Acyl Chain Specificity of Marine Streptomyces klenkii PhosPholipase D and Its Application in Enzymatic Preparation of Phosphatidylserine
Abstract
:1. Introduction
2. Results
2.1. Bioinformatic Analysis of SkPLD
2.2. Expression and Purification of SkPLD
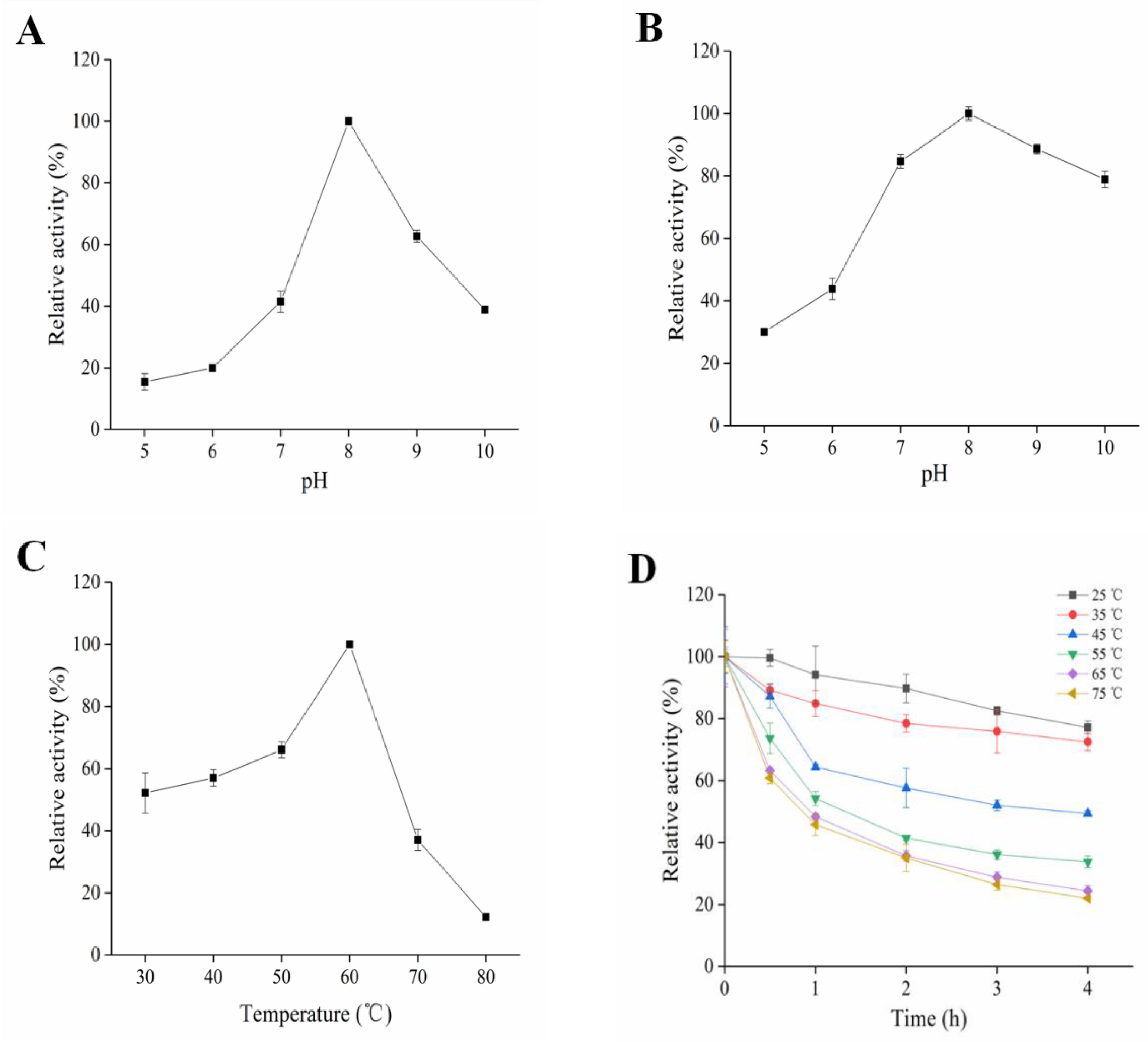
2.3. Enzymatic Characterization of SkPLD
2.4. Kinetic Parameters and Acyl Chain Specificity of SkPLD
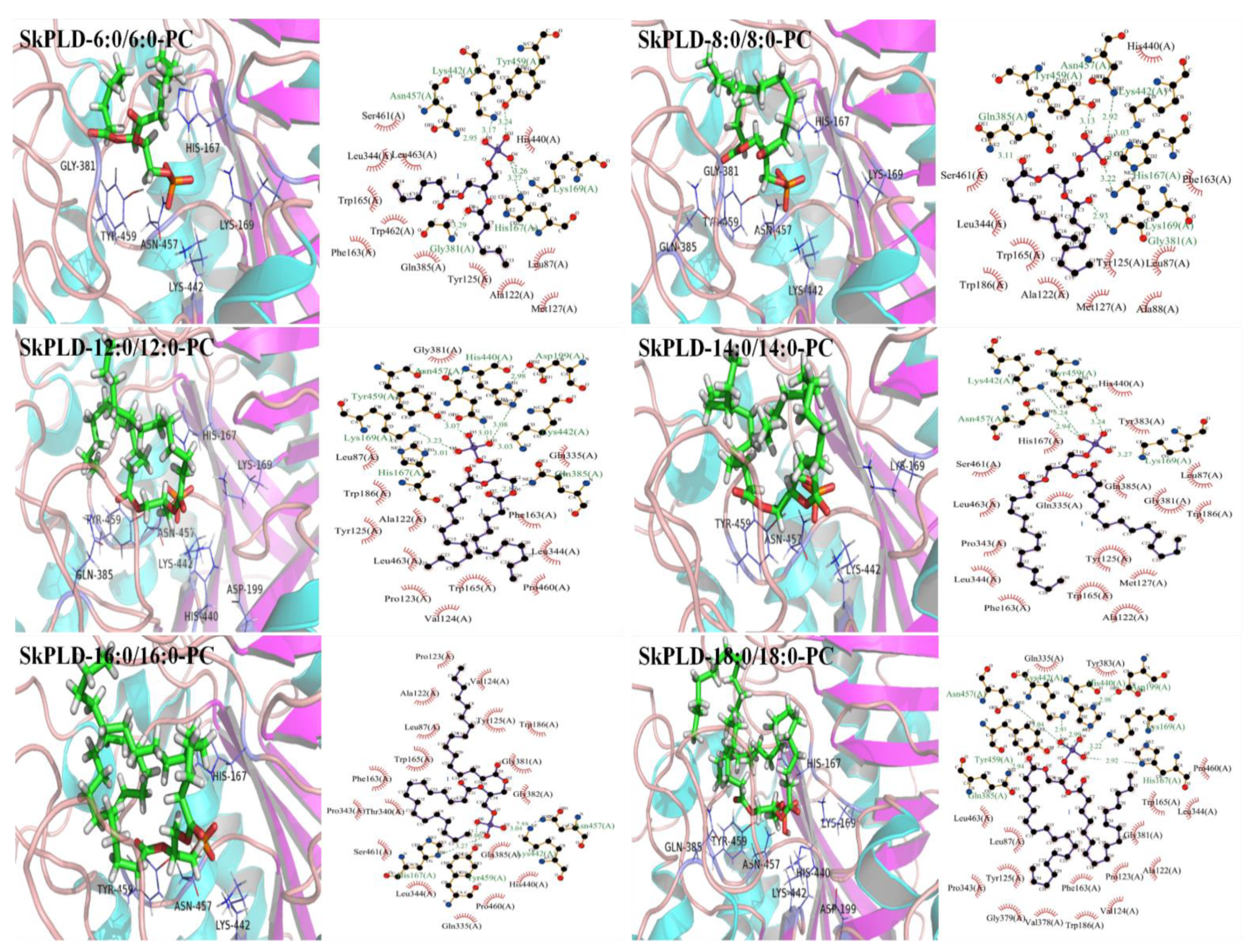
2.5. Structural Feature and Molecular Basis for Chain Length Specificity
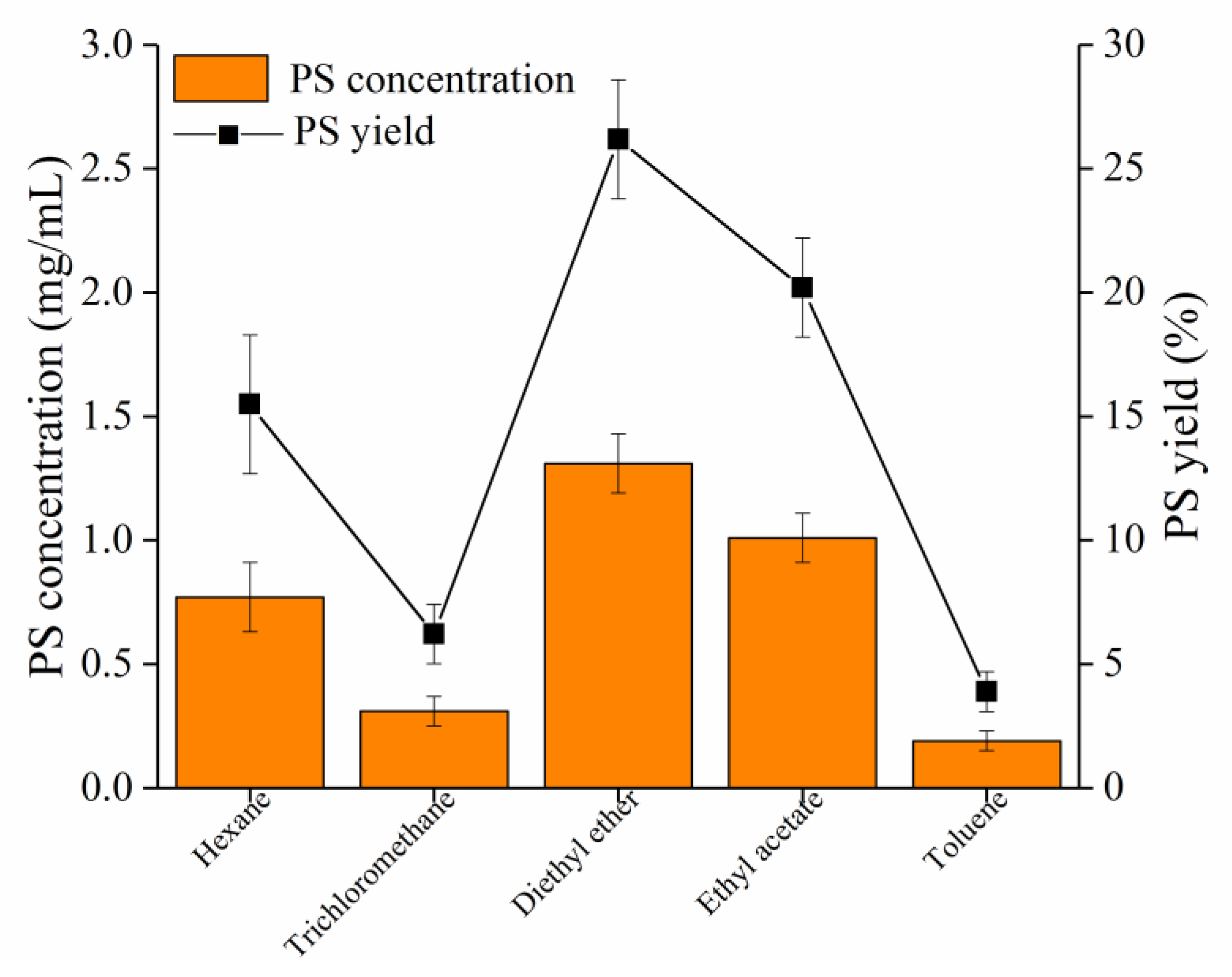
2.6. Enzymatic Synthesis of PS by Recombinant SkPLD
3. Discussion
3.1. Bioinformatic Analysis of SkPLD
3.2. Enzymatic Characterization of SkPLD
3.3. Kinetic Parameters and Acyl Chain Specificity of SkPLD
3.4. Structural Feature and Molecular Basis for Chain Length Specificity
3.5. Enzymatic Synthesis of PS by Recombinant SkPLD
4. Materials and Methods
4.1. Chemicals, Strains, and Materials
4.2. Bioinformatic and Homology Modeling
4.3. Recombinant Expression of SkPLD in Escherichia coli
4.4. Purification of Recombinant SkPLD
4.5. Enzyme Activity Assay
4.6. Enzymatic Characterization of SkPLD
4.7. Application of SkPLD for the PS Synthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, H.-Y.; Huang, B.X.; Spector, A.A. Phosphatidylserine in the brain: Metabolism and function. Prog. Lipid Res. 2014, 56, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Haq, M.; Suraiya, S.; Ahmed, S.; Chun, B.S. Phospholipids from Marine Source: Extractions and Forthcoming Industrial Ap-plications. J. Funct. Foods 2021, 80, 104448. [Google Scholar] [CrossRef]
- Ric, D.; Élodie, B.; Habib, H.; Daniel, B.; Philippe, C.; Line, C.; Nicolas, B.; Sophie, C.; Bernard, D.; Christian, S. Lipid Selectivity, Orientation, and Extent of Membrane Binding of Nonacylated RP2. Biochemistry 2015, 54, 2560–2570. [Google Scholar]
