Abiotic Stress Triggers the Expression of Genes Involved in Protein Storage Vacuole and Exocyst-Mediated Routes
Abstract
:1. Introduction
2. Results
2.1. Arabidopsis thaliana Germination and Growth Are Affected by Stress
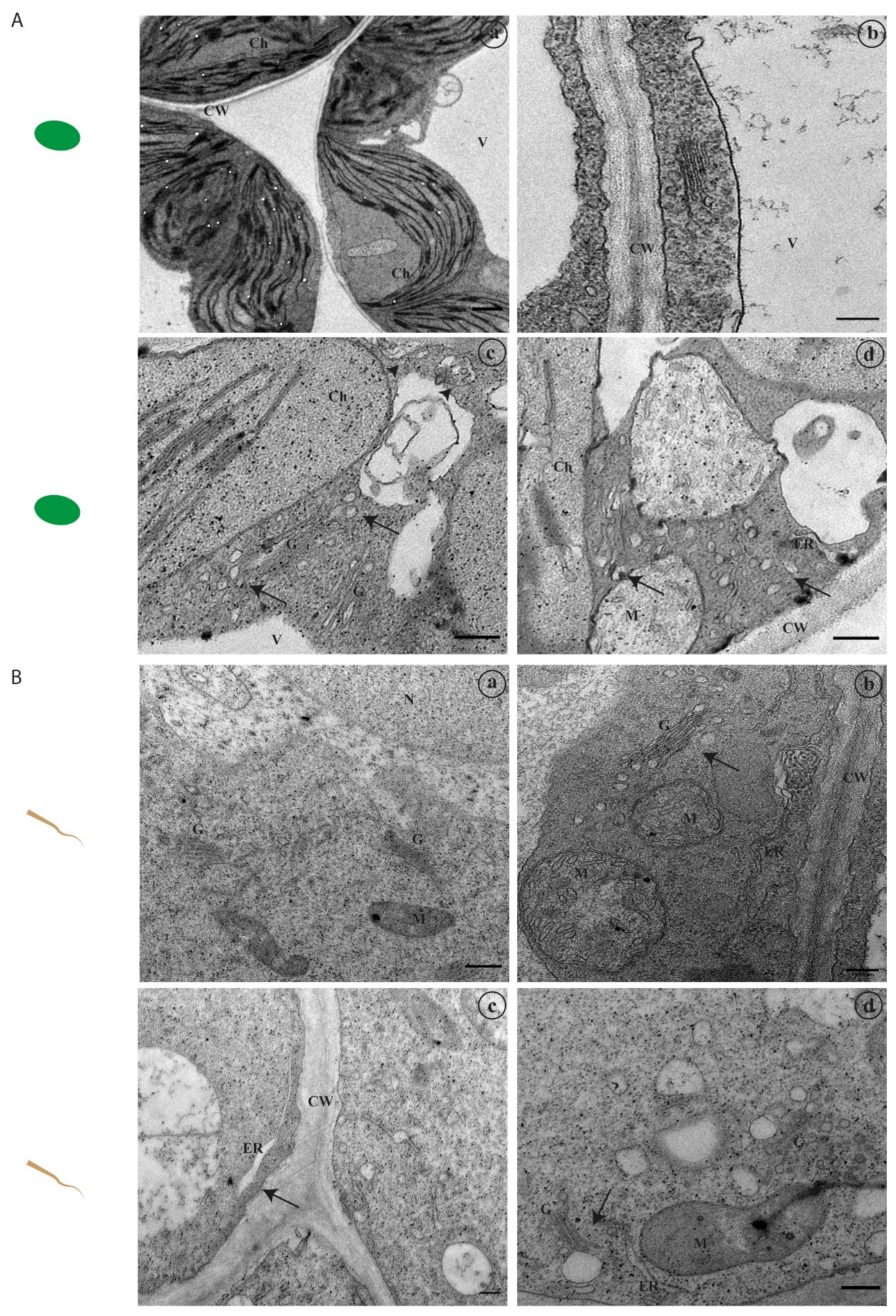
2.2. Abiotic Stress Induces Alterations at the Ultrastructural Level
2.2.1. Ultrastructural Characterisation in Leaf Tissues
2.2.2. Ultrastructural Characterisation of Root Cells
2.3. Endomembrane Trafficking-Related Genes Are Differentially Expressed under Stress
2.3.1. Genes Involved in the Vacuolar Pathway
2.3.2. SNAREs Involved in Plasma Membrane Vesicle Docking and Fusion
2.3.3. Exocyst and Autophagy Pathways Genes
3. Discussion
3.1. Abiotic Stress Induces Endomembrane Rearrangements
3.2. Key Regulators of Endomembrane Trafficking Are Differentially Expressed under Stress
3.2.1. Genes Involved in the Path to the PSV Are Positively Regulated under Stress
3.2.2. SNAREs Involved in Osmotic Regulation at the Plasma Membrane Are Selectively Expressed
3.2.3. Non-Characterised SNAREs, SYP23 and VAMP723, Are Downregulated
3.2.4. Exocytosis and/or Autophagy Mediated by the Exocyst Is Stimulated by Stress
3.3. An Initial Draft of Endomembrane System Response to Stress
4. Materials and Methods
4.1. Biological Material
4.2. Electron Microscopy
4.3. cDNA Preparation
4.4. Gene Selection and Interaction Map
4.5. Quantitative RT-PCR
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Mousavi-Derazmahalleh, M.; Bayer, P.E.; Hane, J.K.; Valliyodan, B.; Nguyen, H.T.; Nelson, M.N.; Erskine, W.; Varshney, R.K.; Papa, R.; Edwards, D. Adapting legume crops to climate change using genomic approaches. Plant Cell Environ. 2019, 42, 6–19. [Google Scholar] [CrossRef] [Green Version]
- Kalinowska, K.; Isono, E. All roads lead to the vacuole—autophagic transport as part of the endomembrane trafficking network in plants. J. Exp. Bot. 2018, 69, 1313–1324. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cushman, J.C.; Bohnert, H.J. Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. 2000, 3, 117–124. [Google Scholar] [CrossRef]
- Moellering, E.R.; Benning, C. Galactoglycerolipid metabolism under stress: A time for remodeling. Trends Plant Sci. 2011, 16, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11. [Google Scholar] [CrossRef]
- Rosquete, M.R.; Davis, D.J.; Drakakaki, G. The plant trans-golgi network: Not just a matter of distinction. Plant Physiol. 2018, 176, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Li, J. Communications between the endoplasmic reticulum and other organelles during abiotic stress response in plants. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Chevalier, A.S.; Chaumont, F. Trafficking of plant plasma membrane aquaporins: Multiple regulation levels and complex sorting signals. Plant Cell Physiol. 2015, 56, 819–829. [Google Scholar] [CrossRef] [Green Version]
- Hachez, C.; Laloux, T.; Reinhardt, H.; Cavez, D.; Degand, H.; Grefen, C.; De Rycke, R.; Inzé, D.; Blatt, M.R.; Russinova, E.; et al. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 2014, 26, 3132–3147. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xu, M.; Gao, C.; Zeng, Y.; Cui, Y.; Shen, W.; Jiang, L. The roles of endomembrane trafficking in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 55–69. [Google Scholar] [CrossRef]
