An ArsR Transcriptional Regulator Facilitates Brucella sp. Survival via Regulating Self and Outer Membrane Protein
Abstract
:1. Introduction
2. Results
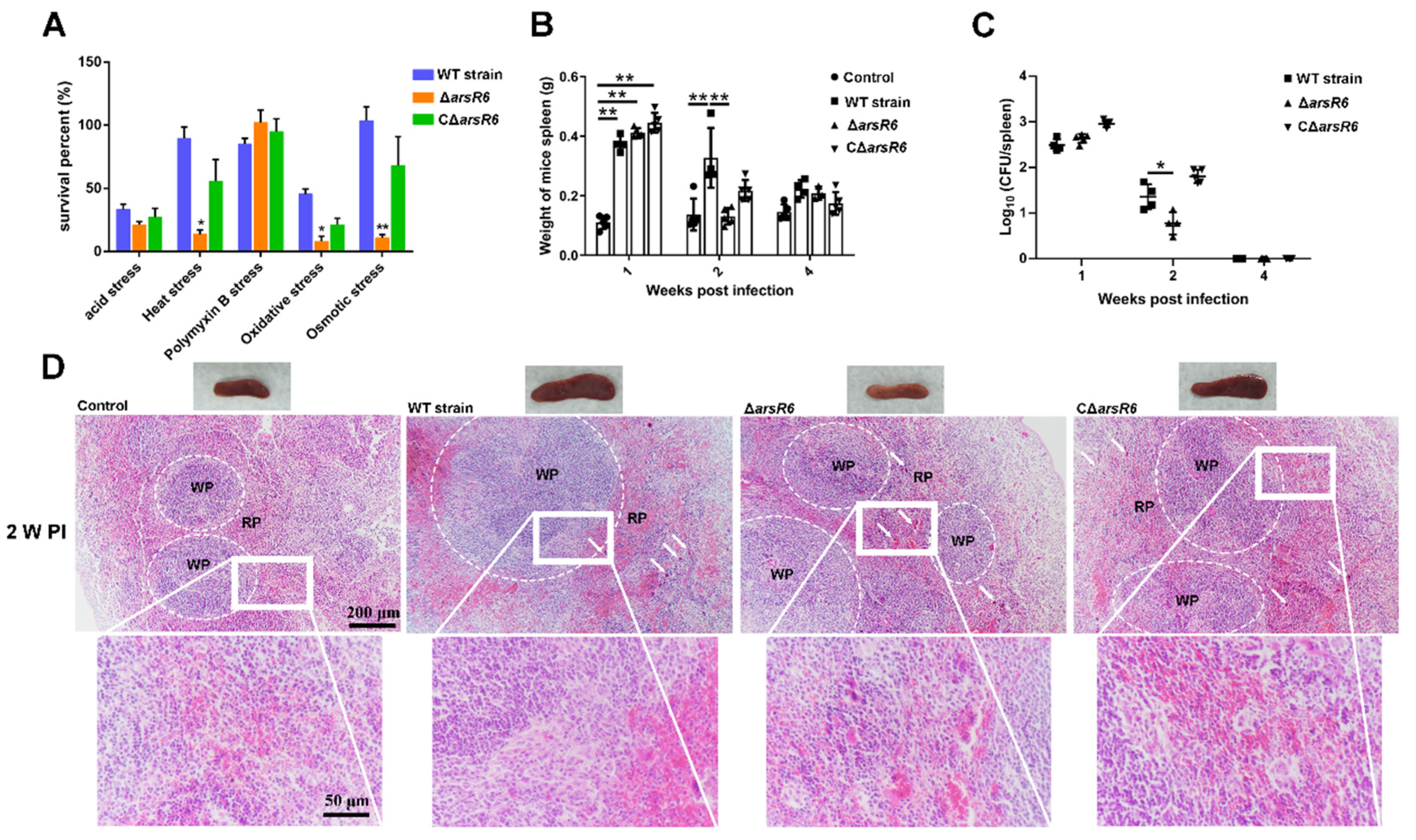
2.1. ArsR6 Is Required for Brucella Survival in a Mouse Model of Infection
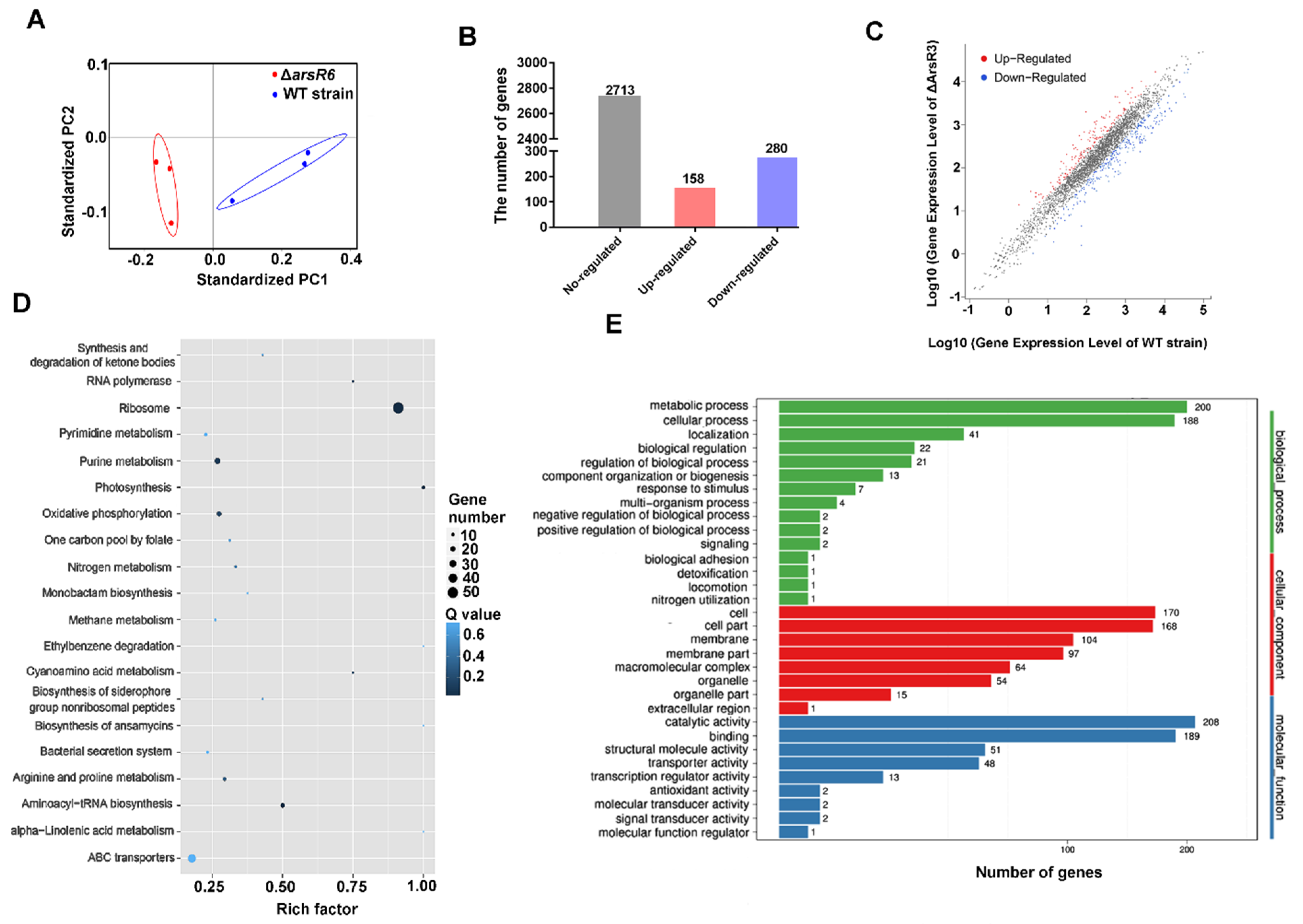
2.2. Identification of ArsR6 Target Genes by RNA-Seq and ChIP–Seq Analyses
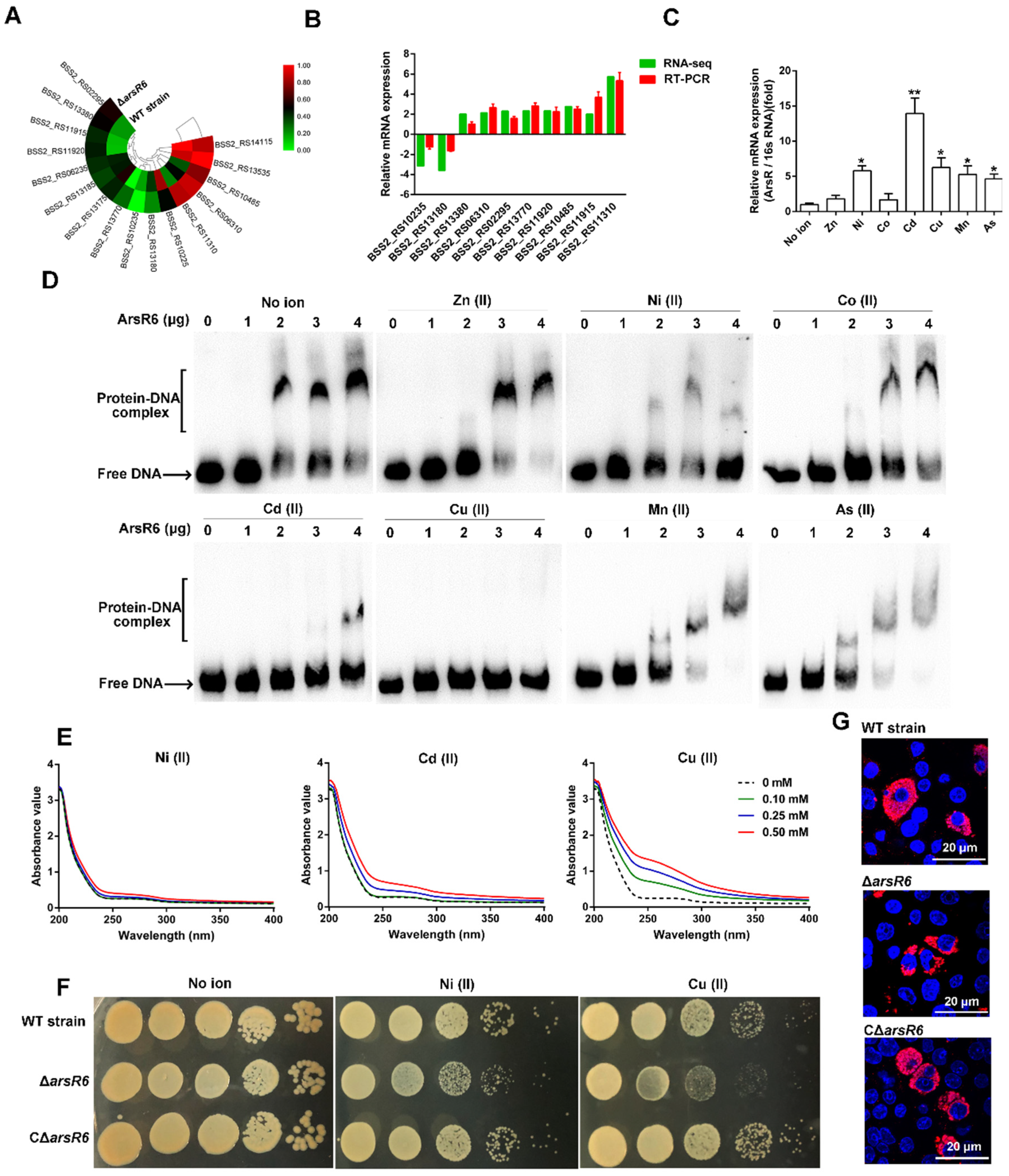
2.3. Transcriptional Autoregulatory Properties of ArsR6
2.4. ArsR6 Is a Metalloregulatory Protein
2.5. The Omp25D Gene Is a Direct Target of Transcriptional Activation by ArsR6
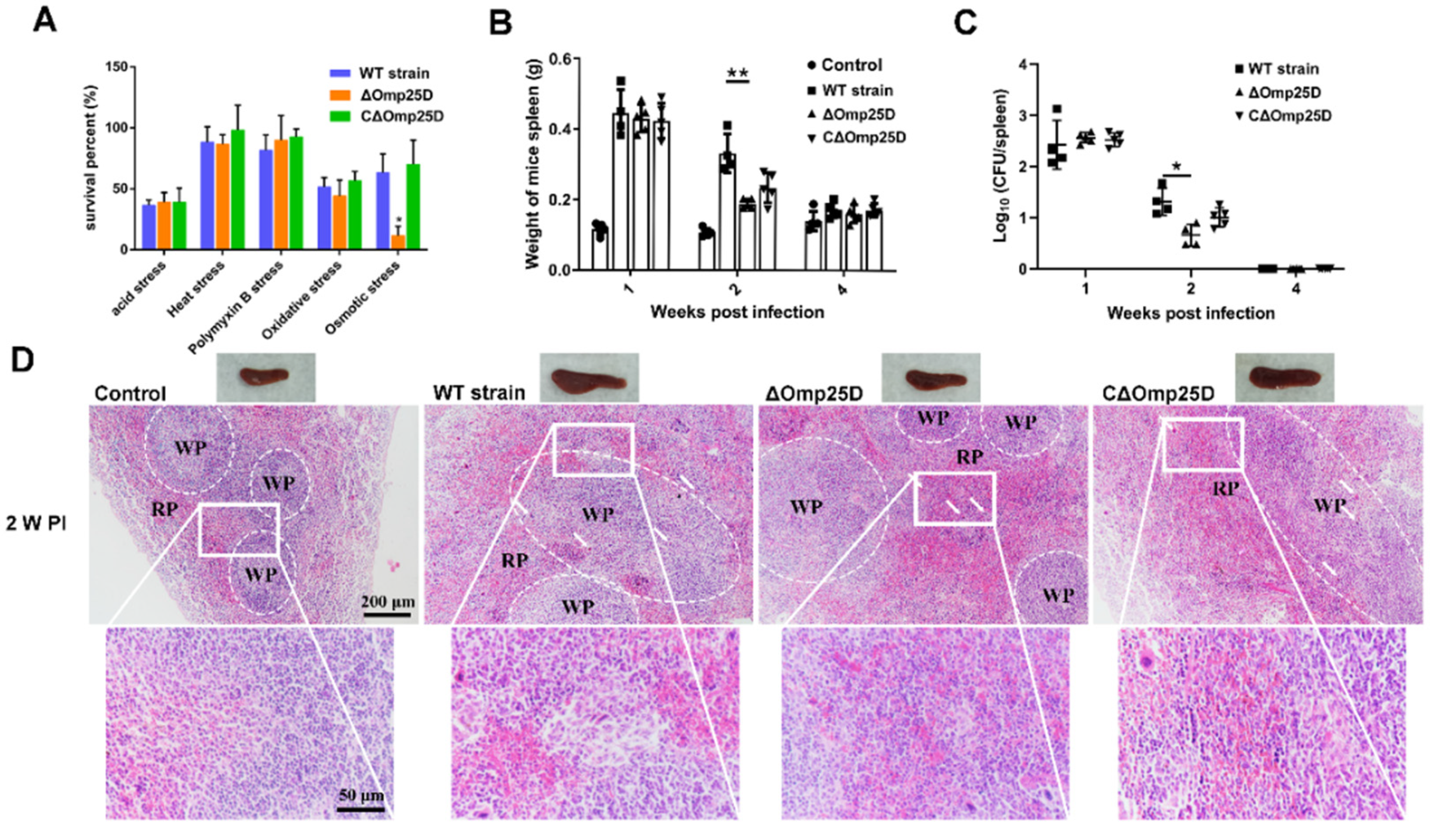
2.6. Omp25D Contributes to Brucella Virulence
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Bacterial Strains and Growth Conditions
4.3. Stress Resistance Assays
4.4. Macrophage Infection
4.5. Real-Time PCR Analysis
4.6. Mouse Infection
4.7. RNA-Sequencing Analysis
4.8. Chromatin Immunoprecipitation Sequencing Analysis
4.9. Protein Expression and Purification
4.10. Electrophoretic Mobility Shift Assay
4.11. DNaseI Footprinting Assay
4.12. Western Blot
4.13. Cell Counting Kit-8 (CCK8) Assay
4.14. Ultraviolet Light Analysis
4.15. β-Galactosidase Activity Assay
4.16. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Boschiroli, M.L.; Foulongne, V.; O’Callaghan, D. Brucellosis: A worldwide zoonosis. Curr. Opin. Microbiol. 2001, 4, 58–64. [Google Scholar] [CrossRef]
- Rambow-Larsen, A.A.; Petersen, E.M.; Gourley, C.R.; Splitter, G.A. Brucella regulators: Self-control in a hostile environment. Trends Microbiol. 2009, 17, 371–377. [Google Scholar] [CrossRef] [Green Version]
