MicroRNAs in Lupus Nephritis–Role in Disease Pathogenesis and Clinical Applications
Abstract
1. Introduction
2. MiRs in the Circulation and Peripheral Blood Mononuclear Cells (PBMCs)
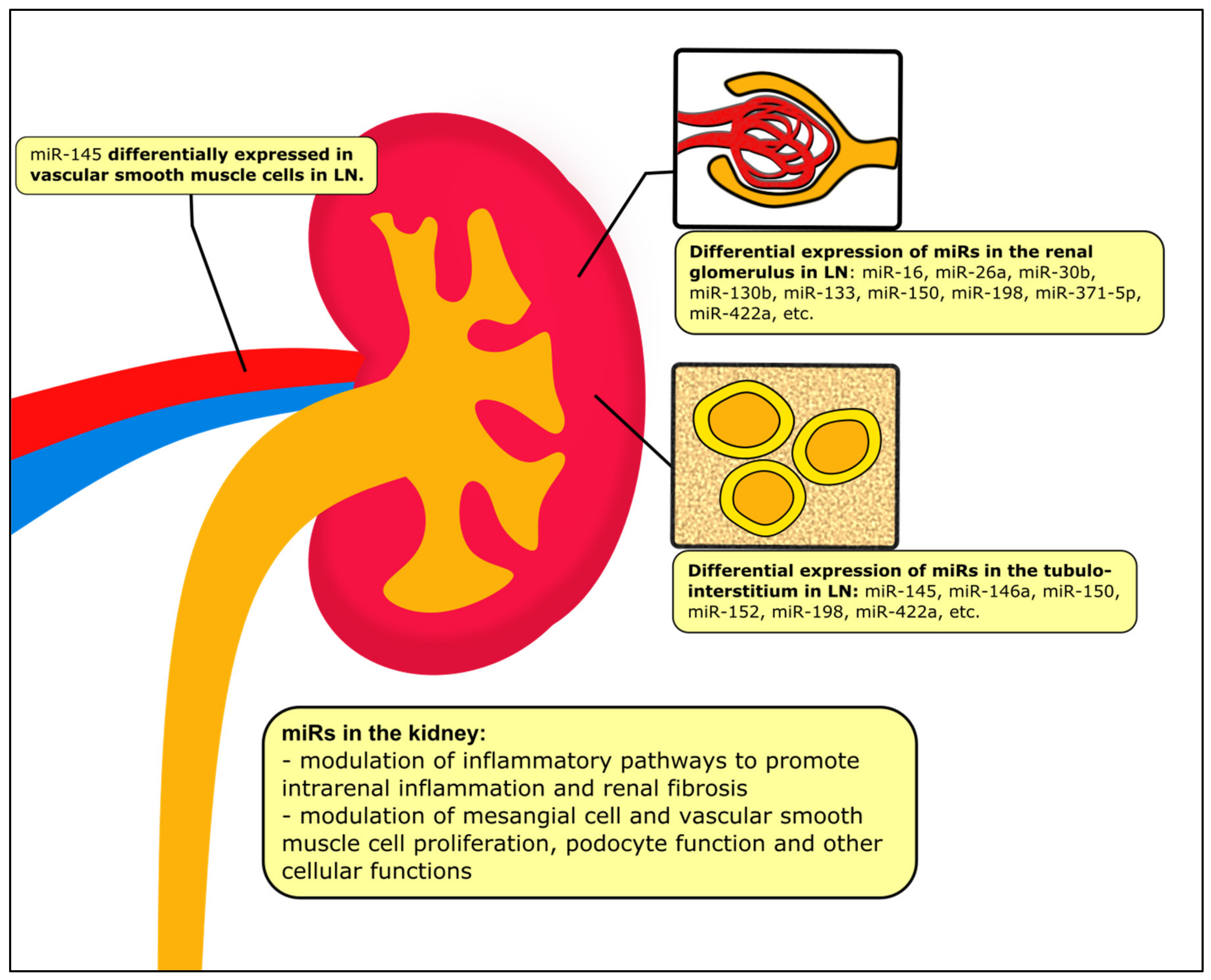
3. MiRs in the Kidney Tissue
4. MiRs in the Urine
5. MiRs and the Treatment of LN
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yap, D.Y.; Tang, C.S.; Ma, M.K.; Lam, M.F.; Chan, T.M. Survival analysis and causes of mortality in patients with lupus nephritis. Nephrol. Dial. Transplant. 2012, 27, 3248–3254. [Google Scholar] [CrossRef] [PubMed]
- Yap, D.Y.; Yung, S.; Chan, T.M. Lupus nephritis: An update on treatments and pathogenesis. Nephrology 2018, 23 (Suppl. 4), 80–83. [Google Scholar] [CrossRef] [PubMed]
- Anders, H.J.; Rovin, B. A pathophysiology-based approach to the diagnosis and treatment of lupus nephritis. Kidney Int. 2016, 90, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, C.M. Epigenetics in SLE. Curr. Rheumatol. Rep. 2017, 19, 58. [Google Scholar] [CrossRef] [PubMed]
- Dziedziejko, V.; Taheri, M. Exploring the Role of Non-Coding RNAs in the Pathophysiology of Systemic Lupus Erythematosus. Int. J. Mol. Sci. 2020, 10, 5050. [Google Scholar] [CrossRef]
- Tsai, C.Y. Aberrant Non-Coding RNA Expression in Patients with Systemic Lupus Erythematosus: Consequences for Immune Dysfunctions and Tissue Damage. Biomolecules 2020, 10, 937. [Google Scholar] [CrossRef]
- Honarpisheh, M.; Köhler, P.; von Rauchhaupt, E.; Lech, M. The Involvement of MicroRNAs in Modulation of Innate and Adaptive Immunity in Systemic Lupus Erythematosus and Lupus Nephritis. J. Immunol. Res. 2018, 2018, 4126106. [Google Scholar] [CrossRef]
- Chafin, C.B.; Reilly, C.M. MicroRNAs implicated in the immunopathogenesis of lupus nephritis. Clin. Dev. Immunol. 2013, 2013, 430239. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Gunaratne, P.H.; Coarfa, C.; Soibam, B.; Tandon, A. miRNA data analysis: Next-gen sequencing. Methods Mol. Biol. 2012, 822, 273–288. [Google Scholar] [CrossRef]
- Nielsen, B.S. MicroRNA in situ hybridization. Methods Mol. Biol. 2012, 822, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Carlsen, A.L.; Schetter, A.J.; Nielsen, C.T.; Lood, C.; Knudsen, S.; Voss, A.; Harris, C.C.; Hellmark, T.; Segelmark, M.; Jacobsen, S.; et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013, 65, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Te, J.L.; Dozmorov, I.M.; Guthridge, J.M.; Nguyen, K.L.; Cavett, J.W.; Kelly, J.A.; Bruner, G.R.; Harley, J.B.; Ojwang, J.O. Identification of unique microRNA signature associated with lupus nephritis. PLoS ONE 2010, 5, e10344. [Google Scholar] [CrossRef]
- Navarro-Quiroz, E.; Pacheco-Lugo, L.; Navarro-Quiroz, R.; Lorenzi, H.; España-Puccini, P.; Díaz-Olmos, Y.; Almendrales, L.; Olave, V.; Gonzalez-Torres, H.; Diaz-Perez, A.; et al. Profiling analysis of circulating microRNA in peripheral blood of patients with class IV lupus nephritis. PLoS ONE 2017, 12, e0187973. [Google Scholar] [CrossRef]
- Zhu, Y.; Xue, Z.; Di, L. Regulation of MiR-146a and TRAF6 in the Diagnose of Lupus Nephritis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017, 23, 2550–2557. [Google Scholar] [CrossRef]
- Tang, Y.; Luo, X.; Cui, H.; Ni, X.; Yuan, M.; Guo, Y.; Huang, X.; Zhou, H.; de Vries, N.; Tak, P.P.; et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009, 60, 1065–1075. [Google Scholar] [CrossRef]