- Ahmad, I.; Devonshire, J.; Mohamed, R.; Schultze, M.; Maathuis, F.J.M. Overexpression of the potassium channel TPK b in small vacuoles confers osmotic and drought tolerance to rice. New Phytol. 2015, 209, 1040–1048. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Pereira, S.; Satiat-Jeunemaitre, B.; Pissarra, J. Cardosin A contains two vacuolar sorting signals using different vacuolar routes in tobacco epidermal cells. Plant J. 2013, 76, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Jürgens, G.; Pimpl, P. Plant membrane trafficking is coming of age. Semin. Cell Dev. Biol. 2018, 80, 83–84. [Google Scholar] [CrossRef]
- Vieira, V.; Peixoto, B.; Costa, M.; Pereira, S.; Pissarra, J.; Pereira, C. N-linked glycosylation modulates golgi-independent vacuolar sorting mediated by the plant specific insert. Plants 2019, 8, 312. [Google Scholar] [CrossRef] [Green Version]
- Pompa, A.; De Marchis, F.; Pallotta, M.T.; Benitez-Alfonso, Y.; Jones, A.; Schipper, K.; Moreau, K.; Žárský, V.; Di Sansebastiano, G.P.; Bellucci, M. Unconventional transport routes of soluble and membrane proteins and their role in developmental biology. Int. J. Mol. Sci. 2017, 18, 703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Sansebastiano, G.P.; Barozzi, F.; Piro, G.; Denecke, J.; de Marcos Lousa, C. Trafficking routes to the plant vacuole: Connecting alternative and classical pathways. J. Exp. Bot. 2018, 69, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.; Brennan, A.; Wei, W.; Guo, G.; Lindsey, K. Vesicle transport in plants: A revised phylogeny of SNARE proteins. Evol. Bioinform. 2020, 16. [Google Scholar] [CrossRef]
- Moshelion, M.; Altman, A. Current challenges and future perspectives of plant and agricultural biotechnology. Trends Biotechnol. 2015, 33, 337–342. [Google Scholar] [CrossRef]
- Xiang, L.; Etxeberria, E.; Van den Ende, W. Vacuolar protein sorting mechanisms in plants. FEBS J. 2013, 280, 979–993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Synek, L.; Schlager, N.; Eliáš, M.; Quentin, M.; Hauser, M.-T.; Žárský, V. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 2006, 48, 54–72. [Google Scholar] [CrossRef] [Green Version]
- Kwon, C.; Bednarek, P.; Schulze-Lefert, P. Secretory pathways in plant immune responses. Plant Physiol. 2008, 147, 1575–1583. [Google Scholar] [CrossRef] [Green Version]
- Kwon, C.; Neu, C.; Pajonk, S.; Yun, H.S.; Lipka, U.; Humphry, M.; Bau, S.; Straus, M.R.; Kwaaitaal, M.; Rampelt, H.; et al. Co-option of a default secretory pathway for plant immune responses. Nature 2008, 451, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Wang, J.; Ding, Y.; Lo, S.W.; Gouzerh, G.; Neuhaus, J.-M.; Jiang, L. The Rice RMR1 associates with a distinct prevacuolar compartment for the protein storage vacuole pathway. Mol. Plant 2011, 4, 854–868. [Google Scholar] [CrossRef] [Green Version]
- Sanmartín, M.; Ordóñez, A.; Sohn, E.J.; Robert, S.; Sánchez-Serrano, J.J.; Surpin, M.A.; Rojo, E. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 3645–3650. [Google Scholar] [CrossRef] [Green Version]