- Qin, W.; Wu, C.J.; Song, W.; Chen, X.L.; Liu, J.; Luo, Q.L.; Liu, L.M. A Novel High-Yield Process of Phospholipase D-Mediated Phosphatidylserine Production with Cyclopentyl Methyl Ethe. Process. Biochem. 2018, 66, 146–149. [Google Scholar] [CrossRef]
- Li, B.; Duan, D.; Wang, J.; Li, H.; Zhang, X.; Zhao, B. Improving phospholipase D activity and selectivity by bio-imprinting-immobilization to produce phosphatidylglycerol. J. Biotechnol. 2018, 281, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Li, B.; Dong, W.; Yang, W.; Zhang, X.; Zhao, B. Synthesis of Phosphatidylethanolamine by Enzymatic Catalysis and Its Substrate Inhibition Kinetics. Chem. Ind. Eng. Prog. 2017, 36, 2601–2606. [Google Scholar]
- Shirouchi, B.; Nagao, K.; Inoue, N.; Furuya, K.; Koga, S.; Matsumoto, H.; Yanagita, T. Dietary Phosphatidylinositol Prevents the Development of Nonalcoholic Fatty Liver Disease in Zucker (fa/fa) Rats. J. Agric. Food Chem. 2008, 56, 2375–2379. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Liu, Y.; Tian, Y.; Zhang, Q.; Cong, P.; Li, H.; Xu, J.; Li, Z.; Wang, J.; Mao, X. Reaction Specificity of Phospholipase D Prepared from Acinetobacter radioresistens a2 in Transphosphatidylation. Lipids 2018, 53, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Laouini, A.; Jaafar-Maalej, C.; Limayem-Blouza, I.; Sfar, S.; Charcosset, C.; Fessi, H. Preparation, characterization and applications of liposomes: State of the art. J. Colloid Sci. Biotechnol. 2012, 1, 147–168. [Google Scholar] [CrossRef]
- Ogino, C.; Kuroda, S.; Tokuyama, S.; Kondo, A.; Shimizu, N.; Tanizawa, K.; Fukuda, H. Phospholipase D from Streptoverti-cillium cinnamoneum: Protein Engineering and Application for Phospholipid Production. J. Mol. Catal. B Enzym. 2003, 23, 107–115. [Google Scholar] [CrossRef]
- Ulbrich-Hofmann, R.; Lerchner, A.; Oblozinsky, M.; Bezakova, L. Phospholipase D and its application in biocatalysis. Biotechnol. Lett. 2005, 27, 535–544. [Google Scholar] [CrossRef]