- Fu, H.X.; Fan, X.P.; Li, M.; Liu, M.J.; Sun, Q.L. MiR-146a relieves kidney injury in mice with systemic lupus erythematosus through regulating NF-κB pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7024–7032. [Google Scholar] [CrossRef]
- Zununi Vahed, S.; Nakhjavani, M.; Etemadi, J.; Jamshidi, H.; Jadidian, N.; Pourlak, T.; Abediazar, S. Altered levels of immune-regulatory microRNAs in plasma samples of patients with lupus nephritis. BioImpacts BI 2018, 8, 177–183. [Google Scholar] [CrossRef]
- Tawfik, N.A.; El-Dydamoni, O.A.; Abozaid, S.Y.; Ebrahem, E.E.; Abd El Rahim, M. Serum miRNA-146a and miRNA-155 as Novel Biomarkers in Lupus Nephritis Activity with Systemic Lupus Erythematosus. Am. J. Biochem. 2019, 9, 21–34. [Google Scholar]
- Su, Y.J.; Lin, I.C.; Wang, L.; Lu, C.H.; Huang, Y.L.; Kuo, H.C. Next generation sequencing identifies miRNA-based biomarker panel for lupus nephritis. Oncotarget 2018, 9, 27911–27919. [Google Scholar] [CrossRef]
- Hashad, D.I.; Abdelmagid, M.H.; Elsherif, S.H. microRNA146a expression in lupus patients with and without renal complications. J. Clin. Lab. Anal. 2012, 26, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Tam, L.S.; Li, E.K.; Kwan, B.C.; Chow, K.M.; Luk, C.C.; Li, P.K.; Szeto, C.C. Serum and urinary cell-free MiR-146a and MiR-155 in patients with systemic lupus erythematosus. Int. J. Rheum. Dis. 2010, 37, 2516–2522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, X.; Si, F. MicroRNA-124 represents a novel diagnostic marker in human lupus nephritis and plays an inhibitory effect on the growth and inflammation of renal mesangial cells by targeting TRAF6. Int. J. Clin. Exp. Pathol. 2019, 12, 1578–1588. [Google Scholar]
- Tu, Y.; Guo, R.; Li, J.; Wang, S.; Leng, L.; Deng, J.; Bucala, R.; Lu, L. MiRNA Regulation of MIF in SLE and Attenuation of Murine Lupus Nephritis With miR-654. Front. Immunol. 2019, 10, 2229. [Google Scholar] [CrossRef]
- Tan, L.; Zhao, M.; Wu, H.; Zhang, Y.; Tong, X.; Gao, L.; Zhou, L.; Lu, Q.; Zeng, J. Downregulated Serum Exosomal miR-451a Expression Correlates With Renal Damage and Its Intercellular Communication Role in Systemic Lupus Erythematosus. Front. Immunol. 2021, 12, 630112. [Google Scholar] [CrossRef]
- Khoshmirsafa, M.; Kianmehr, N.; Falak, R.; Mowla, S.J.; Seif, F.; Mirzaei, B.; Valizadeh, M.; Shekarabi, M. Elevated expression of miR-21 and miR-155 in peripheral blood mononuclear cells as potential biomarkers for lupus nephritis. Int. J. Rheum. Dis. 2019, 22, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Nakhjavani, M.; Etemadi, J.; Pourlak, T.; Mirhosaini, Z.; Zununi Vahed, S.; Abediazar, S. Plasma levels of miR-21, miR-150, miR-423 in patients with lupus nephritis. Iran. J. Kidney Dis. 2019, 13, 198–206. [Google Scholar] [PubMed]
- Pan, W.; Zhu, S.; Yuan, M.; Cui, H.; Wang, L.; Luo, X.; Li, J.; Zhou, H.; Tang, Y.; Shen, N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 2010, 184, 6773–6781. [Google Scholar] [CrossRef] [PubMed]
- You, G.; Cao, H.; Yan, L.; He, P.; Wang, Y.; Liu, B.; Shao, F. MicroRNA-10a-3p mediates Th17/Treg cell balance and improves renal injury by inhibiting REG3A in lupus nephritis. Int. Immunopharmacol. 2020, 88, 106891. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; Lai, N.S.; Chen, H.C.; Yu, H.C.; Huang, K.Y.; Tung, C.H.; Huang, H.B.; Yu, C.L. Decreased microRNA(miR)-145 and increased miR-224 expression in T cells from patients with systemic lupus erythematosus involved in lupus immunopathogenesis. Clin. Exp. Immunol. 2013, 171, 91–99. [Google Scholar] [CrossRef]
