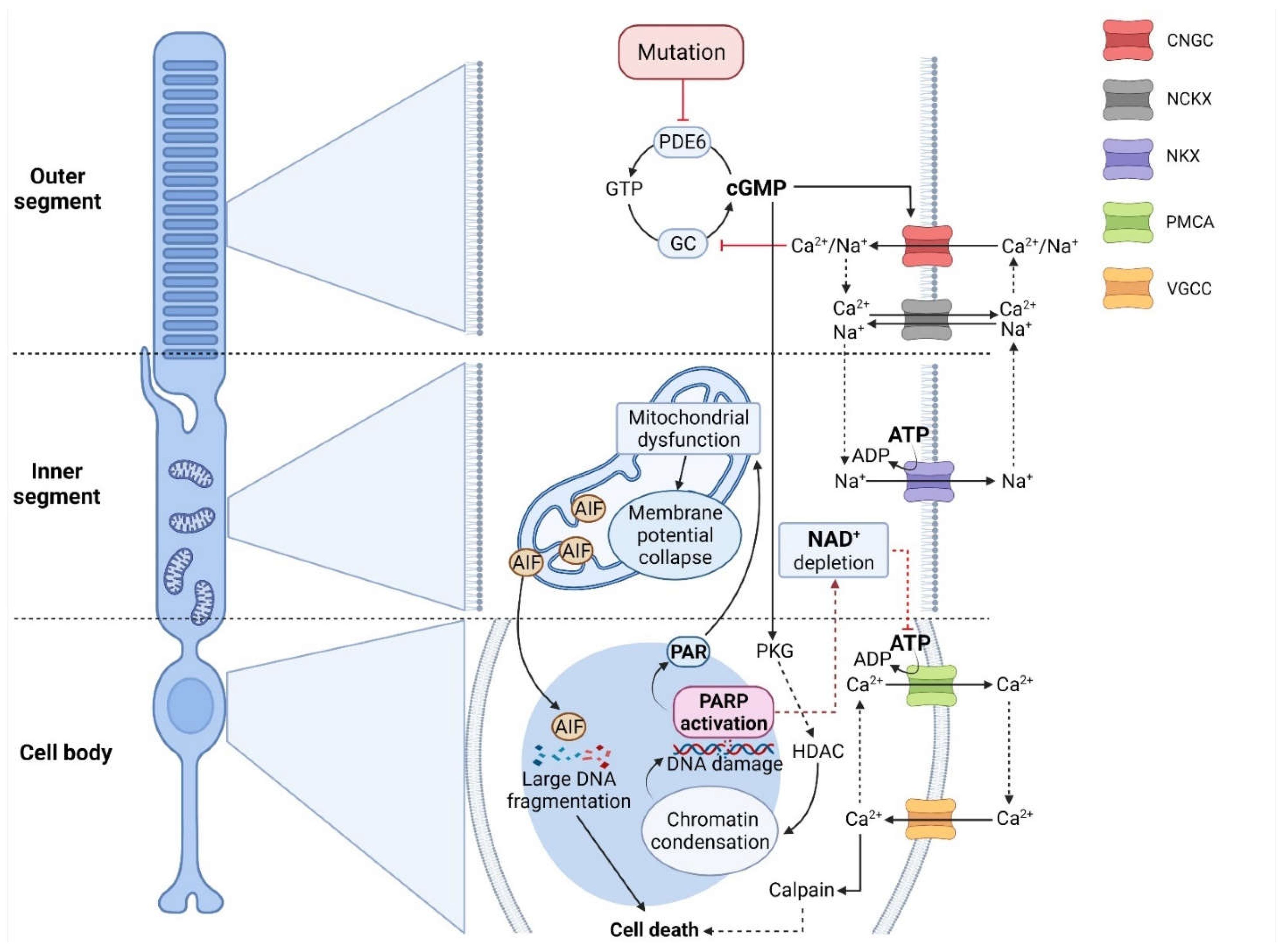
Figure 1.
Crosstalk between cGMP-signaling and PARthanatos in different photoreceptor compartments. In RD-type diseases, photoreceptor degeneration is often initiated by high cGMP levels, caused, for instance, by mutations affecting phosphodiesterase-6 (PDE6). On the one hand, cGMP activates protein kinase G (PKG), which is associated with histone-deacetylase (HDAC), leading to chromatin condensation and DNA damage. This, in turn, may trigger the over-activation of histone deacetylase (HDAC) and indirectly PARP, leading to NAD+/ATP depletion. In addition, PAR polymers produced by PARP can cause mitochondrial dysfunction and translocation of apoptosis-inducing factor (AIF) to the nucleus. On the other hand, cGMP opens the cyclic-nucleotide-gated channel (CNGC), promoting Na+/Ca2+ influx and voltage change in the outer segment. In turn, Ca2+ inhibits guanylyl cyclase (GC), which limits cGMP synthesis from GTP under physiological conditions. The CNGC-dependent voltage change may further open voltage-gated-Ca2+ channels (VGCC) in soma and synapse, leading to more Ca2+ influx. PARP-dependent ATP-depletion may reduce NKX-mediated ion extrusion, resulting in higher intracellular Ca2+ levels. These, in turn, may be linked to calpain-type protease activation, precipitating cell death.
As opposed to other neuronal cell types, photoreceptors are relatively depolarized in their resting state, which in turn leads to a constant synaptic release of the neurotransmitter glutamate. Photon absorption by an opsin protein in the outer segment leads to a conformational change, allowing the opsin to bind and activate the G-protein transducin, which then binds the inhibitory γ-subunit of phosphodiesterase-6 (PDE6), releasing the catalytic PDE6αβ subunits to hydrolyze cGMP. The subsequent decline of cGMP concentrations leads to the closure of CNGCs and the reduction of the dark current. The net decrease in the influx of positively charged Na+ and Ca2+ leads to a hyperpolarization of the photoreceptor cell and the cessation of synaptic glutamate release [22,23].
Low intracellular Ca2+ activates guanylyl cyclase-activating protein (GCAP), stimulating GC, and increasing cGMP synthesis [24,25,26]. Rising cGMP, in turn, re-opens CNGC and facilitates the recovery of the light response [27,28]. As we will see in the next chapter, the very high energy consumption of photoreceptor phototransduction has ramifications for cell death mechanisms.
3. Retinal Energy Metabolism and Cell Death
The retina is one of the most energy-demanding tissues in the body [29], with a large part of its ATP consumption caused by NKX activity and maintenance of the dark current [30,31]. Remarkably, the retina relies mostly on the relatively energy-inefficient glycolysis even in the presence of oxygen, rather than using oxidative phosphorylation, which could provide 18 times as much ATP per mol of glucose [32]. This phenomenon was recognized already by Otto Warburg in the early 1920s and is referred to as “aerobic glycolysis” or as the “Warburg effect” [33]. While the significance of aerobic glycolysis in the retina is, in part, still unclear, several studies have proposed that it may enhance the anabolic activity of photoreceptors [34,35,36].
At any rate, the availability of ATP will determine what kind of cell death mechanism can be operated by a cell. For instance, apoptosis as an active, ATP-dependent process cannot be executed without sufficient ATP, while necrotic forms of cell death require much lower ATP-levels or no ATP at all for their execution [37,38,39,40]. Several different pathological situations may entrain a depletion of ATP. For instance, a cGMP-mediated overactivation of CNG channels may lead to cytosolic Ca2+ overload. High cytosolic Ca2+, together with other causal factors, may lead to an increase in mitochondrial membrane permeability and subsequent membrane depolarization, increased mitochondrial reactive oxygen species (mROS) generation, cytochrome C release, and apoptosis [37,38,41]. In turn, ROS production may deteriorate mitochondrial function even further [42,43]. On one hand, this will decrease photoreceptor ATP production, while on the other hand, ATP consumption will be increased by the extrusion of Ca2+ from the cell soma and synapse via the ATP-driven plasma membrane Ca2+-ATPase (PMCA) [30]. The net effect would be a complete depletion of ATP and then cell death via non-apoptotic, ATP-independent mechanisms.
ATP is also intricately linked to nicotinamide adenine dinucleotide (NAD+), a metabolite that serves as a cofactor for hydrogen transfer. As such NAD+ is vital for the operation of glycolysis and ATP-synthesis in the mitochondria [44]. Thus, if NAD+ levels fall below a critical threshold, key metabolic processes capable of delivering ATP will cease to function. Such a situation may occur in PARthanatos, where the activity of poly(ADP-ribose)polymerase (PARP) may consume excessive amounts of NAD+, thereby indirectly also depleting intracellular ATP [45]. In RD-type diseases, the depletion of ATP consumption and NAD+ may be a significant concern for disease pathogenesis.