- Batut, J.; Andersson, S.G.E.; O’Callaghan, D. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2004, 2, 933–945. [Google Scholar] [CrossRef]
- Jiao, H.W.; Zhou, Z.X.; Li, B.W.; Xiao, Y.; Li, M.J.; Zeng, H.; Guo, X.Y.; Gu, G.J. The Mechanism of Facultative Intracellular Parasitism of Brucella. Int. J. Mol. Sci. 2021, 22, 3673. [Google Scholar] [CrossRef] [PubMed]
- Busenlehner, L.S.; Pennella, M.A.; Giedroc, D.P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 2003, 27, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Ren, S.; Li, Q.; Xie, L.; Xie, J. Molecular Mechanisms Underlying the Function Diversity of ArsR Family Metalloregulator. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Cavet, J.S.; Graham, A.I.; Meng, W.M.; Robinson, N.J. A cadmium-lead-sensing ArsR-SmtB repressor with novel sensory sites-Complementary metal discrimination by NMTR and CMTR in a common cytosol. J. Biol. Chem. 2003, 278, 44560–44566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef]
- Palmer, L.D.; Skaar, E.P. Transition Metals and Virulence in Bacteria. Annu. Rev. Genet. 2016, 50, 67–91. [Google Scholar] [CrossRef] [Green Version]
- Haine, V.; Sinon, A.; Van Steen, F.; Rousseau, S.; Dozot, M.; Lestrate, P.; Lambert, C.; Letesson, J.J.; De Bolle, X. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 2005, 73, 5578–5586. [Google Scholar] [CrossRef] [Green Version]
- Ellermeier, C.D.; Hobbs, E.C.; Gonzalez-Pastor, J.E.; Losick, R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 2006, 124, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Moinier, D.; Slyemi, D.; Byrne, D.; Lignon, S.; Lebrun, R.; Talla, E.; Bonnefoy, V. An ArsR/SmtB Family Member Is Involved in the Regulation by Arsenic of the Arsenite Oxidase Operon in Thiomonas arsenitoxydans. Appl. Environ. Microbiol. 