- Zeng, L.; Wu, J.L.; Liu, L.M.; Jiang, J.Q.; Wu, H.J.; Zhao, M.; Lu, Q.J. Serum miRNA-371b-5p and miRNA-5100 act as biomarkers for systemic lupus erythematosus. Clin. Immunol. 2018, 196, 103–109. [Google Scholar] [CrossRef]
- Hiramatsu-Asano, S.; Sunahori-Watanabe, K.; Zeggar, S.; Katsuyama, E.; Mukai, T.; Morita, Y.; Wada, J. Deletion of Mir223 Exacerbates Lupus Nephritis by Targeting S1pr1 in Fas(lpr/lpr) Mice. Immun. Inflamm. Dis. 2020, 11, 616141. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, X.; Ye, L.; Guo, G.; Li, X.; Chen, C.; Sun, L.; Li, B.; Chen, N.; Xue, X. B Cell-Related Circulating MicroRNAs With the Potential Value of Biomarkers in the Differential Diagnosis, and Distinguishment Between the Disease Activity and Lupus Nephritis for Systemic Lupus Erythematosus. Front. Immunol. 2018, 9, 1473. [Google Scholar] [CrossRef]
- Duroux-Richard, I.; Cuenca, J.; Ponsolles, C.; Piñeiro, A.B.; Gonzalez, F.; Roubert, C.; Areny, R.; Chea, R.; Pefaur, J.; Pers, Y.M.; et al. MicroRNA Profiling of B Cell Subsets from Systemic Lupus Erythematosus Patients Reveals Promising Novel Biomarkers. Int. J. Mol. Sci. 2015, 16, 16953–16965. [Google Scholar] [CrossRef]
- Lin, L.J.; Mai, L.J.; Chen, G.; Zhao, E.N.; Xue, M.; Su, X.D. Expression and diagnostic value of plasma miR-145 and miR-183 in children with lupus nephritis. Zhongguo Dang Dai Er Ke Za Zhi Chin. J. Contemp. Pediatrics 2020, 22, 632–637. [Google Scholar] [CrossRef]
- Ren, D.; Liu, F.; Dong, G.; You, M.; Ji, J.; Huang, Y.; Hou, Y.; Fan, H. Activation of TLR7 increases CCND3 expression via the downregulation of miR-15b in B cells of systemic lupus erythematosus. Cell. Mol. Immunol. 2016, 13, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Yap, D.Y.H.; Yung, S.; Lee, P.; Yam, I.Y.L.; Tam, C.; Tang, C.; Chan, T.M. B Cell Subsets and Cellular Signatures and Disease Relapse in Lupus Nephritis. Front. Immunol. 2020, 11, 1732. [Google Scholar] [CrossRef] [PubMed]
- Segerberg, F.; Lundtoft, C.; Reid, S.; Hjorton, K.; Leonard, D.; Nordmark, G.; Carlsten, M.; Hagberg, N. Autoantibodies to Killer Cell Immunoglobulin-Like Receptors in Patients With Systemic Lupus Erythematosus Induce Natural Killer Cell Hyporesponsiveness. Front. Immunol. 2019, 10, 2164. [Google Scholar] [CrossRef] [PubMed]
- Pesce, S.; Squillario, M.; Greppi, M.; Loiacono, F.; Moretta, L.; Moretta, A.; Sivori, S.; Castagnola, P.; Barla, A.; Candiani, S.; et al. New miRNA Signature Heralds Human NK Cell Subsets at Different Maturation Steps: Involvement of miR-146a-5p in the Regulation of KIR Expression. Front. Immunol. 2018, 9, 2360. [Google Scholar] [CrossRef]
- Bezman, N.A.; Chakraborty, T.; Bender, T.; Lanier, L.L. miR-150 regulates the development of NK and iNKT cells. J. Exp. Med. 2011, 208, 2717–2731. [Google Scholar] [CrossRef]
- Wang, W.; Mou, S.; Wang, L.; Zhang, M.; Shao, X.; Fang, W.; Lu, R.; Qi, C.; Fan, Z.; Cao, Q.; et al. Up-regulation of Serum MiR-130b-3p Level is Associated with Renal Damage in Early Lupus Nephritis. Sci. Rep. 2015, 5, 12644. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, G.; Zhang, X.; Dou, Y.; Dong, Y.; Liu, D.; Xiao, J.; Zhao, Z. Inhibition of microRNA-182-5p contributes to attenuation of lupus nephritis via Foxo1 signaling. Exp. Cell Res. 2018, 373, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Quiroz, E.; Pacheco-Lugo, L.; Lorenzi, H.; Díaz-Olmos, Y.; Almendrales, L.; Rico, E.; Navarro, R.; España-Puccini, P.; Iglesias, A.; Egea, E.; et al. High-Throughput Sequencing Reveals Circulating miRNAs as Potential Biomarkers of Kidney Damage in Patients with Systemic Lupus Erythematosus. PLoS ONE 2016, 11, e0166202. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhao, L.; Li, Y. Comprehensive bioinformatics analysis of mRNA expression profiles and identification of a miRNA-mRNA network associated with lupus nephritis. Lupus 2020, 29, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Tam, L.S.; Kwan, B.C.; Li, E.K.; Chow, K.M.; Luk, C.C.; Li, P.K.; Szeto, C.C. Expression of miR-146a and miR-155 in the urinary sediment of systemic lupus erythematosus. Clin. Rheumatol. 2012, 31, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Kwan, B.C.; Lai, F.M.; Tam, L.S.; Li, E.K.; Chow, K.M.; Wang, G.; Li, P.K.; Szeto, C.C. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology 2012, 17, 346–351. [Google Scholar] [CrossRef]
- Duttagupta, R.; Jiang, R.; Gollub, J.; Getts, R.C.; Jones, K.W. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS ONE 2011, 6, e20769. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Neal, C.S.; Michael, M.Z.; Pimlott, L.K.; Yong, T.Y.; Li, J.Y.; Gleadle, J.M. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol. Dial. Transplant. 2011, 26, 3794–3802. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Atarod, S.; Smith, H.; Dickinson, A.; Wang, X.N. MicroRNA levels quantified in whole blood varies from PBMCs. F1000Research 2014, 3, 183. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Sui, W.; Lan, H.; Yan, Q.; Huang, H.; Huang, Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol. Int. 2009, 29, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Krasoudaki, E.; Banos, A.; Stagakis, E.; Loupasakis, K.; Drakos, E.; Sinatkas, V.; Zampoulaki, A.; Papagianni, A.; Iliopoulos, D.; Boumpas, D.T.; et al. Micro-RNA analysis of renal biopsies in human lupus nephritis demonstrates up-regulated miR-422a driving reduction of kallikrein-related peptidase 4. Nephrol. Dial. Transplant. 2016, 31, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- Malakoutian, T.; Hajian, S.; Ebrahimi, A.; Kamali, K. Assessment of microRNA profile of kidney biopsies of patients with lupus nephritis. J. Nephropathol. 2017, 6, 333–337. [Google Scholar] [CrossRef][Green Version]
- Chan, T.M.; Leung, J.K.; Ho, S.K.; Yung, S. Mesangial cell-binding anti-DNA antibodies in patients with systemic lupus erythematosus. J. Am. Soc. Nephrol. 2002, 13, 1219–1229. [Google Scholar] [CrossRef]
- Yung, S.; Cheung, K.F.; Zhang, Q.; Chan, T.M. Anti-dsDNA antibodies bind to mesangial annexin II in lupus nephritis. J. Am. Soc. Nephrol. 2010, 21, 1912–1927. [Google Scholar] [CrossRef] [PubMed]
- Yung, S.; Ng, C.Y.; Au, K.Y.; Cheung, K.F.; Zhang, Q.; Zhang, C.; Yap, D.Y.; Chau, M.K.; Chan, T.M. Binding of anti-dsDNA antibodies to proximal tubular epithelial cells contributes to renal tubulointerstitial inflammation. F1000Research 2017, 131, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Yung, S.; Tsang, R.C.; Leung, J.K.; Chan, T.M. Increased mesangial cell hyaluronan expression in lupus nephritis is mediated by anti-DNA antibody-induced IL-1beta. Kidney Int. 2006, 69, 272–280. [Google Scholar] [CrossRef]
- Yung, S.; Tsang, R.C.; Sun, Y.; Leung, J.K.; Chan, T.M. Effect of human anti-DNA antibodies on proximal renal tubular epithelial cell cytokine expression: Implications on tubulointerstitial inflammation in lupus nephritis. J. Am. Soc. Nephrol. 2005, 16, 3281–3294. [Google Scholar] [CrossRef]
- Yung, S.; Yap, D.Y.; Chan, T.M. Recent advances in the understanding of renal inflammation and fibrosis in lupus nephritis. F1000Research 2017, 6, 874. [Google Scholar] [CrossRef] [PubMed]