4. Apoptosis
Apoptosis is a form of PCD that occurs not only during development or aging, but also as a defense system. Apoptosis can be activated by intrinsic and extrinsic signaling [46]. The intrinsic pathway, also known as the mitochondrial pathway, is driven by a signal within a cell, inducing the expression of proapoptotic genes and proteins, including those belonging to the BCL-2 family, which form the mitochondrial outer membrane permeabilization (MOMP), allowing for the release of cytochrome C to the cytoplasm where it binds to apoptotic protease-activating factor-1 (APAF1). The resultant multimeric cytochrome-c/APAF1 complex activates caspase-9 [47], which in turn cleaves and activates downstream executioner caspases, such as caspase-3 and -7. The extrinsic pathway is triggered by the activation of cell-surface death receptors, such as the tumor necrosis factor family. Later, caspase-8 gets activated, which may then cleave and activate downstream caspases-3 and -7 directly, or, alternatively, may activate BCL-2 family proteins, executing the same steps as in classical apoptosis [48,49,50].
In the past, apoptosis was regarded as a primary degenerative mechanism in RD [51], yet, in the last decade an increasing amount of evidence pointed to the importance of non-apoptotic mechanisms, including cell death pathways triggered by high cGMP [12,13]. One of the confounding factors that has made it difficult to separate apoptotic PCD from the cell death mechanisms that actually underlie RD is the fact that apoptosis is a prominent feature of retinal development [52] and that many commonly used RD animal models display mutation-induced photoreceptor cell death in the very same time frame [38].
5. PARP Activity and PARthanatos
PARP-type enzymes catalyze the transfer of ADP-ribose to target proteins [53] and can sequentially add ADP-ribose units from NAD+ to form polymeric ADP-ribose chains (PAR) [54]. There are at least 18 PARP family members encoded by different genes and with a shared homology in the conserved catalytic domain [55]. While PARP activity was originally associated with DNA repair enzymes and gene regulation [56,57], it may also be the primary driver for a specific form of cell death, termed PARthanatos [14]. This relatively recently discovered non-apoptotic cell death process is characterized by PARP overactivation, accumulation of PAR, and nuclear translocation of apoptosis-inducing factor (AIF) from the mitochondria. As such, PARthanatos is likely involved in various retinal degenerative diseases [58,59].
5.1. The Core of PARthanatos: PARP and PAR Polymers
PARP-1 is probably a central mediator of this cell death mechanism since the majority (>90%) of PAR polymer synthesis typically stems from PARP-1 [60,61]. PARP-1 can mediate cell death when high levels of DNA damage activate PARP-1 to a degree that depletes cellular NAD+ levels. The subsequent depletion of ATP decreases all energy-dependent functions and leads to cell death before DNA repair can be accomplished [62].
Poly(ADP-ribosyl)ation (PARylation) was first described by Chambon and colleagues more than 50 years ago [63]. For PAR chains formation, NAD+ molecules must be cleaved by PARP, and resulting ADP-ribosyl units must be attached to already existing ones [64,65]. The DNA break-induced activation of PARP-1 triggers PARylation of proteins, including PARP-1 itself (auto-PARylation), to facilitate the recruitment of DNA repairing enzymes that contribute to DNA repair near the DNA damage site. Although PARylation is primarily a survival mechanism, high PARylation activity can cause regulated cell death in cells with excessive DNA damage [60]. Suppressing PARylation rescues cells from PARthanatos, attesting to the important role PARylation plays in PARP-1 and DNA damage-induced cell death [66]. Remarkably, PAR itself exhibits dose-dependent toxicity in neurons when exogenously administered via BioPorter, a cationic lipid formulation that facilitates PAR entry into cells [67].
In physiological conditions, several mechanisms counterbalance excessive PARP-1 activity. (1) PAR molecules are rapidly catabolized by poly-(ADP-ribose) glycohydrolase (PARG) and ADP-ribosyl protein lyase [68,69]; (2) auto-PARylation of PARP-1 downregulates its activity by interfering with interactions between the DNA and DNA binding domain [70]; (3) accumulation of nicotinamide as a by-product of NAD+ consumption inhibits PARP-1 and may act as a negative feedback signal [45,62].
5.2. PAR-Dependent Translocation of AIF
AIF was originally identified as a soluble 57-kDa fragment, which upon dissipation of the mitochondrial membrane potential is released from the mitochondria and translocates to the nucleus in a caspase-independent manner [71]. PAR induces a conformation change in AIF that lowers its affinity to the mitochondrial outer membrane leading to its release [72]. Hence, after PARP-1 overactivation, AIF can be released into the cytoplasm and translocate further to the nucleus, promoting cell death [73]. The protective effect by blocking mitochondrial AIF release or reducing AIF abundance indicates that AIF plays a crucial role in PARthanatos [65,74,75]. Furthermore, AIF can recruit macrophage migration inhibitory factor (MIF) to the nucleus, where MIF cleaves genomic DNA into large-scale fragments via its nuclease activity [73]. Such DNA damage may lead to even more PARP-1 activation, effectively forming a feedback loop that accelerates NAD+ depletion, mitochondrial dysfunction, and DNA degradation.
5.3. Crosstalk between PARthanatos, Ca2+, and Calpain-Type Proteases
As described above (Section 3), ATP depletion caused by excessive PARP-1 activity [76] will compromise a cell’s capability for Ca2+ extrusion via ATP-dependent PMCA (Figure 1). The resulting rise in intracellular Ca2+ may lead to Ca2+-dependent activation of calpain-type proteases [48,53]. While calpain may be involved in AIF release from mitochondria [77], during PARthanatos, AIF release can also happen independently of calpain activity [78]. Still, other than via calpain activation, high intracellular Ca2+ levels, and Ca2+ sequestration into mitochondria can also lead to the generation of reactive oxygen species and mitochondrial dysfunction, further promoting the execution of PARthanatos [79,80].
6. cGMP-Dependent Cell Death in RD
Many genetically distinct forms of RD display a substantial increase in the intracellular photoreceptor concentration of cyclic guanosine monophosphate (cGMP) [12,81,82]. We have previously proposed for this to constitute a new cGMP-dependent cell death pathway for photoreceptor degeneration [48] that would apply to any disease-causing mutation that raises intracellular cGMP levels. In this mechanism, overactivation of the prototypic cGMP targets CNGC and protein kinase G (PKG), produces excessive Ca2+ influx and protein phosphorylation, respectively [12]. Both CNGC and PKG alone or in concert precipitate cell death: CNGC-mediated Na+ and Ca2+ influx may strain the energy metabolism and activate calpain [83,84]. PKG-dependent phosphorylation is associated with histone deacetylase (HDAC) activation [85], which in turn appears to be connected to the activation of PARP [86].
6.1. RD Mutations Associated with High Photoreceptor cGMP
Regarding the genetically very heterogenous group of RD-type diseases, a relevant question concerns the generality of cGMP-dependent cell death. Indeed, excessive accumulation of cGMP in photoreceptors has been observed in various genetically distinct RD mutants [12,81,82,87,88], suggesting cGMP as a near-universal trigger for non-apoptotic PCD mechanisms in RD. Disease-causing mutations may, for instance, result in gain-of-function in genes involved in cGMP synthesis [89,90]. Similarly, loss of function in genes downregulating cGMP can cause RD [91,92,93], and so can mutations in cGMP-signaling targets [19,84,94], such as in CNGC genes. For further details on mutations in RD disease genes that have been connected to exceedingly high levels of cGMP in photoreceptors, please refer to [47,95].
6.2. cGMP-Gated Ion Channels, Ca2+ -Influx, and Cell Death
As mentioned in Section 2, cGMP-activation of CNGCs plays a central role in phototransduction [19]. These channels are heterotetramers consisting of CNGA1 and CNGB1 subunits in rod photoreceptors and CNGA3 and CNGB3 subunits in cones [19]. Elevated photoreceptor cGMP levels may overactivate CNGC and increase Na+ and Ca2+ influx. Knockout of the Cngb1 gene caused rod CNGC dysfunction and significantly delayed photoreceptor degeneration in Pde6b mutant mice [84,87]. On the other hand, loss-of-function mutations in CNGC genes cause RD [95], and even further pharmacological inhibition of CNGC in Pde6b mutant retina accelerated photoreceptor cell death [96]. A possible explanation for these seemingly contradictory outcomes could be that a low concentration of intracellular Ca2+ activates GCAPs to stimulate GCs and increase cGMP synthesis [24,25,26]. In Cngb1-/- mice, the introduction of an additional knockout of the PKG1 gene (i.e., Prkg1-/-) delays photoreceptor degeneration [87], suggesting that photoreceptor death ultimately is mediated by PKG-dependent processes.
The activity of CNGCs depolarizes photoreceptors to the extent that voltage gated Ca2+ channels (VGCC) in the soma and synapse open [97]. This results in further Ca2+ influx that must be extruded by PMCA at the expense of additional ATP and which, otherwise, could lead to the activation of calpain-type proteases, which may promote cell destruction [98,99,100]. Accordingly, the idea that high Ca2+ may be responsible for cell death [48], and by extension for photoreceptor degeneration [101], has motivated several studies attempting to block either CNGC or VGCC for therapeutic purposes. Unfortunately, these attempts generally failed to yield tangible success [102,103,104], which may indicate that Ca2+ influx is, in fact, not as relevant for photoreceptor cell death as previously thought [96]. The lack of therapeutic effect of Ca2+ channel blockers also suggests that the main drivers of cGMP-dependent photoreceptor cell death are mainly independent of Ca2+ [21].