- Eisenach, C.; Chen, Z.-H.; Grefen, C.; Blatt, M.R. The trafficking protein SYP121 of Arabidopsis connects programmed stomatal closure and K+ channel activity with vegetative growth. Plant J. 2011, 69, 241–251. [Google Scholar] [CrossRef]
- Shirakawa, M.; Ueda, H.; Shimada, T.; Koumoto, Y.; Shimada, T.L.; Kondo, M.; Takahashi, T.; Okuyama, Y.; Nishimura, M.; Hara-Nishimura, I. Arabidopsis Qa-SNARE SYP2 proteins localized to different subcellular regions function redundantly in vacuolar protein sorting and plant development. Plant J. 2010, 64, 924–935. [Google Scholar] [CrossRef]
- De Benedictis, M.; Bleve, G.; Faraco, M.; Stigliano, E.; Grieco, F.; Piro, G.; Dalessandro, G.; Di Sansebastiano, G.-P. AtSYP51/52 functions diverge in the post-golgi traffic and differently affect vacuolar sorting. Mol. Plant 2013, 6, 916–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Karnik, R.; Wang, Y.; Wallmeroth, N.; Blatt, M.R.; Grefen, C. The Arabidopsis R-SNARE VAMP721 Interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 2015, 27, 1697–1717. [Google Scholar] [CrossRef] [Green Version]
- Zouhar, J.; Rojo, E.; Bassham, D.C. AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 2009, 149, 1668–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zouhar, J.; Muñoz, A.; Rojo, E. Functional specialization within the vacuolar sorting receptor family: VSR1, VSR3 and VSR4 sort vacuolar storage cargo in seeds and vegetative tissues. Plant J. 2010, 64, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Surpin, M.; Zheng, H.; Morita, M.; Saito, C.; Avila, E.; Blakeslee, J.J.; Bandyopadhyay, A.; Kovaleva, V.; Carter, D.; Murphy, A.; et al. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 2003, 15, 2885–2899. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Jang, M.; Song, K.; Kang, H.; Lee, M.H.; Lee, D.W.; Zouhar, J.; Rojo, E.; Sohn, E.J.; Hwang, I. Functional identification of sorting receptors involved in trafficking of soluble lytic vacuolar proteins in vegetative cells of arabidopsis. Plant Physiol. 2013, 161, 121–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucl. Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Memon, A.R.; Ajktoparakligil, D.; Özdemir, A.; Vertii, A.; Zitka, O.; Krystofova, O.; Hynek, D.; Sobrova, P.; Kaiser, J.; Sochor, J.; et al. Plant defense response. Trop. Agric. Res. 2015, 25, 27–54. [Google Scholar] [CrossRef]
- Zhang, Y.-B.; Yang, S.-L.; Dao, J.-M.; Deng, J.; Shahzad, A.N.; Fan, X.; Li, R.-D.; Quan, Y.-J.; Bukhari, S.A.H.; Zeng, Z.-H. Drought-induced alterations in photosynthetic, ultrastructural and biochemical traits of contrasting sugarcane genotypes. PLoS ONE 2020, 15, e0235845. [Google Scholar] [CrossRef]
- Lamers, J.; van der Meer, T.; Testerink, C. How plants sense and respond to stressful environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [Green Version]
- Borgo, L.; Marur, C.J.; Vieira, L.G.E. Efeitos do alto acúmulo de prolina na ultraestrutura de cloroplastos e mitocôndrias e no ajustamento osmótico de plantas de tabaco. Acta Sci. Agron. 2015, 37, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Venzhik, Y.V.; Shchyogolev, S.Y.; Dykman, L.A. Ultrastructural reorganization of chloroplasts during plant adaptation to abiotic stress factors. Russ. J. Plant Physiol. 2019, 66, 850–863. [Google Scholar] [CrossRef]
