Ions in the Deep Subsurface of Earth, Mars, and Icy Moons: Their Effects in Combination with Temperature and Pressure on tRNA–Ligand Binding
Abstract
:1. Introduction
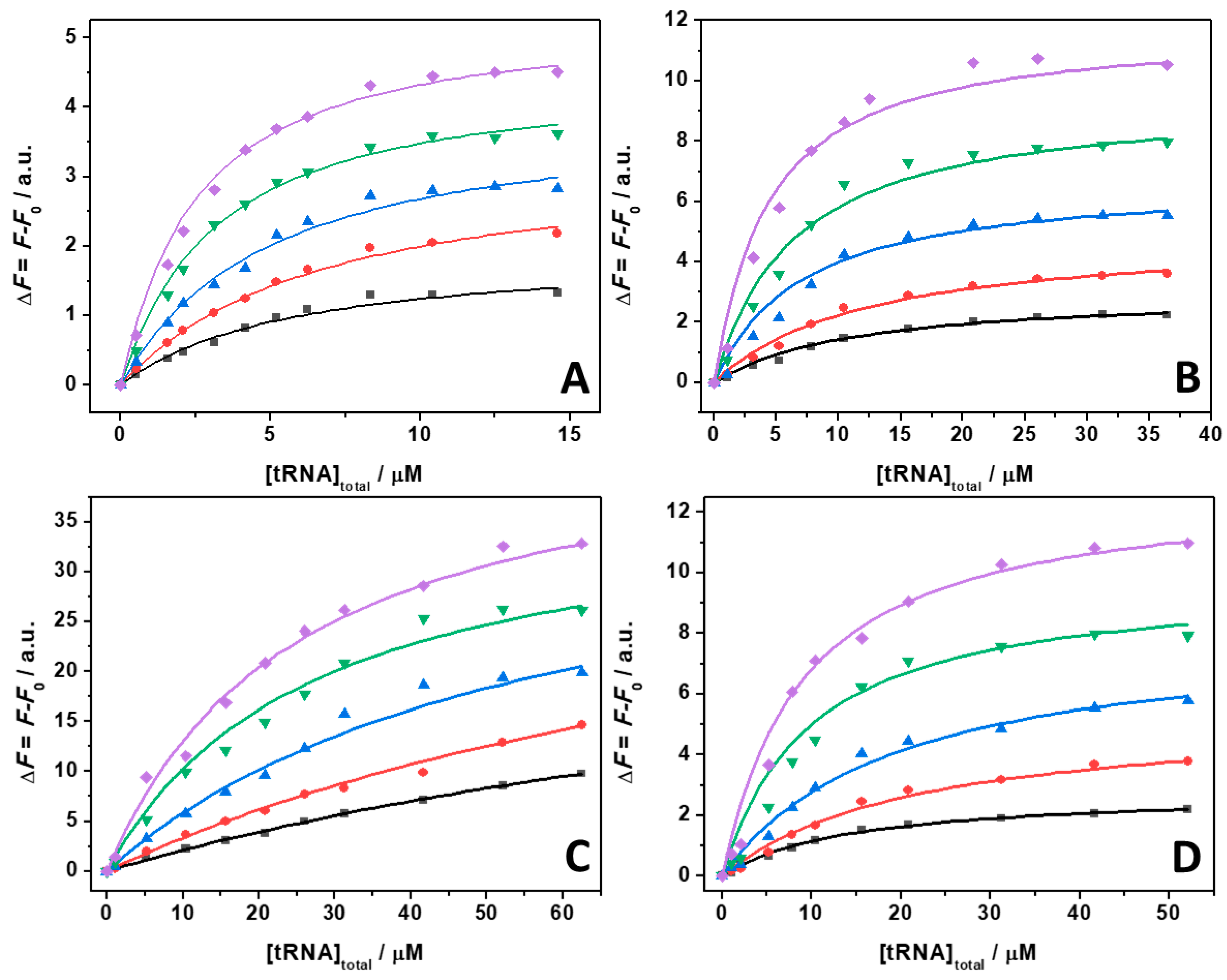
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Sample Preparation
4.3. Steady-State Fluorescence Spectroscopy
4.4. Circular Dichroism Spectroscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daniel, I.; Oger, P.; Winter, R. Origins of Life and Biochemistry under High-Pressure Conditions. Chem. Soc. Rev. 2006, 35, 858. [Google Scholar] [CrossRef]
- Meersman, F.; Daniel, I.; Bartlett, D.H.; Winter, R.; Hazael, R.; McMillan, P.F. High-Pressure Biochemistry and Biophysics. Rev. Mineral. Geochem. 2013, 75, 607–648. [Google Scholar] [CrossRef] [Green Version]
- Payler, S.J.; Biddle, J.F.; Sherwood Lollar, B.; Fox-Powell, M.G.; Edwards, T.; Ngwenya, B.T.; Paling, S.M.; Cockell, C.S. An Ionic Limit to Life in the Deep Subsurface. Front. Microbiol. 2019, 10, 426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegler, S.; Herzog, M.; Oliva, R.; Gault, S.; Cockell, C.S.; Winter, R. Structural Responses of Model Biomembranes to Mars-Relevant Salts. Phys. Chem. Chem. Phys. 2021, 23, 14212–14223. [Google Scholar] [CrossRef]
- Akasaka, K. (Ed.) High Pressure Bioscience: Basic Concepts, Applications and Frontiers; Subcellular biochemistry; Springer: Dordrecht, The Netherlands, 2015; ISBN 978-94-017-9917-1. [Google Scholar]
- Winter, R. Interrogating the Structural Dynamics and Energetics of Biomolecular Systems with Pressure Modulation. Annu. Rev. Biophys. 2019, 48, 441–463. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Caro, J.A.; Norberto, D.R.; Barthe, P.; Roumestand, C.; Schlessman, J.L.; Garcia, A.E.; Garcia-Moreno, E.B.; Royer, C.A. Cavities Determine the Pressure Unfolding of Proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 6945–6950. [Google Scholar] [CrossRef] [Green Version]