6.3. Protein Kinase G: A Link between cGMP-Signaling and Cell Death?
PKG is a major downstream effector of cGMP-signaling pathways [105]. Conventionally, cGMP-PKG signaling is often seen as protective, especially in a neuronal cell context [106,107,108]. However, overactivation of PKG can also cause cell death [109,110,111], and in the retina and in photoreceptors, a link between excessive cGMP-signaling, PKG activity, and cell death has been well established [81,112,113]. Moreover, application of PKG inhibitors in vivo in RD retina resulted in photoreceptor protection [81,114], and abolishing Prkg1 expression in the mouse promoted rod photoreceptor survival [87]. Conversely, knockout of the Prkg2 gene in the mouse protected cone photoreceptors in a model for hereditary cone degeneration [115]. However, it is worth noting that the exact PKG expression patterns in the retina are still partially unresolved and that in situ hybridization [116,117] and immunostaining studies [118] are not in entire agreement with each other as to which PKG isoform is expressed in which retinal cell type.
Another open question is the nature of the protein targets that PKG may phosphorylate in the retina and how such phosphorylation could bring about photoreceptor cell death. A recent study used protein phosphorylation array technology to identify PKG targets in retinal tissue lysates. While some of the previously known PKG targets—such as vasodilation-stimulated phosphoprotein (VASP) and cyclic AMP response element-binding (CREB)—were confirmed, many novel substrates were also found, including ryanodine receptor-1 (RYR1) and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) [119]. The exact significance of these and other PKG targets for photoreceptor degeneration will have to be elucidated in future studies.
6.4. Histone Deacetylase Activity as an Event Downstream of PKG
Modification of histones by the activity of histone deacetylases (HDACs) plays a key role in epigenetic regulation of gene expression by changing the structure of chromatin and by modulating the accessibility for transcription factors to their target DNA sequences [120]. Notably, excessive activation of HDACs in photoreceptors has been observed in connection with the accumulation of cGMP and activation of PKG [12,86,121], and inhibition of HDAC protects the retina from cGMP-induced neurodegeneration in several RD mutants [122,123,124,125,126,127,128]. Nonetheless, the mechanism of HDAC activation remains mysterious. While in C. elegans, PKG-dependent phosphorylation appears to activate HDACs [85], it is unclear whether such direct PKG-HDAC interactions also happen in higher vertebrates, even though HDAC1 can serve as a substrate for PKG in vitro [129]. Overall, HDAC activity may have both positive and negative effects on cell survival, and a PKG-dependent disturbance in the equilibrium of different HDAC functions—whether direct or indirect—could play an important role in neurodegeneration.
6.5. PARP: A Link between PARthanatos and cGMP-Dependent Cell Death
As discussed above, PARP-1 is the central mediator of PARthanatos due to its ability to synthesize PAR polymers [60,61]. In addition, PARP activity is strongly increased in photoreceptors during the progression of RD [12,130,131]. Remarkably, the inhibition of HDAC activity with trichostatin A led to a decrease in photoreceptor PAR accumulation, indicating that PARP activity may occur downstream of HDAC [86,125]. This may be linked to HDAC-mediated removal of acetylated residues from histones, leading to chromatin condensation and transcriptional repression. Chromatin condensation, in turn, impairs the recruitment of DNA repair factors and results in the accumulation of DNA breaks [132]. Due to continuous exposure to endogenous and exogenous DNA-damaging insults, cells accumulate DNA damage such as single-strand DNA breaks (SSBs) and double-strand DNA breaks (DSBs). This requires constant surveillance and activation of the DNA repair response [132] that is facilitated by PARP-1 [52]. Paradoxically, inhibition of PARP significantly delays photoreceptor loss in cGMP-dependent cell death [133,134,135,136], and PARP-1 gene knockout increases resistance to RD [137], indicating that PARP activity, perhaps through its consumption of NAD+, is an important driver of photoreceptor degeneration.
7. Therapy Developments Targeting Programmed Cell Death
The degenerative photoreceptor pathways discussed thus far may provide targets for the rational design of therapeutic interventions that could prevent or slow down the progression of RD. Numerous studies have attempted to exploit mechanistic insights for therapy development purposes, and many of these works have focused on apoptosis as the presumed causative mechanism (Table 1).

Table 1.
An overview of current therapeutical targets in three different PCDs.
7.1. Apoptosis as a Target for Therapeutic Intervention in RD
RD was initially thought to be driven by apoptosis [51], motivating many interventional studies that aimed to block different steps of the apoptotic cascade [138,139,140]. Unfortunately, these approaches were mostly ineffective, even though virtually the whole apoptotic cascade has been targeted. For instance, neither the pharmacological inhibition of caspase-type proteases [139], nor the genetic manipulation of Bcl-2 [141], Bcl-XL [142], c-fos [146], caspase-3 [140], or caspase-7 [147] promoted long-term photoreceptor survival. On the other hand, proapoptotic protein Bcl-2-associated X protein (BAX) activation was observed in three different animal models for RD [148]. However, the role of BAX in RD also seems ambiguous as its gene knockout may delay cell death of rod photoreceptors but not that of cones in Rpe65 KO animals [138]. Together, most of the available evidence does not suggest a major role for apoptosis in RD and hence makes it seem unlikely that therapeutic interventions targeting apoptosis can be successful.
7.2. Targeting PARthanatos and PARP Activity
PARthanatos occurs in a highly choreographed, multistep fashion, and several steps in the cascade could serve as therapeutic targets for managing diseases associated with cell death [143]. Since PARthanatos is characterized by overactivation of PARP-1 [60,61], PARP inhibitors should prioritize saving photoreceptor degeneration. Currently, PARP inhibitors are mostly used for cancer therapy due to their ability to prevent DNA repair, and several PARP inhibitors are being tested clinically or have already been approved for clinical use [144]. Thus, it could be economically efficient to repurpose PARP inhibitors for applications beyond oncology, including in RD [136].
Olaparib became the first PARP inhibitor to be approved by the FDA to treat metastatic breast cancer in January 2018 [149]. In the Pde6b mutant rd1 mouse, Olaparib significantly delayed photoreceptor loss [133] and affected the release of extracellular vesicles associated with immune modulation, tumor invasion, regeneration, degenerative processes, cellular communication, cell homeostasis, and neovascularization [134]. In addition, other PARP inhibitors also showed its neuroprotective effects in the retina. For instance, BMN-673, 3-aminobenzamide [136], and PJ34 [145] decreased PARylation and reduced photoreceptor cell death. While these data confirm PARP in general as a therapeutic target for RD, it is still uncertain whether PARP-1 is the main target or other PARP isoforms also need to be considered. Moreover, a better understanding of how PARP activity precipitates cell death will likely be needed to develop a truly effective therapy.
7.3. Novel Therapeutic Approaches Targeting cGMP-Dependent Cell Death
In a large subset of RD patients, cGMP-dependent cell death may be the prevalent pathogenic mechanism, highlighting this pathway for gene- and mutation-independent therapeutic purposes [12,48]. The most likely targets for high cGMP levels in the photoreceptor are PKG and/or CNGC (Figure 1) [87]. cGMP-induced overactivation of CNGC, presumably causing a dangerous inflow of Ca2+, has been proposed as a key event in photoreceptor degeneration [91,113], but as we discussed above, singly inhibiting Ca2+ might not rescue photoreceptors [96,103,104,150], due to feedback of low-Ca2+ levels increasing cGMP [24,25,26]. On the other hand, PKG appears as an essential mediator of cGMP-dependent cell death [81,112]. Accordingly, an inhibitory cGMP analogue delivered via a nanosized liposomal carrier to facilitate transport across the blood-retinal barrier was highly effective at protecting photoreceptors in three different in vivo RD models [114].
Still, interfering with PKG activity would not per se reduce the high intracellular cGMP levels caused by RD mutations. Thus, an alternative approach could be to target the cGMP synthesis pathway. Here, the enzyme inosine monophosphate dehydrogenase (IMPDH), which catalyzes the rate-limiting step of GTP production [151] stands out as an exciting novel target. Yang et al. [82] demonstrated that mycophenolate mofetil, a prodrug of mycophenolic acid that reversibly inhibits IMPDH [152], reduced photoreceptor cGMP and preserved photoreceptors in Pde6b mouse mutants.
Altogether, these data make cGMP-signaling appear as a lovely pathway for RD therapy development. As a note of caution, it is crucial to consider that cGMP-signaling covers essential functions in many cells of the body. Thus, a prospective drug will need to be either highly specific for its target in photoreceptors or delivered with a system that supplies the drug specifically to the retina and the photoreceptors.
8. The Future Therapeutic Research in RD
Although many novel therapies have been discovered (Section 7), there are still challenges in clinical translation. For instance, RD-type diseases show an enormous genetic heterogeneity with disease-causing mutations in more than 270 genes [48]. Since each of these disease genes can carry from several dozens to several hundred or more individual mutations [153,154], we may, at present, estimate the total number of disease mutations to amount to several tens of thousands. This situation severely hinders the design of clinical trials as the numbers of patients carrying a specific disease-causing mutation will be small, even in a best-case scenario. However, a careful choice of the patients to be included in a clinical trial with precisely known genotypes is critical for success during clinical testing.