- Zechmann, B. Ultrastructure of plastids serves as reliable abiotic and biotic stress marker. PLoS ONE 2019, 14, e0214811. [Google Scholar] [CrossRef]
- Carter, C.J.; Bednarek, S.Y.; Raikhel, N.V. Membrane trafficking in plants: New discoveries and approaches. Curr. Opin. Plant Biol. 2004, 7, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Pereira, S.; Pissarra, J. Delivering of proteins to the plant vacuole—An update. Int. J. Mol. Sci. 2014, 15, 7611–7623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, T.; Fuji, K.; Ichino, T.; Teh, O.; Koumoto, Y.; Hara-nishimura, I. GREEN FLUORESCENT SEED, to Evaluate Vacuolar Trafficking in Arabidopsis Seeds. Plant Vacuolar Trafficking—Methods and Protocols 2018, 1789, 1–7. [Google Scholar] [CrossRef]
- Zhu, D.; Zhang, M.; Gao, C.; Shen, J. Protein trafficking in plant cells: Tools and markers. Sci. China Life Sci. 2020, 63, 343–363. [Google Scholar] [CrossRef] [Green Version]
- Rosquete, M.R.; Drakakaki, G. Plant TGN in the stress response: A compartmentalized overview. Curr. Opin. Plant Biol. 2018, 46, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-X.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- De Caroli, M.; Furini, A.; DalCorso, G.; Rojas, M.; Di Sansebastiano, G.-P. Endomembrane reorganization induced by heavy metals. Plants 2020, 9, 482. [Google Scholar] [CrossRef] [Green Version]
- Stigliano, E.; Di Sansebastiano, G.-P.; Neuhaus, J.-M. Contribution of chitinase A’s C-terminal vacuolar sorting determinant to the study of soluble protein compartmentation. Int. J. Mol. Sci. 2014, 15, 11030–11039. [Google Scholar] [CrossRef] [Green Version]
- Di Sansebastiano, G.-P.; Faraco, M.; Zouhar, J.; Dalessandro, G. The study of plant SNAREs specificityin vivo. Plant Biosyst. 2009, 143, 621–629. [Google Scholar] [CrossRef]
- Barozzi, F.; Papadia, P.; Stefano, G.; Renna, L.; Brandizzi, F.; Migoni, D.; Fanizzi, F.P.; Piro, G.; Di Sansebastiano, G.-P. Variation in Membrane Trafficking Linked to SNARE AtSYP51 Interaction with aquaporin NIP1;1. Front. Plant Sci. 2019, 9, 1949. [Google Scholar] [CrossRef] [Green Version]
- Ebine, K.; Okatani, Y.; Uemura, T.; Goh, T.; Shoda, K.; Niihama, M.; Morita, M.; Spitzer, C.; Otegui, M.; Nakano, A.; et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of arabidopsis thaliana. Plant Cell 2008, 20, 3006–3021. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Rogers, J.C.; Jiang, L. Plant RMR proteins: Unique vacuolar sorting receptors that couple ligand sorting with membrane internalization. FEBS J. 2011, 278, 59–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besserer, A.; Burnotte, E.; Bienert, G.; Chevalier, A.S.; Errachid, A.; Grefen, C.; Blatt, M.R.; Chaumont, F. Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121. Plant Cell 2012, 24, 3463–3481. [Google Scholar] [CrossRef] [Green Version]
- Kulich, I.; Žárský, V. Autophagy-related direct membrane import from ER/Cytoplasm into the vacuole or apoplast: A hidden gateway also for secondary metabolites and phytohormones? Int. J. Mol. Sci. 2014, 15, 7462–7474. [Google Scholar] [CrossRef] [Green Version]