2014, 80, 6413–6426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Nadar, V.S.; Rosen, B.P. A novel MAs(III)-selective ArsR transcriptional repressor. Mol. Microbiol. 2017, 106, 469–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonucci, I.; Gallo, G.; Limauro, D.; Contursi, P.; Ribeiro, A.L.; Blesa, A.; Berenguer, J.; Bartolucci, S.; Fiorentino, G. An ArsR/SmtB family member regulates arsenic resistance genes unusually arranged in Thermus thermophilus HB27. Microb. Biotechnol. 2017, 10, 1690–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, S.; Kumar, A.; Singhal, A.; Tyagi, J.S.; Prasad, H.K. CmtR, a cadmium-sensing ArsR-SmtB repressor, cooperatively interacts with multiple operator sites to autorepress its transcription in Mycobacterium tuberculosis. FEBS J. 2009, 276, 3428–3439. [Google Scholar] [CrossRef] [PubMed]
- Cavet, J.S.; Meng, W.; Pennella, M.A.; Applehoff, R.J.; Giedroc, D.P.; Robinson, N.J. A nickel-cobalt-sensing ArsR-SmtB family repressor-Contributions of cytosol and effector binding sites to metal selectivity. J. Biol. Chem. 2002, 277, 38441–38448. [Google Scholar] [CrossRef] [Green Version]
- Osman, D.; Cavet, J.S. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 2010, 27, 668–680. [Google Scholar] [CrossRef]
- Edmonds, M.D.; Cloeckaert, A.; Elzer, P.H. Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 2002, 88, 205–221. [Google Scholar] [CrossRef]