- Zerrifi, S.E.A.; Redouane, E.M.; Mugani, R.; Ribeiro, I.; Carvalho, M.F.; Campos, A.; Barakate, M.; Vasconcelos, V.; Oudra, B.; El Khalloufi, F. Moroccan actinobacteria with promising activity against toxic cyanobacteria Microcystis aeruginosa. Environ. Sci. Pollut. Res. 2020, 28, 235–245. [Google Scholar] [CrossRef]
- Yuan, D.J.; Lan, D.M.; Xin, R.P.; Yang, B.; Wang, Y.H. Screening and Characterization of a Thermostable Lipase from Marine Streptomyces sp. strain W007. Biotechnol. Appl. Biochem. 2016, 63, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.-B.; Gong, J.-S.; Hou, H.-J.; Li, H.; Lu, Z.-M.; Xu, H.-Y.; Xu, Z.-H.; Shi, J.-S. Mining of a phospholipase D and its application in enzymatic preparation of phosphatidylserine. Bioengineered 2017, 9, 80–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waite, M. The PLD superfamily: Insights into catalysis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 1999, 1439, 187–197. [Google Scholar] [CrossRef]
- Wang, J.; Li, B.; Zhang, X.; Hu, Q.; Yu, W.; Wang, H.; Duan, D.; Li, J.; Zhao, B. Docking and molecular dynamics studies on the mechanism of phospholipase D-mediated transphosphatidylation to construct the reaction kinetic model: Application in phosphatidylserine production. J. Taiwan Inst. Chem. Eng. 2018, 96, 82–92. [Google Scholar] [CrossRef]
- Ogino, C.; Daido, H.; Ohmura, Y.; Takada, N. Remarkable enhancement in PLD activity from Streptoverticillium cinnamoneum by substituting serine residue into the GG/GS motif. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2007, 1774, 671–678. [Google Scholar] [CrossRef]
- Leiros, I.; Secundo, F.; Zambonelli, C.; Servi, S.; Hough, E. The first crystal structure of a phospholipase D. Structure 2000, 8, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Chen, M.; Xu, W.; Zhang, W.; Zhang, T.; Guang, C.; Mu, W. Microbial phospholipase D: Identification, modification and application. Trends Food Sci. Technol. 2020, 96, 145–156. [Google Scholar] [CrossRef]