- Yung, S.; Zhang, Q.; Chau, M.K.; Chan, T.M. Distinct effects of mycophenolate mofetil and cyclophosphamide on renal fibrosis in NZBWF1/J mice. Autoimmunity 2015, 48, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Yung, S.; Zhang, Q.; Zhang, C.Z.; Chan, K.W.; Lui, S.L.; Chan, T.M. Anti-DNA antibody induction of protein kinase C phosphorylation and fibronectin synthesis in human and murine lupus and the effect of mycophenolic acid. Arthritis Rheum. 2009, 60, 2071–2082. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Su, B.; Ni, H.; Li, L.; Chen, X.; You, X.; Zhang, H. microRNA-199a may be involved in the pathogenesis of lupus nephritis via modulating the activation of NF-κB by targeting Klotho. Mol. Immunol. 2018, 103, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, L.; Xie, G.L.; Bao, J.F.; Yu, Q. Let-7 miRNAs Modulate the Activation of NF-κB by Targeting TNFAIP3 and Are Involved in the Pathogenesis of Lupus Nephritis. PLoS ONE 2015, 10, e0121256. [Google Scholar] [CrossRef]
- Chafin, C.B.; Regna, N.L.; Dai, R.; Caudell, D.L.; Reilly, C.M. MicroRNA-let-7a expression is increased in the mesangial cells of NZB/W mice and increases IL-6 production in vitro. Autoimmunity 2013, 46, 351–362. [Google Scholar] [CrossRef]
- Wang, W.; Gao, J.; Wang, F. MiR-663a/MiR-423-5p are involved in the pathogenesis of lupus nephritis via modulating the activation of NF-κB by targeting TNIP2. Am. J. Transl. Res. 2017, 9, 3796–3803. [Google Scholar] [PubMed]
- Han, X.; Wang, Y.; Zhang, X.; Qin, Y.; Qu, B.; Wu, L.; Ma, J.; Zhou, Z.; Qian, J.; Dai, M.; et al. MicroRNA-130b Ameliorates Murine Lupus Nephritis Through Targeting the Type I Interferon Pathway on Renal Mesangial Cells. Arthritis Rheumatol. 2016, 68, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wang, J.; Li, F. Dysregulation of PTEN caused by the underexpression of microRNA-130b is associated with the severity of lupus nephritis. Mol. Med. Rep. 2018, 17, 7966–7972. [Google Scholar] [CrossRef] [PubMed]
- Costa-Reis, P.; Russo, P.A.; Zhang, Z.; Colonna, L.; Maurer, K.; Gallucci, S.; Schulz, S.W.; Kiani, A.N.; Petri, M.; Sullivan, K.E. The Role of MicroRNAs and Human Epidermal Growth Factor Receptor 2 in Proliferative Lupus Nephritis. Arthritis Rheumatol. 2015, 67, 2415–2426. [Google Scholar] [CrossRef]
- Cui, D.; Zhu, D.; Ren, H.; Lin, J.; Lai, W.; Huang, Q.; Zhao, J.; Yang, M. MicroRNA-198 contributes to lupus nephritis progression by inhibition of phosphatase and tensin homology deleted on chromosome ten expression. J. Nephrol. 2017, 16, 7813–7820. [Google Scholar] [CrossRef]
- Luan, J.; Fu, J.; Chen, C.; Jiao, C.; Kong, W.; Zhang, Y.; Chang, Q.; Wang, Y.; Li, D.; Illei, G.G.; et al. LNA-anti-miR-150 ameliorated kidney injury of lupus nephritis by inhibiting renal fibrosis and macrophage infiltration. Arthritis Res. Ther. 2019, 21, 276. [Google Scholar] [CrossRef]
- Zhou, H.; Hasni, S.A.; Perez, P.; Tandon, M.; Jang, S.I.; Zheng, C.; Kopp, J.B.; Austin, H., 3rd; Balow, J.E.; Alevizos, I.; et al. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. J. Am. Soc. Nephrol. 2013, 24, 1073–1087. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, N.; Zhang, J.; Zhao, H.; Wang, X. miR-410 suppresses the expression of interleukin-6 as well as renal fibrosis in the pathogenesis of lupus nephritis. Clin. Exp. Pharmacol. Physiol. 2016, 43, 616–625. [Google Scholar] [CrossRef]