- Martin-Martin, A.I.; Caro-Hernandez, P.; Orduna, A.; Vizcaino, N.; Fernandez-Lago, L. Importance of the Omp25/Omp31 family in the internalization and intracellular replication of virulent B. ovis in murine macrophages and HeLa cells. Microbes Infect. 2008, 10, 706–710. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, X.; Wu, X.; Yang, X.; Han, C.; Wang, Z.; Du, Q.; Zhao, X.; Liu, S.-L.; Tong, D.; et al. Brucella Downregulates Tumor Necrosis Factor-alpha to Promote Intracellular Survival via Omp25 Regulation of Different MicroRNAs in Porcine and Murine Macrophages. Front. Immunol. 2017, 8, 2013. [Google Scholar] [CrossRef] [Green Version]
- Fernández, L.V.; Oropeza-Navarro, R.; Ortiz-Rico, A.; Robles-Pesina, G.; Ramírez-Lezama, J.; Castañeda-Ramírez, A.; Verdugo-Rodríguez, A. Brucella melitensis omp31 Mutant Is Attenuated and Confers Protection Against Virulent Brucella melitensis Challenge in BALB/c Mice. J. Microbiol. Biotechnol. 2020, 30, 497–504. [Google Scholar] [CrossRef]
- Lapaque, N.; Moriyon, I.; Moreno, E.; Gorvel, J.P. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 2005, 8, 60–66. [Google Scholar] [CrossRef]
- Tibor, A.; Wansard, V.; Bielartz, V.; Delrue, R.M.; Danese, I.; Michel, P.; Walravens, K.; Godfroid, J.; Letesson, J.J. Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 2002, 70, 5540–5546. [Google Scholar] [CrossRef] [Green Version]
- Murphy, J.N.; Saltikov, C.W. The ArsR Repressor Mediates Arsenite-Dependent Regulation of Arsenate Respiration and Detoxification Operons of Shewanella sp. Strain ANA-3. J. Bacteriol. 2009, 191, 6722–6731. [Google Scholar] [CrossRef] [Green Version]
- Saha, R.P.; Samanta, S.; Patra, S.; Sarkar, D.; Saha, A.; Singh, M.K. Metal homeostasis in bacteria: The role of ArsR-SmtB family of transcriptional repressors in combating varying metal concentrations in the environment. Biometals 2017, 30, 459–503. [Google Scholar] [CrossRef]