Another problem for clinical translation is a lack of in vivo biomarkers due to the incomplete understanding of the underlying photoreceptor cell death mechanisms. Ideally, the biomarker could be used for the rapid assessment of treatment efficacy and should be allowed for live non-invasive visualization of cell death in the retina. Recent developments in magnetic resonance imaging (MRI) suggest that it may be possible to non-invasively observe oxidative stress or the production of free radicals in the retina of animals [155]. In addition, cGMP, a potential blood-based parameter, is increased in blood from RP patients compared to healthy counterparts [156,157]. This may be related to the excessive cGMP levels in the photoreceptors of many RP types, i.e., the phenomenon discussed in several sections above, especially since at least some of the patients had mutations in the PDE6A gene [157].
9. Concluding Remarks
The question as to what type of PCD mechanism governs cell death in RD-type diseases has been investigated for at least three decades, and many findings have been contentious for many years. It is thus worth noting that this review is neither exhaustive nor unbiased. While the recent decade has yielded a large body of evidence pointing at cGMP-dependent cell death and PARthanatos as likely causative mechanisms in RD, future research will have to confirm this, hopefully delivering effective RD therapies to patients.
Author Contributions
Conceptualization, J.Y. and F.P.-D.; writing—original draft preparation, J.Y., Y.C. and Y.Z.; writing—review and editing, J.Y., Y.C. and F.P.-D.; visualization, J.Y.; supervision, F.P.-D.; project administration, F.P.-D. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by Charlotte and Tistou Kerstan Foundation, Baden-Württemberg Foundation (BWST_WSF_006), and China Scholarship Council.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The figure was created with https://biorender.com/, accessed on 11 August 2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Maghsoudi, N.; Zakeri, Z.; Lockshin, R.A. Programmed cell death and apoptosis—Where it came from and where it is going: From Elie Metchnikoff to the control of caspases. Exp. Oncol. 2012, 34, 146–152. [Google Scholar]
- Ameisen, J.C. On the origin, evolution, and nature of programmed cell death: A timeline of four billion years. Cell Death Differ. 2002, 9, 367–393. [Google Scholar] [CrossRef] [PubMed]
- Berman-Frank, I.; Bidle, K.D.; Haramaty, L.; Falkowski, P.G. The demise of the marine cyanobacterium, Trichodesmium spp., via an autocatalyzed cell death pathway. Limnol. Oceanogr. 2004, 49, 997–1005. [Google Scholar] [CrossRef]
- Duncan, J.L.; Pierce, E.A.; Laster, A.M.; Daiger, S.P.; Birch, D.G.; Ash, J.D.; Iannaccone, A.; Flannery, J.G.; Sahel, J.A.; Zack, D.J.; et al. Inherited Retinal Degenerations: Current Landscape and Knowledge Gaps. Transl. Vis. Sci. Technol. 2018, 7, 6. [Google Scholar] [CrossRef]
- Bertelsen, M.; Jensen, H.; Bregnhøj, J.F.; Rosenberg, T. Prevalence of Generalized Retinal Dystrophy in Denmark. Ophthalmic Epidemiol. 2014, 21, 217–223. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, T.B.; Luther, E.E. Retinitis Pigmentosa. StatPearls [Internet]. 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK519518/ (accessed on 11 August 2021).
- Hamel, C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006, 1, 1–12. [Google Scholar] [CrossRef]
- Sahel, J.-A.; Marazova, K.; Audo, I. Clinical Characteristics and Current Therapies for Inherited Retinal Degenerations. Cold Spring Harb. Perspect. Med. 2015, 5, a017111. [Google Scholar] [CrossRef]
- Marigo, V. Programmed Cell Death in Retinal Degeneration: Targeting Apoptosis in Photoreceptors as Potential Therapy for Retinal Degeneration. Cell Cycle 2007, 6, 652–655. [Google Scholar] [CrossRef]
- Sanges, D.; Comitato, A.; Tammaro, R.; Marigo, V. Apoptosis in retinal degeneration involves cross-talk between apoptosis-inducing factor (AIF) and caspase-12 and is blocked by calpain inhibitors. PNAS 2006, 103, 17366–17371. [Google Scholar] [CrossRef]
- Doonan, F.; Donovan, M.; Cotter, T.G. Caspase-Independent Photoreceptor Apoptosis in Mouse Models of Retinal Degeneration. J. Neurosci. Res. 2003, 23, 5723–5731. [Google Scholar] [CrossRef]
- Arango-Gonzalez, B.; Trifunovic, D.; Sahaboglu, A.; Kranz, K.; Michalakis, S.; Farinelli, P.; Koch, S.; Koch, F.; Cottet, S.; Janssen-Bienhold, U.; et al. Identification of a Common Non-Apoptotic Cell Death Mechanism in Hereditary Retinal Degeneration. PLoS ONE 2014, 9, e112142. [Google Scholar] [CrossRef]
- Power, M.J.; Rogerson, L.E.; Schubert, T.; Berens, P.; Euler, T.; Paquet-Durand, F. Systematic spatiotemporal mapping reveals divergent cell death pathways in three mouse models of hereditary retinal degeneration. J. Comp. Neurol. 2020, 528, 1113–1139. [Google Scholar] [CrossRef]
- David, K.K.; Andrabi, S.A.; Dawson, T.M.; Dawson, V.L. Parthanatos, a messenger of death. Front. Biosci. 2009, 14, 1116. [Google Scholar] [CrossRef]
- Lamb, T.D.; Hunt, D.M. Evolution of the vertebrate phototransduction cascade activation steps. Dev. Biol. 2017, 431, 77–92. [Google Scholar] [CrossRef]
- Yau, K.W.; Baylor, D.A. Cyclic GMP-Activated Conductance of Retinal Photoreceptor Cells. Annu. Rev. Neurosci. 1989, 12, 289–327. [Google Scholar] [CrossRef] [PubMed]
- Yau, K.-W.; Nakatani, K. Cation selectivity of light-sensitive conductance in retinal rods. Nature 1984, 309, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Biel, M.; Michalakis, S. Function and Dysfunction of CNG Channels: Insights from Channelopathies and Mouse Models. Mol. Neurobiol. 2007, 35, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Biel, M.; Michalakis, S. Cyclic Nucleotide-Gated Channels. In cGMP: Generators, Effectors and Therapeutic Implications; Springer: Berlin/Heidelberg, Germany, 2009; Volume 191, pp. 111–136. [Google Scholar] [CrossRef]
- Donato, L.; Scimone, C.; Alibrandi, S.; Abdalla, E.M.; Nabil, K.M.; D’Angelo, R.; Sidoti, A. New Omics–Derived Perspectives on Retinal Dystrophies: Could Ion Channels-Encoding or Related Genes Act as Modifier of Pathological Phenotype? Int. J. Mol. Sci. 2021, 22, 70. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Chen, Y.; Yan, J.; Christensen, G.; Belhadj, S.; Tolone, A.; Paquet-Durand, F. The role of cGMP-signalling and calcium-signalling in photoreceptor cell death: Perspectives for therapy development. Pflugers Arch 2021, 473, 1411–1421. [Google Scholar] [CrossRef] [PubMed]
- Arshavsky, V.Y.; Lamb, T.D.; Pugh, E.N., Jr. G Proteins and Phototransduction. Annu. Rev. Physiol. 2002, 64, 153–187. [Google Scholar] [CrossRef]
- Fain, G.L.; Hardie, R.; Laughlin, S.B. Phototransduction and the Evolution of Photoreceptors. Curr. Biol. 2010, 20, R114–R124. [Google Scholar] [CrossRef] [PubMed]
- Dizhoor, A.M.; Olshevskaya, E.V.; Henzel, W.J.; Wong, S.C.; Stults, J.T.; Ankoudinova, I.; Hurley, J.B. Cloning, Sequencing, and Expression of a 24-kDa Ca2+-binding Protein Activating Photoreceptor Guanylyl Cyclase. J. Biol. Chem. 1995, 270, 25200–25206. [Google Scholar] [CrossRef]
- Gorczyca, W.A.; Gray-Keller, M.P.; Detwiler, P.B.; Palczewski, K. Purification and physiological evaluation of a guanylate cyclase activating protein from retinal rods. PNAS 1994, 91, 4014–4018. [Google Scholar] [CrossRef]