- Zambonellia, C.; Morandib, P.; Vanonib, M.A.; Tedeschic, G.; Servia, S.; Curtib, B. Cloning and expression in Escherichia coli of the gene encoding Streptomyces PMF PLD, a phospholipase D with high transphosphatidylation activity. Enzym. Microb. Technol. 2003, 33, 676–688. [Google Scholar] [CrossRef]
- Simkhada, J.R.; Cho, S.S.; Lee, H.J.; Yoo, J.C. Purification and biochemical properties of phospholipase d (PLD57) produced by Streptomyces sp. CS-57. Arch. Pharmacal Res. 2007, 30, 1302–1308. [Google Scholar] [CrossRef]
- Hatanaka, T.; Negishi, T.; Kubota-Akizawa, M.; Hagishita, T. Study on thermostability of phospholipase D from Streptomyces sp. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2002, 1598, 156–164. [Google Scholar] [CrossRef]
- Yasmeen Yousif Ahmed, E.; Kazusa, M.; Katsuhiko, S.; Jiro, A. Effect of Active Site Pocket Structure Modification of D-Stereospecific Amidohydrolase on the Recognition of Stereospecific and Hydrophobic Substrates. Mol. Biotechnol. 2018, 60, 690–697. [Google Scholar]
- Gaskin, D.J.H.; Romojaro, A.; Turner, N.A.; Jenkins, J.; Vulfson, E.N. Alteration of lipase chain length specificity in the hydrolysis of esters by random mutagenesis. Biotechnol. Bioeng. 2001, 73, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Jasmina, D.; Hideo, N.; Yugo, I. Acyl chain that matters: Introducing sn-2 acyl chain preference to a phospholipase D by protein engineering. Protein Eng. Des. Sel. 2019, 32, 1–11. [Google Scholar]
- Lee, J.S. Binding energies of hydrogen-bonded complexes from extrapolation with localized basis sets. J. Chem. Phys. 2007, 127, 08B616. [Google Scholar] [CrossRef]
- Pleiss, J.; Fischer, M.; Schmid, R.D. Anatomy of lipase binding sites: The scissile fatty acid binding site. Chem. Phys. Lipids 1998, 93, 67–80. [Google Scholar] [CrossRef]