- Wang, Y.; Pennella, A.; Giedroc, D.P. Structural and functional characterization of M-tuberculosis CmtR, a Pb(II)/Cd(II) sensing SmtB/ArsR metalloregulatory repressor. In Biophysical Journal (Vol. 88, No. 1, pp. 407A-407A); Biophysical Society: Bethesda, MD, USA, 2005. [Google Scholar]
- Xu, C.; Rosen, B.P. Dimerization is essential for DNA binding and repression by the ArsR metalloregulatory protein of Escherichia coli. J. Biol. Chem. 1997, 272, 15734–15738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.X.; McClure, R.; Nudel, K.; Daou, N.; Genco, C.A. Characterization of the Neisseria gonorrhoeae Iron and Fur Regulatory Network. J. Bacteriol. 2016, 198, 2180–2191. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, T.B.; Amrhein, N.; Macheroux, P. Characterization of YqjM, an old yellow enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J. Biol. Chem. 2003, 278, 19891–19897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.L.; Singh, A.K.; Heo, L.; Seok, C.; Roe, J.H. Factors affecting redox potential and differential sensitivity of SoxR to redox-active compounds. Mol. Microbiol. 2015, 97, 808–821. [Google Scholar] [CrossRef] [PubMed]
- Gee, J.M.; Valderas, M.W.; Kovach, M.E.; Grippe, V.K.; Robertson, G.T.; Ng, W.L.; Richardson, J.M.; Winkler, M.E.; Roop, R.M. The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect. Immun. 2005, 73, 2873–2880. [Google Scholar] [CrossRef] [Green Version]