- Palczewski, K.; Subbaraya, I.; Gorczyca, W.A.; Helekar, B.S.; Ruiz, C.C.; Ohguro, H.; Huang, J.; Zhao, X.; Crabb, J.W.; Johnson, R.S. Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase-activating protein. Neuron 1994, 13, 395–404. [Google Scholar] [CrossRef]
- Méndez, A.; Burns, M.E.; Sokal, I.; Dizhoor, A.M.; Baehr, W.; Palczewski, K.; Baylor, D.A.; Chen, J. Role of guanylate cyclase-activating proteins (GCAPs) in setting the flash sensitivity of rod photoreceptors. PNAS 2001, 98, 9948–9953. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.E.; Méndez, A.; Chen, J.; Baylor, D.A. Dynamics of Cyclic GMP Synthesis in Retinal Rods. Neuron 2002, 36, 81–91. [Google Scholar] [CrossRef]
- Hurley, J.B.; Lindsay, K.J.; Du, J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 2015, 93, 1079–1092. [Google Scholar] [CrossRef]
- Ames, A., III. Energy requirements of CNS cells as related to their function and to their vulnerability to ischemia: A commentary based on studies on retina. Can. J. Physiol. Pharmacol. 1992, 70, S158–S164. [Google Scholar] [CrossRef] [PubMed]
- Okawa, H.; Sampath, A.P.; Laughlin, S.B.; Fain, G.L. ATP Consumption by Mammalian Rod Photoreceptors in Darkness and in Light. Curr. Biol. 2008, 18, 1917–1921. [Google Scholar] [CrossRef] [PubMed]
- Kanow, M.A.; Giarmarco, M.M.; Jankowski, C.S.; Tsantilas, K.; Engel, A.L.; Du, J.; Linton, J.D.; Farnsworth, C.C.; Sloat, S.R.; Rountree, A.; et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. eLife 2017, 6, e28899. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in The Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Venkatesh, A.; Ma, S.; Le, Y.Z.; Hall, M.N.; Rüegg, M.A.; Punzo, C. Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J. Clin. Investig. 2015, 125, 1446–1458. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Du, J.; Justus, S.; Hsu, C.-W.; Bonet-Ponce, L.; Wu, W.-H.; Tsai, Y.-T.; Wu, W.-P.; Jia, Y.; Duong, J.K.; et al. Reprogramming metabolism by targeting sirtuin 6 attenuates retinal degeneration. J. Clin. Investig. 2016, 126, 4659–4673. [Google Scholar] [CrossRef] [PubMed]
- Chinchore, Y.; Begaj, T.; Wu, D.; Drokhlyansky, E.; Cepko, C.L. Glycolytic reliance promotes anabolism in photoreceptors. eLife 2017, 6, e25946. [Google Scholar] [CrossRef]
- Pivovarova, N.B.; Andrews, S.B. Calcium-dependent mitochondrial function and dysfunction in neurons. FEBS J. 2010, 277, 3622–3636. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Pelluz, J.; Arango-Gonzalez, B.; Kustermann, S.; Romero, F.J.; van Veen, T.; Zrenner, E.; Ekström, P.; Paquet-Durand, F. Photoreceptor Cell Death Mechanisms in Inherited Retinal Degeneration. Mol. Neurobiol. 2008, 38, 253–269. [Google Scholar] [CrossRef]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.; Blagosklonny, M.; El-Deiry, W.S.; Golstein, P.; Green, D.; et al. Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2008. Cell Death Differ. 2008, 16, 3–11. [Google Scholar] [CrossRef]
- Di Lisa, F.; Bernardi, P. A CaPful of mechanisms regulating the mitochondrial permeability transition. J. Mol. Cell. Cardiol. 2009, 46, 775–780. [Google Scholar] [CrossRef]
- Rinaldi, C.; Donato, L.; Alibrandi, S.; Scimone, C.; D’Angelo, R.; Sidoti, A. Oxidative Stress and the Neurovascular Unit. Life 2021, 11, 767. [Google Scholar] [CrossRef] [PubMed]
- Scimone, C.; Donato, L.; Alibrandi, S.; Vadalà, M.; Giglia, G.; Sidoti, A.; D’Angelo, R. N-retinylidene-N-retinylethanolamine adduct induces expression of chronic inflammation cytokines in retinal pigment epithelium cells. Exp. Eye Res. 2021, 29, 108641. [Google Scholar] [CrossRef]
- Xie, N.; Zhang, L.; Gao, W.; Huang, C.; Huber, P.E.; Zhou, X.; Li, C.; Shen, G.; Zou, B. NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 2020, 5, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, S.H.; Pierce, E.A. Parthanatos as a Cell Death Pathway Underlying Retinal Disease. Retin. Degener. Dis. 2019, 1185, 323–327. [Google Scholar] [CrossRef]
- Orrenius, S.; Zhivotovsky, B.; Nicotera, P. Regulation of cell death: The calcium–apoptosis link. Nat. Rev. Mol. Cell Biol. 2003, 4, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Akey, C.W. Apoptosome Structure, Assembly, and Procaspase Activation. Structure 2013, 21, 501–515. [Google Scholar] [CrossRef]
- Power, M.; Das, S.; Schütze, K.; Marigo, V.; Ekström, P.; Paquet-Durand, F. Cellular mechanisms of hereditary photoreceptor degeneration—Focus on cGMP. Prog. Retin. Eye Res. 2020, 74, 100772. [Google Scholar] [CrossRef]
- Fricker, M.; Tolkovsky, A.M.; Borutaite, V.; Coleman, M.; Brown, G.C. Neuronal Cell Death. Physiol. Rev. 2018, 98, 813–880. [Google Scholar] [CrossRef]
- Weinlich, R.; Oberst, A.; Beere, H.M.; Green, D.R. Necroptosis in development, inflammation and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 127–136. [Google Scholar] [CrossRef]
- Portera-Cailliau, C.; Sung, C.; Nathans, J.; Adler, R. Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. PNAS 1994, 91, 974–978. [Google Scholar] [CrossRef]
- Mervin, K.; Stone, J. Developmental Death of Photoreceptors in the C57BL/6JMouse: Association with Retinal Function and Self-protection. Exp. Eye Res. 2002, 75, 703–713. [Google Scholar] [CrossRef]
- Morales, J.; Li, L.; Fattah, F.J.; Dong, Y.; Bey, E.A.; Patel, M.; Gao, J.; Boothman, D.A. Review of Poly (ADP-ribose) Polymerase (PARP) Mechanisms of Action and Rationale for Targeting in Cancer and Other Diseases. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Teloni, F.; Altmeyer, M. Readers of poly(ADP-ribose): Designed to be fit for purpose. Nucleic Acids Res. 2015, 44, 993–1006. [Google Scholar] [CrossRef]
- Amé, J.C.; Spenlehauer, C.; de Murcia, G. The PARP superfamily. Bioessays 2004, 26, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.L.; Ren, E.C. Functional Aspects of PARP1 in DNA Repair and Transcription. Biomolecules 2012, 2, 524–548. [Google Scholar] [CrossRef] [PubMed]
- Hageman, G.J.; Stierum, R.H. Niacin, poly(ADP-ribose) polymerase-1 and genomic stability. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2001, 475, 45–56. [Google Scholar] [CrossRef]
- Pan, Y.-R.; Song, J.-Y.; Fan, B.; Wang, Y.; Che, L.; Zhang, S.-M.; Chang, Y.-X.; He, C.; Li, G.-Y. mTOR may interact with PARP-1 to regulate visible light-induced parthanatos in photoreceptors. Cell Commun. Signal. 2020, 18, 1–17. [Google Scholar] [CrossRef]
- Andrabi, S.A.; Dawson, T.M.; Dawson, V.L. Mitochondrial and Nuclear Cross Talk in Cell Death. Ann. N. Y. Acad. Sci. 2008, 1147, 233–241. [Google Scholar] [CrossRef]
- Robinson, N.; Ganesan, R.; Hegedűs, C.; Kovács, K.; Kufer, T.A.; Virág, L. Programmed necrotic cell death of macrophages: Focus on pyroptosis, necroptosis, and parthanatos. Redox Biol. 2019, 26, 101239. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Kraus, W.L. The PARP Side of the Nucleus: Molecular Actions, Physiological Outcomes, and Clinical Targets. Mol. Cell 2010, 39, 8–24. [Google Scholar] [CrossRef]
- Murata, M.M.; Kong, X.; Moncada, E.; Chen, Y.; Imamura, H.; Wang, P.; Berns, M.W.; Yokomori, K.; Digman, M.A. NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. Mol. Biol. Cell 2019, 30, 2584–2597. [Google Scholar] [CrossRef]
- Chambon, P.; Weill, J.D.; Mandel, P. Nicotinamide mononucleotide activation of a new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963, 11, 39–43. [Google Scholar] [CrossRef]