- Traul, K.; Driedger, A.; Ingle, D.; Nakhasi, D. Review of the toxicologic properties of medium-chain triglycerides. Food Chem. Toxicol. 2000, 38, 79–98. [Google Scholar] [CrossRef]
- Qin, X.; Zhong, J.; Wang, Y. A mutant T1 lipase homology modeling, and its molecular docking and molecular dynamics simulation with fatty acids. J. Biotechnol. 2021, 337, 24–34. [Google Scholar] [CrossRef]
- Chen, S.; Xu, L.; Li, Y.; Hao, N.; Yan, M. Bioconversion of Phosphatidylserine by Phospholipase D from Streptomyces racemochromogenes in a Microaqueous Water-Immiscible Organic Solvent. J. Agric. Chem. Soc. Jpn. 2013, 77, 1939–1941. [Google Scholar]
- Qian, J.; Yang, P.; Wang, X. Preparation of phospholipase D and catalytic synthesis of phosphatidylserine from phosphatidylcholine. China Oil Fat. 2017, 42, 62–71. [Google Scholar]
- Saovanee, C.; Uwe, T.B.; Apichat, U.; Aran, H.K. Efficient phosphatidylserine synthesis by a phospholipase D from Strepto-myces sp. SC734 isolated from soil-contaminated palm oil. Eur. Lipid Sci. Technol. 2016, 118, 803–813. [Google Scholar]
- Li, B.; Lu, F.P.; Tian, L.; Li, Y.; Du, L. Cloning and expression of phospholipase D gene pld from Streptomyces chromofuscus. Ann. Microbiol. 2008, 58, 227–231. [Google Scholar] [CrossRef]
- Han, H.X.; Cao, D.; Shi, S.J.; Wang, Q. Optimization of fermentation conditions of phospholipase D from Streptomyces sp. CA-1. Sci. Technol. Food Ind. 2014, 35, 176–180. [Google Scholar]
- Hosokawa, M.; Shimatani, T.; Kanada, T.; Inoue, Y.; Takahashi, K. Conversion to docosahexaenoic acid-containing phospha-tidylserine from squid skin lecithin by phospholipase D-mediated transphosphatidylation. J. Agric. Food Chem. 2000, 48, 4550–4554. [Google Scholar] [CrossRef]