- Byndloss, M.X.; Tsolis, R.M. Brucella spp. Virulence Factors and Immunity. Annu. Rev. Anim. Biosci. 2016, 4, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Nakashima, S.; Hirose, K.; Shibasaka, M.; Katsuhara, M.; Ezaki, B.; Giedroc, D.P.; Kasamo, K. A novel cyanobacterial SmtB/ArsR family repressor regulates the expression of a CPx-ATPase and a metallothionein in response to both Cu(I)/Ag(I) and Zn(II)/Cd(II). J. Biol. Chem. 2004, 279, 17810–17818. [Google Scholar] [CrossRef] [Green Version]
- Becker, K.W.; Skaar, E.P. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 2014, 38, 1235–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seleem, M.N.; Boyle, S.M.; Sriranganathan, N. Brucella: A pathogen without classic virulence genes. Vet. Microbiol. 2008, 129, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Roop, R.M.; Barton, I.S.; Hopersberger, D.; Martin, D.W. Uncovering the Hidden Credentials of Brucella Virulence. Microbiol. Mol. Biol. Rev. 2021, 85, e00021-19. [Google Scholar] [CrossRef]
- Caro-Hernandez, P.; Fernandez-Lago, L.; de Miguel, M.-J.; Martin-Martin, A.I.; Cloeckaert, A.; Grillo, M.-J.; Vizcaino, N. Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect. Immun. 2007, 75, 4050–4061. [Google Scholar] [CrossRef] [Green Version]
- Salhi, I.; Boigegrain, R.A.; Machold, J.; Weise, C.; Cloeckaert, A.; Rouot, B. Characterization of new members of the group 3 outer membrane protein family of Brucella spp. Infect. Immun. 2003, 71, 4326–4332. [Google Scholar] [CrossRef] [Green Version]
- Pasquevich, K.A.; Carabajal, M.V.; Guaimas, F.F.; Bruno, L.; Roset, M.S.; Coria, L.M.; Serrantes, D.A.R.; Comerci, D.J.; Cassataro, J. Omp19 Enables Brucella abortus to Evade the Antimicrobial Activity From Host’s Proteolytic Defense System. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.C.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Ruan, J.; Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008, 18, 1851–1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.X.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.J.; Liu, T.; Manrai, A.K.; Liu, X.S. CEAS: Cis-regulatory element annotation system. Bioinformatics 2009, 25, 2605–2606. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.G.; Wang, T.T.; Hua, C.F.; Sun, W.J.; Li, X.Y.; Grunwald, L.; Liu, J.G.; Wu, N.; Shao, X.L.; Yin, Y.M.; et al. A compendium of DNA-binding specificities of transcription factors in Pseudomonas syringae. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhi, F.; Zhou, D.; Chen, J.; Fang, J.; Zheng, W.; Li, J.; Hao, M.; Shi, Y.; Jin, Y.; Wang, A. An ArsR Transcriptional Regulator Facilitates Brucella sp. Survival via Regulating Self and Outer Membrane Protein. Int. J. Mol. Sci. 2021, 22, 10860. https://doi.org/10.3390/ijms221910860
Zhi F, Zhou D, Chen J, Fang J, Zheng W, Li J, Hao M, Shi Y, Jin Y, Wang A. An ArsR Transcriptional Regulator Facilitates Brucella sp. Survival via Regulating Self and Outer Membrane Protein. International Journal of Molecular Sciences. 2021; 22(19):10860. https://doi.org/10.3390/ijms221910860
Chicago/Turabian StyleZhi, Feijie, Dong Zhou, Jialu Chen, Jiaoyang Fang, Weifang Zheng, Junmei Li, Mingyue Hao, Yong Shi, Yaping Jin, and Aihua Wang. 2021. "An ArsR Transcriptional Regulator Facilitates Brucella sp. Survival via Regulating Self and Outer Membrane Protein" International Journal of Molecular Sciences 22, no. 19: 10860. https://doi.org/10.3390/ijms221910860