- Žárský, V.; Kulich, I.; Fendrych, M.; Pecenkova, T. Exocyst complexes multiple functions in plant cells secretory pathways. Curr. Opin. Plant Biol. 2013, 16, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Pečenková, T.; Potocká, A.; Potocký, M.; Ortmannová, J.; Drs, M.; Janková Drdová, E.; Pejchar, P.; Synek, L.; Soukupová, H.; Žárský, V.; et al. Redundant and diversified roles among selected arabidopsis thaliana EXO70 paralogs during biotic stress responses. Front. Plant Sci. 2020, 11, 960. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]






| Gene | Identifier | Role and Localisation | References |
|---|---|---|---|
| AtVSR2 | AT1G71980.1 | Involved in the trafficking of vacuolar proteins. May function as a sorting receptor for protein trafficking to the protein storage vacuole (PSV). | [20] |
| AtATG8 | AT1G62040.2 | Ubiquitin-like modifier involved in autophagosome formation. May mediate the delivery of the autophagosomes to the vacuole via the microtubule cytoskeleton; belongs to the ATG8 family | [21] |
| AtEXO70 | AT5G61010.1 | Essential component for the formation and the recruitment of exocyst subunits to the exocyst-positive organelle (EXPO), a secreted double membrane structure also called extracellular exosome. | [22] |
| AtVAMP722 | AT2G33120.2 | Involved in the targeting and/or fusion of transport vesicles to their target membrane. Associated with biotic stress response. Defines a complex with SYP121. | [23,24] |
| AtRMR1 | AT5G66160.1 | Involved in the trafficking of vacuolar proteins. Functions probably as a sorting receptor for protein trafficking to the protein storage vacuole (PSV) by binding the C-terminal vacuolar sorting determinant (VSD) of vacuolar-sorted proteins | [25] |
| AtVTI11 | AT5G39510.1 | May function as a v-SNARE responsible for targeting AtELP-containing vesicles from the trans-Golgi network (TGN) to the prevacuolar compartment (PVC). Promotes the formation of vacuolar membrane ‘bulbs’. | [26] |
| AtSYP121 | AT3G11820.1 | Encodes a syntaxin localized at the plasma membrane. Functions in positioning anchoring of the KAT1 K+ channel protein at the plasma membrane. Additionally, functions in non-host resistance against barley powdery mildew. Forms a complex with VTI11 and SYP51. | [10,27] |
| AtSYP23 | AT4G17730.2 | Cytosolic syntaxin with no transmembrane domain. May function in the docking or fusion of transport vesicles with the prevacuolar membrane. | [28] |
| AtSYP51 | AT1G16240.1 | Involved in the transport of chitinase to the PSV. Interacts with VTI12. | [29] |
| AtSYP52 | AT1G79590.2 | Involved in the route to the Lytic vacuole. Forms a complex with VTI11. | [29] |
| AtSYP61 | AT1G28490.1 | Vesicle trafficking protein that functions in the secretory pathway. Involved in osmotic stress tolerance and in abscisic acid (ABA) regulation of stomatal responses. Forms a complex with VTI12. | [10] |
| AtSYP22 | AT5G46860.1 | Syntaxin-related protein required for vacuolar assembly. Localized in the vacuolar membranes. | [28] |
| AtVAMP723 | AT2G33110.1 | Localized at the endoplasmic reticulum. Involved in the targeting and/or fusion of transport vesicles to their target membrane. | [30] |