- Bai, P. Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance. Mol. Cell 2015, 58, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.L. PARPs and ADP-Ribosylation: 50 …Years and Counting. Mol. Cell 2015, 58, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Kondratova, A.A.; Cheon, H.; Dong, B.; Holvey-Bates, E.G.; Hasipek, M.; Taran, I.; Gaughan, C.; Jha, B.K.; Silverman, R.H.; Stark, G.R. Suppressing PAR ylation by 2′,5′-oligoadenylate synthetase 1 inhibits DNA damage-induced cell death. EMBO J. 2020, 39, e101573. [Google Scholar] [CrossRef] [PubMed]
- Andrabi, S.A.; Kim, N.S.; Yu, S.-W.; Wang, H.; Koh, D.W.; Sasaki, M.; Klaus, J.A.; Otsuka, T.; Zhang, Z.; Koehler, R.C.; et al. Poly(ADP-ribose) (PAR) polymer is a death signal. PNAS 2006, 103, 18308–18313. [Google Scholar] [CrossRef]
- De Murcia, G.; de Murcia, J.M. Poly(ADP-ribose) polymerase: A molecular nick-sensor. Trends Biochem. Sci. 1994, 19, 172–176. [Google Scholar] [CrossRef]
- Barkauskaite, E.; Jankevicius, G.; Ahel, I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol. Cell 2015, 58, 935–946. [Google Scholar] [CrossRef]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef]
- Bano, D.; Prehn, J.H. Apoptosis-Inducing Factor (AIF) in Physiology and Disease: The Tale of a Repented Natural Born Killer. EBioMedicine 2018, 30, 29–37. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, N.S.; Haince, J.-F.; Kang, H.C.; David, K.K.; Andrabi, S.A.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Poly(ADP-Ribose) (PAR) Binding to Apoptosis-Inducing Factor Is Critical for PAR Polymerase-1-Dependent Cell Death (Parthanatos). Sci. Signal. 2011, 4, ra20. [Google Scholar] [CrossRef]
- Park, H.; Kam, T.-I.; Dawson, T.M.; Dawson, V.L. Poly (ADP-ribose) (PAR)-dependent cell death in neurodegenerative diseases. Int. Rev. Cell Mol. Biol. 2020, 353, 1–29. [Google Scholar] [CrossRef]
- Wang, Y.; Dawson, V.L.; Dawson, T.M. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Exp. Neurol. 2009, 218, 193–202. [Google Scholar] [CrossRef]
- Cheung, E.C.; Melanson-Drapeau, L.; Cregan, S.P.; Vanderluit, J.L.; Ferguson, K.L.; McIntosh, W.C.; Park, D.S.; Bennett, S.A.; Slack, R.S. Apoptosis-Inducing Factor Is a Key Factor in Neuronal Cell Death Propagated by BAX-Dependent and BAX-Independent Mechanisms. J. Neurosci. 2005, 25, 1324–1334. [Google Scholar] [CrossRef]
- Berger, N.; Sims, J.; Catino, D.; Berger, S. Poly(ADP-ribose) Polymerase Mediates the Suicide Response to Massive DNA Damage: Studies in Normal and DNA-repair Defective Cells. In ADP-Ribosylation, DNA Repair and Cancer; CRC Press: Boka Raton, FL, USA, 2020; pp. 219–226. [Google Scholar]
- Ozaki, T.; Yamashita, T.; Ishiguro, S.-I. Mitochondrial m-calpain plays a role in the release of truncated apoptosis-inducing factor from the mitochondria. Biochim. Biophys. Acta (BBA) Bioenerg. 2009, 1793, 1848–1859. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, N.S.; Li, X.; Greer, P.A.; Koehler, R.C.; Dawson, V.L.; Dawson, T.M. Calpain activation is not required for AIF translocation in PARP-1-dependent cell death (parthanatos). J. Neurochem. 2009, 110, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Song, R.; Pang, Q.; Liu, Y.; Zhuang, J.; Chen, Y.; Hu, J.; Hu, J.; Liu, Y.; Liu, Z.; et al. Propofol inhibits parthanatos via ROS–ER–calcium–mitochondria signal pathway in vivo and vitro. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Jakhar, R.; Bhardwaj, M.; Chauhan, A.K.; Kang, S.C. Fumonisin B1 induces poly (ADP-ribose) (PAR) polymer-mediated cell death (parthanatos) in neuroblastoma. Food Chem. Toxicol. 2021, 154, 112326. [Google Scholar] [CrossRef] [PubMed]
- Paquet-Durand, F.; Hauck, S.M.; Van Veen, T.; Ueffing, M.; Ekström, P. PKG activity causes photoreceptor cell death in two retinitis pigmentosa models. J. Neurochem. 2009, 108, 796–810. [Google Scholar] [CrossRef]
- Yang, P.; Lockard, R.; Titus, H.; Hiblar, J.; Weller, K.; Wafai, D.; Weleber, R.G.; Duvoisin, R.M.; Morgans, C.W.; Pennesi, M.E. Suppression of cGMP-Dependent Photoreceptor Cytotoxicity with Mycophenolate Is Neuroprotective in Murine Models of Retinitis Pigmentosa. Investig. Opthalmol. Vis. Sci. 2020, 61, 25. [Google Scholar] [CrossRef]
- Kulkarni, M.; Trifunovic, D.; Schubert, T.; Euler, T.; Paquet-Durand, F. Calcium dynamics change in degenerating cone photoreceptors. Hum. Mol. Genet. 2016, 25, 3729–3740. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Beck, S.; Michalakis, S.; Goldmann, T.; Huber, G.; Mühlfriedel, R.; Trifunović, D.; Fischer, M.D.; Fahl, E.; Duetsch, G.; et al. A key role for cyclic nucleotide gated (CNG) channels in cGMP-related retinitis pigmentosa. Hum. Mol. Genet. 2011, 20, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Xu, N.; Box, A.C.; Schaefer, L.; Kannan, K.; Zhang, Y.; Florens, L.; Seidel, C.; Washburn, M.P.; Wiegraebe, W.; et al. Nuclear cGMP-Dependent Kinase Regulates Gene Expression via Activity-Dependent Recruitment of a Conserved Histone Deacetylase Complex. PLoS Genet. 2011, 7, e1002065. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Pelluz, J.; Alavi, M.V.; Sahaboglu, A.; Kustermann, S.; Farinelli, P.; Azadi, S.; Van Veen, T.; Romero, F.J.; Paquet-Durand, F.; Ekström, P. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell Death Dis. 2010, 1, e24. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Tsang, S.H.; Chen, J. Two pathways of rod photoreceptor cell death induced by elevated cGMP. Hum. Mol. Genet. 2017, 26, 2299–2306. [Google Scholar] [CrossRef]
- Trifunovic, D.; Dengler, K.; Michalakis, S.; Zrenner, E.; Wissinger, B.; Paquet-Durand, F. cGMP-dependent cone photoreceptor degeneration in the cpfl1 mouse retina. J. Comp. Neurol. 2010, 518, 3604–3617. [Google Scholar] [CrossRef]
- Sato, S.; Peshenko, I.V.; Olshevskaya, E.V.; Kefalov, V.J.; Dizhoor, A.M. GUCY2D Cone–Rod Dystrophy-6 Is a “Phototransduction Disease” Triggered by Abnormal Calcium Feedback on Retinal Membrane Guanylyl Cyclase 1. J. Neurosci. Res. 2018, 38, 2990–3000. [Google Scholar] [CrossRef]
- Peshenko, I.V.; Olshevskaya, E.V.; Dizhoor, A.M. Functional Study and Mapping Sites for Interaction with the Target Enzyme in Retinal Degeneration 3 (RD3) Protein. J. Biol. Chem. 2016, 291, 19713–19723. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.E.; Sandberg, M.A.; Berson, E.L.; Dryja, T.P. Recessive mutations in the gene encoding the β–subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat. Genet. 1993, 4, 130–134. [Google Scholar] [CrossRef]
- Dryja, T.P.; Finn, J.T.; Peng, Y.W.; McGee, T.L.; Berson, E.L.; Yau, K.W. Mutations in the gene encoding the alpha subunit of the rod cGMP-gated channel in autosomal recessive retinitis pigmentosa. PNAS 1995, 92, 10177–10181. [Google Scholar] [CrossRef]
- Tsang, S.H.; Gouras, P.; Yamashita, C.K.; Kjeldbye, H.; Fisher, J.; Farber, D.B.; Goff, S.P. Retinal Degeneration in Mice Lacking the gamma Subunit of the Rod cGMP Phosphodiesterase. Science 1996, 272, 1026–1029. [Google Scholar] [CrossRef]
- Reuter, P.; Koeppen, K.; Ladewig, T.; Kohl, S.; Baumann, B.; Wissinger, B. Mutations in CNGA3 impair trafficking or function of cone cyclic nucleotide-gated channels, resulting in achromatopsia. Hum. Mutat. 2008, 29, 1228–1236. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Marigo, V.; Ekström, P. RD Genes Associated with High Photoreceptor cGMP-Levels (Mini-Review). Retin. Degener. Dis. 2019, 1185, 245–249. [Google Scholar] [CrossRef]