- Li, X.; Luo, F.; Li, J.; Luo, C. MiR-183 delivery attenuates murine lupus nephritis-related injuries via targeting mTOR. Arthritis Res. Ther. 2019, 90, e12810. [Google Scholar] [CrossRef]
- Qi, H.; Cao, Q.; Liu, Q. MicroRNA-183 exerts a protective role in lupus nephritis through blunting the activation of TGF-β/Smad/TLR3 pathway via reducing Tgfbr1. Exp. Cell Res. 2020, 394, 112138. [Google Scholar] [CrossRef]
- Huang, Z.; Pang, G.; Huang, Y.G.; Li, C. miR-133 inhibits proliferation and promotes apoptosis by targeting LASP1 in lupus nephritis. Exp. Mol. Pathol. 2020, 114, 104384. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Cao, Q.; Liu, Q. MicroRNA-16 directly binds to DEC2 and inactivates the TLR4 signaling pathway to inhibit lupus nephritis-induced kidney tissue hyperplasia and mesangial cell proliferation. Int. Immunopharmacol. 2020, 88, 106859. [Google Scholar] [CrossRef]
- Yao, F.; Sun, L.; Fang, W.; Wang, H.; Yao, D.; Cui, R.; Xu, J.; Wang, L.; Wang, X. Hsa-miR-371-5p inhibits human mesangial cell proliferation and promotes apoptosis in lupus nephritis by directly targeting hypoxia-inducible factor 1α. Mol. Med. Rep. 2016, 14, 5693–5698. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liao, W.; Yi, Z.; Xiang, W.; He, X. Association of miRNA-145 expression in vascular smooth muscle cells with vascular damages in patients with lupus nephritis. Int. J. Clin. Exp. Pathol. 2015, 8, 12646–12656. [Google Scholar] [PubMed]
- Zheng, J.; Guo, R.; Tang, Y.; Fu, Q.; Chen, J.; Wu, L.; Leng, L.; Bucala, R.; Song, Y.; Lu, L. miR-152 Attenuates the Severity of Lupus Nephritis Through the Downregulation of Macrophage Migration Inhibitory Factor (MIF)-Induced Expression of COL1A1. Front. Immunol. 2019, 10, 158. [Google Scholar] [CrossRef]
- Cai, Z.; Xiang, W.; Peng, X.; Ding, Y.; Liao, W.; He, X. MicroRNA-145 Involves in the Pathogenesis of Renal Vascular Lesions and May Become a Potential Therapeutic Target in Patients with Juvenile Lupus Nephritis. Kidney Blood Press. Res. 2019, 44, 643–655. [Google Scholar] [CrossRef]
- Adhya, Z.; El Anbari, M.; Anwar, S. Soluble TNF-R1, VEGF and other cytokines as markers of disease activity in systemic lupus erythematosus and lupus nephritis. Lupus 2019, 28, 713–721. [Google Scholar] [CrossRef]
- Ciprandi, G.; Murdaca, G.; Colombo, B.M.; De Amici, M.; Marseglia, G.L. Serum vascular endothelial growth factor in allergic rhinitis and systemic lupus erythematosus. Hum. Immunol. 2008, 69, 510–512. [Google Scholar] [CrossRef]
- Mortimer, A.; Marr, N.; Karim, M.Y.; Frieri, M. Accelerated atherosclerosis in systemic lupus erythematosus: Role of proinflammatory cytokines and therapeutic approaches. Lupus 2012, 12, 25–32. [Google Scholar] [CrossRef]
- Doleshal, M.; Magotra, A.A.; Choudhury, B.; Cannon, B.D.; Labourier, E.; Szafranska, A.E. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2008, 10, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Nakajima, G.; Gavin, E.; Morris, C.G.; Kudo, K.; Hayashi, K.; Ju, J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 2007, 13, 1668–1674. [Google Scholar] [CrossRef] [PubMed]
- Hoefig, K.P.; Heissmeyer, V. Measuring microRNA expression in size-limited FACS-sorted and microdissected samples. Methods Mol. Biol. 2010, 667, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.L.; Cao, Y.; Liu, D.; Xu, M.; Liu, H.; Tang, R.N.; Ma, K.L.; Liu, B.C. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int. J. Biol. Sci. 2013, 9, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- van Balkom, B.W.; Pisitkun, T.; Verhaar, M.C.; Knepper, M.A. Exosomes and the kidney: Prospects for diagnosis and therapy of renal diseases. Kidney Int. 2011, 80, 1138–1145. [Google Scholar] [CrossRef]