- Lee, J.S.; Bat-Ochir, M.; Demirev, A.V.; Nam, D.H. Molecular cloning of the phospholipase D gene from Streptomyces sp. YU100 and its expression in Escherichia coli. J. Microb. 2009, 47, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-L.; Duan, Z.-Q. Insight into enzymatic synthesis of phosphatidylserine in deep eutectic solvents. Catal. Commun. 2016, 82, 16–19. [Google Scholar] [CrossRef]
- Li, B.; Wang, J.; Zhang, X.; Zhao, B.; Niu, L. Aqueous–Solid System for Highly Efficient and Environmentally Friendly Transphosphatidylation Catalyzed by Phospholipase D To Produce Phosphatidylserine. J. Agric. Food Chem. 2016, 64, 7555–7560. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Wang, J.; Li, H.; Zhao, B. High-Yield and Sustainable Production of Phosphatidylserine in Purely Aqueous Solutions via Adsorption of Phosphatidylcholine on Triton-X-100-Modified Silica. J. Agric. Food Chem. 2017, 65, 10767–10774. [Google Scholar] [CrossRef]
- Ribitsch, D.; Karl, W.; Wehrschütz-Sigl, E.; Tutz, S.; Remler, P.; Weber, H.; Gruber, K.; Stehr, R.; Bessler, C.; Hoven, N.; et al. Heterologous Expression and Characterization of Choline Oxidase from the Soil Bacterium Arthrobacter nicotianae. Appl. Microbiol. Biotechnol. 2008, 81, 875–886. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Florence, C. Multiple Sequence Alignment with Hierarchical Clustering. Nucleic Acids Res. 1998, 16, 10881–10890. [Google Scholar]
- Andrew, W.; Martino, B.; Stefan, B.; Gabriel, S.; Gerardo, T.; Rafal, G.; Florian, T.H.; de Tjaart, A.P.B.; Christine, R.; Lorenza, B.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar]
- Suzuki, A.; Kakuno, K.; Satio, R.; Iwasaki, Y.; Yamane, T. Crystal Structure of Phospholipase D from Streptomyces antibioticus. Acta Crystallogr. Sect. A Found. Crystallogr. 2000, 56, s242. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2010, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Sussman, J.L.; Abola, E.E.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J. The Protein Data Bank. Genetica 1999, 106, 149–158. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; Delano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Shimbo, K.; Yano, H.; Miyamoto, Y. Purification and Properties of Phospholipase D from Streptomyces lydicus. Agric. Biol. Chem. 1990, 54, 1189–1193. [Google Scholar] [CrossRef] [Green Version]

| Substrates | Km (mM) a | kcat (S−1) b | kcat/Km (S−1 mM−1) c |
|---|---|---|---|
| 6:0/6:0-PC | 2.01 ± 0.18 | 100.85 ± 4.42 | 50.27 |
| 8:0/8:0-PC | 2.59 ± 0.28 | 110.54 ± 6.30 | 42.63 |
| 12:0/12:0-PC | 1.09 ± 0.10 | 73.29 ± 2.68 | 67.13 |
| 14:0/14:0-PC | 1.07 ± 0.18 | 61.65 ± 4.05 | 57.51 |
| 16:0/16:0-PC | 2.01 ± 0.19 | 32.97 ± 1.48 | 16.43 |
| 18:0/18:0-PC | 1.27 ± 0.14 | 15.45 ± 0.69 | 12.12 |
| Complex | Binding Energy (kcal/mol) |
|---|---|
| SkPLD-6:0/6:0-PC | –7.10 |
| SkPLD-8:0/8:0-PC | −7.13 |
| SkPLD-12:0/12:0-PC | −7.54 |
| SkPLD-14:0/14:0-PC | −7.29 |
| SkPLD-16:0/16:0-PC | −6.79 |
| SkPLD-18:0/18:0-PC | −7.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, R.; Cui, R.; Lan, D.; Wang, F.; Wang, Y. Acyl Chain Specificity of Marine Streptomyces klenkii PhosPholipase D and Its Application in Enzymatic Preparation of Phosphatidylserine. Int. J. Mol. Sci. 2021, 22, 10580. https://doi.org/10.3390/ijms221910580
Hu R, Cui R, Lan D, Wang F, Wang Y. Acyl Chain Specificity of Marine Streptomyces klenkii PhosPholipase D and Its Application in Enzymatic Preparation of Phosphatidylserine. International Journal of Molecular Sciences. 2021; 22(19):10580. https://doi.org/10.3390/ijms221910580
Chicago/Turabian StyleHu, Rongkang, Ruiguo Cui, Dongming Lan, Fanghua Wang, and Yonghua Wang. 2021. "Acyl Chain Specificity of Marine Streptomyces klenkii PhosPholipase D and Its Application in Enzymatic Preparation of Phosphatidylserine" International Journal of Molecular Sciences 22, no. 19: 10580. https://doi.org/10.3390/ijms221910580