- Das, S.; Popp, V.; Power, M.; Groeneveld, K.; Melle, C.; Rogerson, L.; Achury, M.; Schwede, F.; Strasser, T.; Euler, T. Redefining the role of Ca2+-permeable channels in hereditary photoreceptor degeneration using the D-and L-cis enantiomers of diltiazem. bioRxiv 2020. [Google Scholar] [CrossRef]
- Catterall, W.A. Voltage-Gated Calcium Channels. Cold Spring Harb. Perspect. Biol. 2011, 3, a003947. [Google Scholar] [CrossRef]
- Shinkai-Ouchi, F.; Shindo, M.; Doi, N.; Hata, S.; Ono, Y. Calpain-2 participates in the process of calpain-1 inactivation. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Goll, D.E.; Thompson, V.F.; Li, H.; Wei, W.; Cong, J. The calpain system. Physiol. Rev. 2003, 83, 731–801. [Google Scholar] [CrossRef] [PubMed]
- Baudry, M.; Bi, X. Calpain-1 and Calpain-2: The Yin and Yang of Synaptic Plasticity and Neurodegeneration. Trends Neurosci. 2016, 39, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Fox, D.A.; Poblenz, A.T.; He, L. Calcium Overload Triggers Rod Photoreceptor Apoptotic Cell Death in Chemical-Induced and Inherited Retinal Degenerations. Ann. N. Y. Acad. Sci. 1999, 893, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Barabas, P.; Peck, C.C.; Krizaj, D. Do Calcium Channel Blockers Rescue Dying Photoreceptors in the Pde6brd1 Mouse? Retin. Degener. Dis. 2009, 664, 491–499. [Google Scholar] [CrossRef]
- Schön, C.; Paquet-Durand, F.; Michalakis, S. Cav1.4 L-Type Calcium Channels Contribute to Calpain Activation in Degenerating Photoreceptors of rd1 Mice. PLoS ONE 2016, 11, e0156974. [Google Scholar] [CrossRef]
- Read, D.S.; McCallbc, M.A.; Gregg, R.G. Absence of Voltage-dependent Calcium Channels Delays Photoreceptor Degeneration in rd Mice. Exp. Eye Res. 2002, 75, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, F.; Bernhard, D.; Lukowski, R.; Weinmeister, P. cGMP regulated protein kinases (cGK). In cGMP: Generators, Effectors and Therapeutic Implications; Springer: Berlin/Heidelberg, Germany, 2009; pp. 137–162. [Google Scholar] [CrossRef]
- Fiscus, R.R. Involvement of Cyclic GMP and Protein Kinase G in the Regulation of Apoptosis and Survival in Neural Cells. Neurosignals 2002, 11, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Frankenreiter, S.; Bednarczyk, P.; Kniess, A.; Bork, N.I.; Straubinger, J.; Koprowski, P.; Wrzosek, A.; Mohr, E.; Logan, A.; Murphy, M.P.; et al. cGMP-Elevating Compounds and Ischemic Conditioning Provide Cardioprotection Against Ischemia and Reperfusion Injury via Cardiomyocyte-Specific BK Channels. Circulation 2017, 136, 2337–2355. [Google Scholar] [CrossRef] [PubMed]
- Jaumann, M.; Dettling, J.; Gubelt, M.; Zimmermann, U.; Gerling, A.; Paquet-Durand, F.; Feil, S.; Wolpert, S.; Franz, C.; Varakina, K.; et al. cGMP-Prkg1 signaling and Pde5 inhibition shelter cochlear hair cells and hearing function. Nat. Med. 2012, 18, 252–259. [Google Scholar] [CrossRef]
- Canzoniero, L.M.; Adornetto, A.; Secondo, A.; Magi, S.; Dell’Aversano, C.; Scorziello, A.; Amoroso, S.; Di Renzo, G. Involvement of the nitric oxide/protein kinase G pathway in polychlorinated biphenyl-induced cell death in SH-SY 5Y neuroblastoma cells. J. Neurosci. Res. 2006, 84, 692–697. [Google Scholar] [CrossRef]
- Brunetti, M.; Mascetra, N.; Manarini, S.; Martelli, N.; Cerletti, C.; Musiani, P.; Aiello, F.B.; Evangelista, V. Inhibition of cGMP-dependent protein kinases potently decreases neutrophil spontaneous apoptosis. Biochem. Biophys. Res. Commun. 2002, 297, 498–501. [Google Scholar] [CrossRef]
- Vighi, E.; Rentsch, A.; Henning, P.; Comitato, A.; Hoffmann, D.; Bertinetti, D.; Bertolotti, E.; Schwede, F.; Herberg, F.W.; Genieser, H.-G.; et al. New cGMP analogues restrain proliferation and migration of melanoma cells. Oncotarget 2018, 9, 5301–5320. [Google Scholar] [CrossRef]
- Mencl, S.; Trifunović, D.; Zrenner, E.; Paquet-Durand, F. PKG-Dependent Cell Death in 661W Cone Photoreceptor-like Cell Cultures (Experimental Study). In Retinal Degenerative Diseases; Springer: Cham, Switzerland, 2018; Volume 1074, pp. 511–517. [Google Scholar] [CrossRef]
- Farber, D.B.; Lolley, R.N. Cyclic Guanosine Monophosphate: Elevation in Degenerating Photoreceptor Cells of the C3H Mouse Retina. Science 1974, 186, 449–451. [Google Scholar] [CrossRef]
- Vighi, E.; Trifunovic, D.; Veiga-Crespo, P.; Rentsch, A.; Hoffmann, D.; Sahaboglu, A.; Strasser, T.; Kulkarni, M.; Bertolotti, E.; Heuvel, A.V.D.; et al. Combination of cGMP analogue and drug delivery system provides functional protection in hereditary retinal degeneration. PNAS 2018, 115, E2997–E3006. [Google Scholar] [CrossRef]
- Koch, M.; Scheel, C.; Ma, H.; Yang, F.; Stadlmeier, M.; Glück, A.F.; Murenu, E.; Traube, F.R.; Carell, T.; Biel, M.; et al. The cGMP-Dependent Protein Kinase 2 Contributes to Cone Photoreceptor Degeneration in the Cnga3-Deficient Mouse Model of Achromatopsia. Int. J. Mol. Sci. 2021, 22, 52. [Google Scholar] [CrossRef]
- Gamm, D.M.; Barthel, L.K.; Raymond, P.A.; Uhler, M.D. Localization of cGMP-dependent protein kinase isoforms in mouse eye. Investig. Ophthalmol. Vis. Sci. 2000, 41, 2766–2773. [Google Scholar] [CrossRef]
- Feil, S.; Zimmermann, P.; Knorn, A.; Brummer, S.; Schlossmann, J.; Hofmann, F.; Feil, R. Distribution of cGMP-dependent protein kinase type I and its isoforms in the mouse brain and retina. Neuroscience 2005, 135, 863–868. [Google Scholar] [CrossRef]
- Ekstrom, P.; Ueffing, M.; Zrenner, E.; Paquet-Durand, F. Novel in situ activity assays for the quantitative molecular analysis of neurodegenerative processes in the retina. Curr. Med. Chem. 2014, 21, 3478–3493. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Groten, J.; Marigo, V.; Tomar, T.; Hilhorst, R. Identification of Novel Substrates for cGMP Dependent Protein Kinase (PKG) through Kinase Activity Profiling to Understand Its Putative Role in Inherited Retinal Degeneration. Int. J. Mol. Sci. 2021, 22, 1180. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.Y.; Allis, C.D. Histone Acetylation and Chromatin Assembly: A Single Escort, Multiple Dances? Cell 1996, 87, 5–8. [Google Scholar] [CrossRef]
- Jaliffa, C.; Ameqrane, I.; Dansault, A.; Leemput, J.; Vieira, V.; Lacassagne, E.; Provost, A.; Bigot, K.; Masson, C.; Menasche, M.; et al. Sirt1 Involvement in rd10 Mouse Retinal Degeneration. PNAS 2009, 50, 3562–3572. [Google Scholar] [CrossRef] [PubMed]
- Samardzija, M.; Masarini, K.; Ueffing, M.; Trifunović, D. HDAC Inhibition Prevents Primary Cone Degeneration Even After the Onset of Degeneration. In Retinal Degenerative Diseases; Springer: Cham, Switzerland, 2019; pp. 383–387. [Google Scholar] [CrossRef]
- Trifunović, D.; Petridou, E.; Comitato, A.; Marigo, V.; Ueffing, M.; Paquet-Durand, F. Primary Rod and Cone Degeneration Is Prevented by HDAC Inhibition. In Retinal Degenerative Diseases; Springer: Cham, Switzerland, 2018; pp. 367–373. [Google Scholar] [CrossRef]
- Trifunović, D.; Arango-Gonzalez, B.; Comitato, A.; Barth, M.; del Amo, E.M.; Kulkarni, M.; Sahaboglu, A.; Hauck, S.M.; Urtti, A.; Arsenijevic, Y.; et al. HDAC inhibition in thecpfl1mouse protects degenerating cone photoreceptors in vivo. Hum. Mol. Genet. 2016, 25, 4462–4472. [Google Scholar] [CrossRef]
- Sancho-Pelluz, J.; Paquet-Durand, F. HDAC Inhibition Prevents Rd1 Mouse Photoreceptor Degeneration. In Retinal Degenerative Diseases; Springer: Boston, MA, USA, 2012; Volume 723, pp. 107–113. [Google Scholar] [CrossRef]