| AtVPS45 | AT1G77140.1 | A peripheral membrane protein that associates with microsomal membranes, likely to function in the transport of proteins to the vacuole. Has been described as a positive regulator for PSV route. Forms a complex with VTI12 and SYP61. | [31] |
| AtVSR1 | AT3G52850.1 | Vacuolar-sorting receptor (VSR) involved in clathrin-coated vesicles sorting from Golgi apparatus to vacuoles. Required for the sorting of 12S globulin, 2S albumin and maybe other seed storage proteins to protein storage vacuoles (PSVs) in seeds. | [32] |
| AtVTI12 | AT1G26670.1 | Normally localizes to the trans-golgi network and plasma membrane. Involved in protein trafficking to protein storage vacuoles | [26,33] |
| AtVSR2 | AT2G14720.1 | Vacuolar sorting receptor (VSR) involved in clathrin-coated vesicles sorting from Golgi apparatus to vacuoles; belongs to the VSR (BP-80) family. | [34] |
| Stress Type | Nomenclature | Additive | Concentration |
|---|---|---|---|
| Control | Control | - | - |
| Saline | S1 | Sodium Chloride | 50 mM |
| S2 | Sodium Chloride | 100 mM | |
| Hydric | H1 | Mannitol | 50 mM |
| H2 | Mannitol | 100 mM | |
| Oxidative | Ox | Hydrogen Peroxide | 0.5 mM |
| Heavy Metal | Zn | Zinc Sulphate | 150 µM |
| Genes | Primer Forward | Primer Reverse |
|---|---|---|
| AtSYP23 | GCAGCGTGCCCTTCTTGTGG | TCCTTGGGCAGTTGCAGCGTA |
| AtSYP121 | TCCTCCGATCGAACCAGGACCTC | TTCTCGCCGGTGACGGTGAA |
| AtVAMP723 | CCCGTGGTGTGATATGTGAG | CCACAAACCGAGAGGATGAT |
| AtVTI12 | GCAATGTCCGTGGAGAGGCTTGA | TGCGCATGAAGGAGGGTTTGG |
| AtBP-80 | GGGAGCGGCGCAGATTCTTG | GCCGGTTTCATTCGCCACCTT |
| AtRMR1 | GCGAGGGAGGCACACCAGGA | TTTCCCCGGCCTTGTGGTGA |
| AtEXO70 | TCCCCGATGAAACAGGCTCGTC | GCCTCCATGAAAGGGGCGTGT |
| AtUBC91 | TCACAATTTCCAAGGTGCTGC | TCATCTGGGTTTGGATCCGT |
| AtSAND-11 | AACTCTATGCAGCATTTGATCCACT | TGATTGCATATCTTTATCGCCATC |
| AtVTI11 | GATTTTCCGACTCCGACGTA | CAAACGCGTCACTCATTGTT |
| AtSYP52 | ATGTGGTGGCAACTTGTGAA | CTTTGCCTCACAGACACGAA |
| AtVPS45 | CAGTTGTCGATCCCTCTGGT | TTTCAACGCCAGAAACACTG |
| AtSYP61 | TTGAAAAACGGAGGAGATGG | TTCACTTGCATGACCTGCTC |
| AtVAMP722 | CAATTTGTGGGGGATTCAAC | GATCTTGGGAAGCACAGAGC |
| AtVSR1 | GATGTGGACGAGTGCAAAGA | GCTCACGCATGTAAAGCAAA |
| AtRMR2 | CTTCCTTGGGCTCATTACCA | GGTGATCGACGGTTAGAGGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neves, J.; Sampaio, M.; Séneca, A.; Pereira, S.; Pissarra, J.; Pereira, C. Abiotic Stress Triggers the Expression of Genes Involved in Protein Storage Vacuole and Exocyst-Mediated Routes. Int. J. Mol. Sci. 2021, 22, 10644. https://doi.org/10.3390/ijms221910644
Neves J, Sampaio M, Séneca A, Pereira S, Pissarra J, Pereira C. Abiotic Stress Triggers the Expression of Genes Involved in Protein Storage Vacuole and Exocyst-Mediated Routes. International Journal of Molecular Sciences. 2021; 22(19):10644. https://doi.org/10.3390/ijms221910644
Chicago/Turabian StyleNeves, João, Miguel Sampaio, Ana Séneca, Susana Pereira, José Pissarra, and Cláudia Pereira. 2021. "Abiotic Stress Triggers the Expression of Genes Involved in Protein Storage Vacuole and Exocyst-Mediated Routes" International Journal of Molecular Sciences 22, no. 19: 10644. https://doi.org/10.3390/ijms221910644