- Tangtanatakul, P.; Klinchanhom, S.; Sodsai, P.; Sutichet, T.; Promjeen, C.; Avihingsanon, Y.; Hirankarn, N. Down-regulation of let-7a and miR-21 in urine exosomes from lupus nephritis patients during disease flare. Asian Pac. J. Allergy Immunol. 2019, 37, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Perez-Hernandez, J.; Martinez-Arroyo, O.; Ortega, A.; Galera, M.; Solis-Salguero, M.A.; Chaves, F.J.; Redon, J.; Forner, M.J.; Cortes, R. Urinary exosomal miR-146a as a marker of albuminuria, activity changes and disease flares in lupus nephritis. Int. J. Rheum. Dis. 2021, 34, 1157–1167. [Google Scholar] [CrossRef]
- Guan, J.; Wang, G.; Tam, L.S.; Kwan, B.C.; Li, E.K.; Chow, K.M.; Li, P.K.; Szeto, C.C. Urinary sediment ICAM-1 level in lupus nephritis. Lupus 2012, 21, 1190–1195. [Google Scholar] [CrossRef]
- Cardenas-Gonzalez, M.; Srivastava, A.; Pavkovic, M.; Bijol, V.; Rennke, H.G.; Stillman, I.E.; Zhang, X.; Parikh, S.; Rovin, B.H.; Afkarian, M.; et al. Identification, Confirmation, and Replication of Novel Urinary MicroRNA Biomarkers in Lupus Nephritis and Diabetic Nephropathy. Lupus 2017, 63, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, X.; Tang, X.; Bian, X.; Shen, B.; Zhao, H.; Luo, S.; Chen, Z.; Zhang, K. MicroRNA expression profile of urinary exosomes in Type IV lupus nephritis complicated by cellular crescent. J. Biol. Res. 2018, 25, 16. [Google Scholar] [CrossRef]
- Solé, C.; Cortés-Hernández, J.; Felip, M.L.; Vidal, M.; Ordi-Ros, J. miR-29c in urinary exosomes as predictor of early renal fibrosis in lupus nephritis. Nephrol. Dial. Transplant. 2015, 30, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Solé, C.; Moliné, T.; Vidal, M.; Ordi-Ros, J.; Cortés-Hernández, J. An Exosomal Urinary miRNA Signature for Early Diagnosis of Renal Fibrosis in Lupus Nephritis. Cells 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Perez-Hernandez, J.; Forner, M.J.; Pinto, C.; Chaves, F.J.; Cortes, R.; Redon, J. Increased Urinary Exosomal MicroRNAs in Patients with Systemic Lupus Erythematosus. PLoS ONE 2015, 10, e0138618. [Google Scholar] [CrossRef] [PubMed]
- Yap, D.Y.; Ma, M.K.; Tang, C.S.; Chan, T.M. Proliferation signal inhibitors in the treatment of lupus nephritis: Preliminary experience. Nephrology 2012, 17, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Yap, D.Y.H.; Tang, C.; Chan, G.C.W.; Kwan, L.P.Y.; Ma, M.K.M.; Mok, M.M.Y.; Chan, T.M. Longterm Data on Sirolimus Treatment in Patients with Lupus Nephritis. J. Rheumatol. 2018, 45, 1663–1670. [Google Scholar] [CrossRef]
- Jin, H.Y.; Gonzalez-Martin, A.; Miletic, A.V.; Lai, M.; Knight, S.; Sabouri-Ghomi, M.; Head, S.R.; Macauley, M.S.; Rickert, R.C.; Xiao, C. Transfection of microRNA Mimics Should Be Used with Caution. Front. Genet. 2015, 6, 340. [Google Scholar] [CrossRef]
- Tang, Q.; Yang, Y.; Zhao, M.; Liang, G.; Wu, H.; Liu, Q.; Xie, Y.; Li, D.; Dai, Y.; Yung, S.; et al. Mycophenolic acid upregulates miR-142-3P/5P and miR-146a in lupus CD4+T cells. Lupus 2015, 24, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Gutierrez, P.R.; Ceribelli, A.; Satoh, M.; Sobel, E.S.; Reeves, W.H.; Chan, E.K. Reduced levels of CCL2 and CXCL10 in systemic lupus erythematosus patients under treatment with prednisone, mycophenolate mofetil, or hydroxychloroquine, except in a high STAT1 subset. Arthritis Res. Ther. 2014, 16, R23. [Google Scholar] [CrossRef]