- Samardzija, M.; Corna, A.; Gomez-Sintes, R.; Jarboui, M.A.; Armento, A.; Roger, J.E.; Petridou, E.; Haq, W.; Paquet-Durand, F.; Zrenner, E.; et al. HDAC inhibition ameliorates cone survival in retinitis pigmentosa mice. Cell Death Differ. 2020, 28, 1317–1332. [Google Scholar] [CrossRef]
- Sundaramurthi, H.; Roche, S.L.; Grice, G.L.; Moran, A.; Dillion, E.T.; Campiani, G.; Nathan, J.A.; Kennedy, B.N. Selective Histone Deacetylase 6 Inhibitors Restore Cone Photoreceptor Vision or Outer Segment Morphology in Zebrafish and Mouse Models of Retinal Blindness. Front. Cell Dev. Biol. 2020, 8, 689. [Google Scholar] [CrossRef]
- Perron, N.R.; Nasarre, C.; Bandyopadhyay, M.; Beeson, C.C.; Rohrer, B. SAHA is neuroprotective in in vitro and in situ models of retinitis pigmentosa. Mol. Vis. 2021, 27, 151–160. [Google Scholar]
- Tsai, S.-C.; Seto, E. Regulation of Histone Deacetylase 2 by Protein Kinase CK2. J. Biol. Chem. 2002, 277, 31826–31833. [Google Scholar] [CrossRef]
- Kaur, J.; Mencl, S.; Sahaboglu, A.; Farinelli, P.; Van Veen, T.; Zrenner, E.; Ekström, P.; Paquet-Durand, F.; Arango-Gonzalez, B. Calpain and PARP Activation during Photoreceptor Cell Death in P23H and S334ter Rhodopsin Mutant Rats. PLoS ONE 2011, 6, e22181. [Google Scholar] [CrossRef] [PubMed]
- Paquet-Durand, F.; Silva, J.; Talukdar, T.; Johnson, L.E.; Azadi, S.; Van Veen, T.; Ueffing, M.; Hauck, S.M.; Ekström, P.A.R. Excessive Activation of Poly(ADP-Ribose) Polymerase Contributes to Inherited Photoreceptor Degeneration in the Retinal Degeneration 1 Mouse. J. Neurosci. Res. 2007, 27, 10311–10319. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, R.D.; Gajjar, M.K.; Jensen, K.E.; Hamerlik, P. Enhanced efficacy of combined HDAC and PARP targeting in glioblastoma. Mol. Oncol. 2016, 10, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Sahaboglu, A.; Barth, M.; Secer, E.; del Amo, E.M.; Urtti, A.; Arsenijevic, Y.; Zrenner, E.; Paquet-Durand, F. Olaparib significantly delays photoreceptor loss in a model for hereditary retinal degeneration. Sci. Rep. 2016, 6, 39537. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Gil, L.; Sancho-Pelluz, J.; Zrenner, E.; Oltra, M.; Sahaboglu, A. Poly ADP ribosylation and extracellular vesicle activity in rod photoreceptor degeneration. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Olivares-González, L.; Velasco, S.; Millán, J.M.; Rodrigo, R. Intravitreal administration of adalimumab delays retinal degeneration in rd10 mice. FASEB J. 2020, 34, 13839–13861. [Google Scholar] [CrossRef]
- Sahaboglu, A.; Miranda, M.; Canjuga, D.; Avci-Adali, M.; Savytska, N.; Secer, E.; Feria-Pliego, J.A.; Kayık, G.; Durdagi, S. Drug repurposing studies of PARP inhibitors as a new therapy for inherited retinal degeneration. Cell. Mol. Life Sci. 2020, 77, 2199–2216. [Google Scholar] [CrossRef]
- Sahaboglu, A.; Tanimoto, N.; Kaur, J.; Sancho-Pelluz, J.; Huber, G.; Fahl, E.; Arango-Gonzalez, B.; Zrenner, E.; Ekström, P.; Löwenheim, H.; et al. PARP1 Gene Knock-Out Increases Resistance to Retinal Degeneration without Affecting Retinal Function. PLoS ONE 2010, 5, e15495. [Google Scholar] [CrossRef]
- Hamann, S.; Schorderet, D.F.; Cottet, S. Bax-Induced Apoptosis in Leber’s Congenital Amaurosis: A Dual Role in Rod and Cone Degeneration. PLoS ONE 2009, 4, e6616. [Google Scholar] [CrossRef]
- Yoshizawa, K.; Kiuchi, K.; Nambu, H.; Yang, J.; Senzaki, H.; Kiyozuka, Y.; Shikata, N.; Tsubura, A. Caspase-3 inhibitor transiently delays inherited retinal degeneration in C3H mice carrying the rd gene. Graefe’s Arch. Clin. Exp. Ophthalmol. 2002, 240, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Zeiss, C.J.; Neal, J.; Johnson, E.A. Caspase-3 in postnatal retinal development and degeneration. IOVS 2004, 45, 964–970. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.; Zeng, Y.; Bajwa, R.; Klatt, L.; Li, Y.; Maguire, A.M. Adenovirus-mediated delivery of rhodopsin-promoted bcl-2 results in a delay in photoreceptor cell death in the rd/rd mouse. Gene Ther. 1998, 5, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Joseph, R.M.; Li, T. Overexpression of Bcl-2 or Bcl-XL transgenes and photoreceptor degeneration. Investig. Ophthalmol. Vis. Sci. 1996, 37, 2434–2446. [Google Scholar]
- Fatokun, A.A.; Dawson, V.L.; Dawson, T.M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 2014, 171, 2000–2016. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.-Y.; Wu, N.; Chen, Y.-C.; Cheng, Q.; Wang, J. PARP inhibitor resistance: The underlying mechanisms and clinical implications. Mol. Cancer 2020, 19, 1–16. [Google Scholar] [CrossRef]
- Sahaboglu, A.; Sharif, A.; Feng, L.; Secer, E.; Zrenner, E.; Paquet-Durand, F. Temporal progression of PARP activity in the Prph2 mutant rd2 mouse: Neuroprotective effects of the PARP inhibitor PJ34. PLoS ONE 2017, 12, e0181374. [Google Scholar] [CrossRef]
- Hafezi, F.; Abegg, M.; Grimm, C.; Wenzel, A.; Munz, K.; Stürmer, J.; Farber, D.; Remé, C.E. Retinal degeneration in the rd mouse in the absence of c-fos. IOVS 1998, 39, 2239–2244. [Google Scholar]
- Venkatesh, A.; Cheng, S.-Y.; Punzo, C. Loss of the cone-enriched caspase-7 does not affect secondary cone death in retinitis pigmentosa. Mol. Vis. 2017, 23, 944–951. [Google Scholar]
- Comitato, A.; Sanges, D.; Rossi, A.; Humphries, M.M.; Marigo, V. Activation of Bax in Three Models of Retinitis Pigmentosa. IOVS 2014, 55, 3555–3561. [Google Scholar] [CrossRef]
- Bixel, K.; Hays, J.L. Olaparib in the management of ovarian cancer. Pharm. Pers. Med. 2015, 8, 127–135. [Google Scholar] [CrossRef]
- Hüttl, S.; Michalakis, S.; Seeliger, M.; Luo, D.-G.; Acar, N.; Geiger, H.; Hudl, K.; Mader, R.; Haverkamp, S.; Moser, M.; et al. Impaired Channel Targeting and Retinal Degeneration in Mice Lacking the Cyclic Nucleotide-Gated Channel Subunit CNGB1. J. Neurosci. Res. 2005, 25, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Aherne, A.; Kennan, A.; Kenna, P.F.; McNally, N.; Lloyd, D.G.; Alberts, I.L.; Kiang, A.-S.; Humphries, M.M.; Ayuso, C.; Engel, P.C.; et al. On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa. Hum. Mol. Genet. 2004, 13, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.C.; Kowalski, W.J.; Muller, C.D.; Eugui, E.M. Mechanisms of Action of Mycophenolic Acid. Ann. N. Y. Acad. Sci. 1993, 696, 63–87. [Google Scholar] [CrossRef] [PubMed]
- Athanasiou, D.; Aguila, M.; Bellingham, J.; Li, W.; McCulley, C.; Reeves, P.J.; Cheetham, M.E. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog. Retin. Eye Res. 2018, 62, 1–23. [Google Scholar] [CrossRef]
- Messchaert, M.; Haer-Wigman, L.; Khan, M.I.; Cremers, F.P.M.; Collin, R.W.J. EYSmutation update: In silico assessment of 271 reported and 26 novel variants in patients with retinitis pigmentosa. Hum. Mutat. 2018, 39, 177–186. [Google Scholar] [CrossRef]
- Berkowitz, B.A. Oxidative stress measured in vivo without an exogenous contrast agent using QUEST MRI. J. Magn. Reson. 2018, 291, 94–100. [Google Scholar] [CrossRef]
- De la Camara, C.; Salom, D.; Sequedo, M.D.; Hervas, D.; Marin-Lambies, C.; Aller, E.; Jaijo, T.; Díaz-Llopis, M.; Millán, J.M.; Rodrigo, R. Altered Antioxidant-Oxidant Status in the Aqueous Humor and Peripheral Blood of Patients with Retinitis Pigmentosa. PLoS ONE 2013, 8, e74223. [Google Scholar] [CrossRef]
- Kjellström, U.; Veiga-Crespo, P.; Andréasson, S.; Ekström, P. Increased Plasma cGMP in a Family with Autosomal Recessive Retinitis Pigmentosa Due to Homozygous Mutations in the PDE6A Gene. IOVS 2016, 57, 6048–6057. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).