- Cecchi, I.; Perez-Sanchez, C.; Sciascia, S.; Radin, M.; Arias de la Rosa, I.; Barbarroja Puerto, N.; Scudeler, L.; Perez-Sanchez, L.; Patiño Trives, A.M.; Aguirre Zamorano, M.A.; et al. Circulating microRNAs as potential biomarkers for monitoring the response to in vivo treatment with Rituximab in systemic lupus erythematosus patients. Autoimmun. Rev. 2020, 19, 102488. [Google Scholar] [CrossRef]
- Yang, J.; Yang, X.; Yang, J.; Li, M. Hydroxychloroquine Inhibits the Differentiation of Th17 Cells in Systemic Lupus Erythematosus. J. Rheumatol. 2018, 45, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Chafin, C.B.; Regna, N.L.; Hammond, S.E.; Reilly, C.M. Cellular and urinary microRNA alterations in NZB/W mice with hydroxychloroquine or prednisone treatment. Int. Immunopharmacol. 2013, 17, 894–906. [Google Scholar] [CrossRef] [PubMed]
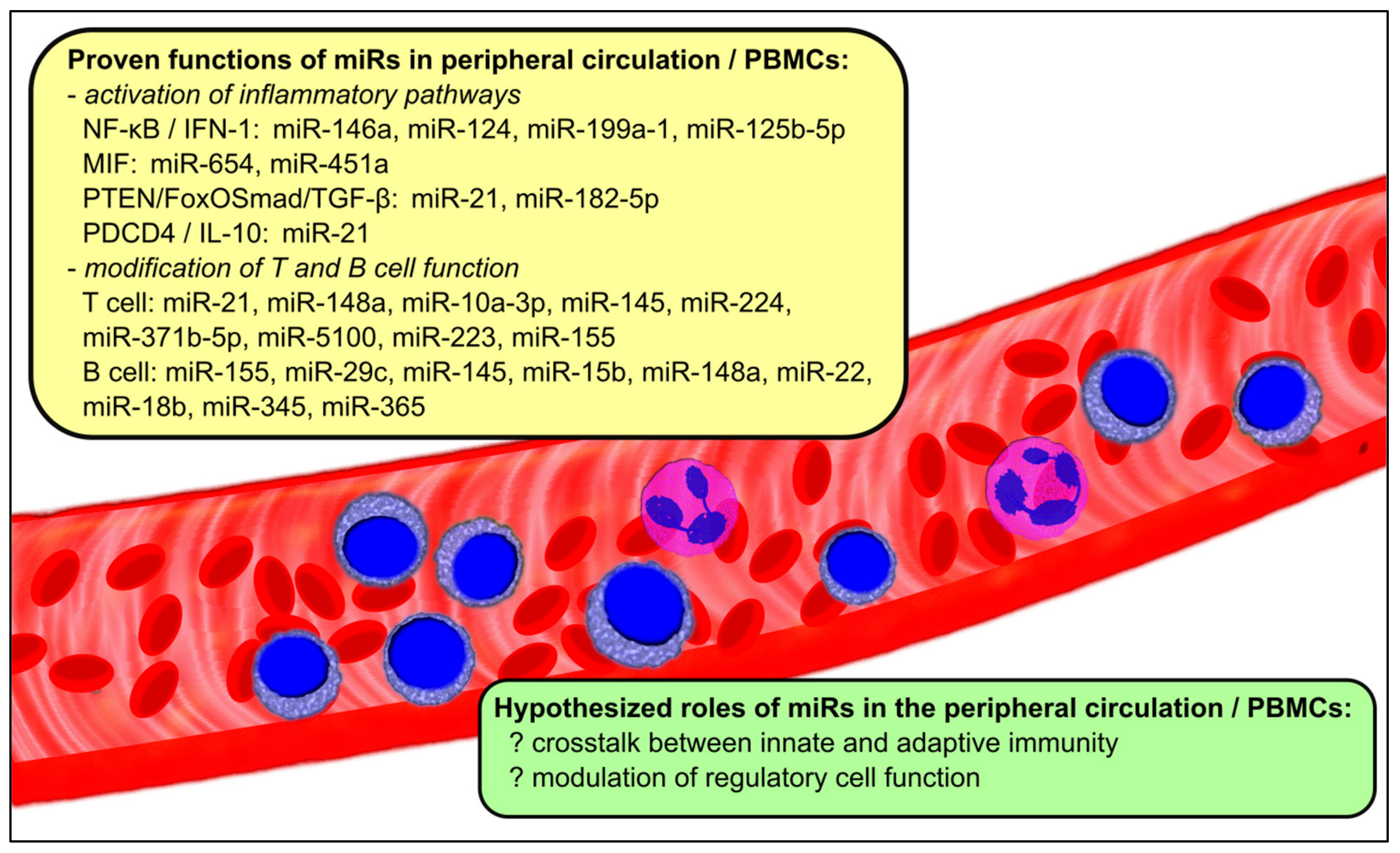
| Associated Pathways | Relevant miRs in the Circulation and PBMCs | Change in miR Expression |
|---|---|---|
| NF-κB/IFN-1 | miR-146a [20,21,22,24,26,27,50,51] miR-124 [28] miR-199a-1 [19] miR-125b-5p [52] | conflicting decrease increase increase |
| MIF | miR-654 [29] miR-451a [30] | decrease decrease |
| PTEN/FoxO Smad/TGF-β | miR-21 [31,32,33] miR-182-5p | increase increase |
| PDCD4/IL-10 | miR-21 [31,32,33] | increase |
| T cell function/differentiation | miR-21 [33] miR-148a [33] miR-10a-3p [34] miR-224 [35] miR-371b-5p [36] miR-5100 [36] miR-223 [37] miR-155 [17,23,27,31,38] | increase increase decrease increase increase increase decrease decrease |
| B cell | miR-155 [17,23,27,31,38] miR-29c [39] miR-145 [39,40] miR-15b [38,41] miR-148a [43] miR-22 [38,39] miR-18b [38,39] miR-345 [38,39] miR-365 [38,39] | decrease increase increase decrease increase increase increase increase increase |
| Associated Pathways | Relevant miRs in the Kidney Tissue | Change in miR Expression |
|---|---|---|
| NF-κB | miR-199a [69] miR-let-7a [70,71] miR-let-7e [70] miR-663a/miR-423-5p [72] | increase increase increase increase |
| IFN-1 | miR-130b [73,74] miR-26a [75] miR-30b [75] | conflicting decrease decrease |
| PTEN | miR-130b [73,74] miR-198 [51,76] | conflicting increase |
| TGF-β1 | miR-150 [77,78] miR-410 [79] miR-183 [80,81] | increase decrease decrease |
| Other cellular proliferative and inflammatory pathways | miR-133 [82] miR-16 [83] miR-371-5p [84] miR-146a [51] miR-422a [59] miR-145 [85] | decrease decrease decrease increase increase decrease |
| Other fibrotic pathways | miR-152 [86] | decrease |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
So, B.Y.F.; Yap, D.Y.H.; Chan, T.M. MicroRNAs in Lupus Nephritis–Role in Disease Pathogenesis and Clinical Applications. Int. J. Mol. Sci. 2021, 22, 10737. https://doi.org/10.3390/ijms221910737
So BYF, Yap DYH, Chan TM. MicroRNAs in Lupus Nephritis–Role in Disease Pathogenesis and Clinical Applications. International Journal of Molecular Sciences. 2021; 22(19):10737. https://doi.org/10.3390/ijms221910737
Chicago/Turabian StyleSo, Benjamin Y. F., Desmond Y. H. Yap, and Tak Mao Chan. 2021. "MicroRNAs in Lupus Nephritis–Role in Disease Pathogenesis and Clinical Applications" International Journal of Molecular Sciences 22, no. 19: 10737. https://doi.org/10.3390/ijms221910737
APA StyleSo, B. Y. F., Yap, D. Y. H., & Chan, T. M. (2021). MicroRNAs in Lupus Nephritis–Role in Disease Pathogenesis and Clinical Applications. International Journal of Molecular Sciences, 22(19), 10737. https://doi.org/10.3390/ijms221910737