- Silva, J.L.; Foguel, D.; Royer, C.A. Pressure Provides New Insights into Protein Folding, Dynamics and Structure. Trends Biochem. Sci. 2001, 26, 612–618. [Google Scholar] [CrossRef]
- Smeller, L.; Rubens, P.; Heremans, K. Pressure Effect on the Temperature-Induced Unfolding and Tendency to Aggregate of Myoglobin. Biochemistry 1999, 38, 3816–3820. [Google Scholar] [CrossRef]
- Akasaka, K. Probing Conformational Fluctuation of Proteins by Pressure Perturbation. Chem. Rev. 2006, 106, 1814–1835. [Google Scholar] [CrossRef]
- Dubins, D.N.; Lee, A.; Macgregor, R.B.; Chalikian, T.V. On the Stability of Double Stranded Nucleic Acids. J. Am. Chem. Soc. 2001, 123, 9254–9259. [Google Scholar] [CrossRef]
- Collins, K.D.; Washabaugh, M.W. The Hofmeister Effect and the Behaviour of Water at Interfaces. Q. Rev. Biophys. 1985, 18, 323–422. [Google Scholar] [CrossRef]
- Lo Nostro, P.; Ninham, B.W. Hofmeister Phenomena: An Update on Ion Specificity in Biology. Chem. Rev. 2012, 112, 2286–2322. [Google Scholar] [CrossRef]
- Luisi, P.L. The Emergence of Life: From Chemical Origins to Synthetic Biology, 2nd ed.; Cambridge University Press: Cambridge, UK, 2016; ISBN 978-1-316-13599-0. [Google Scholar]
- Lingam, M.; Loeb, A. Life in the Cosmos: From Biosignatures to Technosignatures; Harvard University Press: Cambridge, MA, USA, 2021; ISBN 978-0-674-98757-9. [Google Scholar]
- Wilton, D.J.; Ghosh, M.; Chary, K.V.A.; Akasaka, K.; Williamson, M.P. Structural Change in a B-DNA Helix with Hydrostatic Pressure. Nucleic Acids Res. 2008, 36, 4032–4037. [Google Scholar] [CrossRef] [Green Version]
- Girard, E.; Prange, T.; Dhaussy, A.-C.; Migianu-Griffoni, E.; Lecouvey, M.; Chervin, J.-C.; Mezouar, M.; Kahn, R.; Fourme, R. Adaptation of the Base-Paired Double-Helix Molecular Architecture to Extreme Pressure. Nucleic Acids Res. 2007, 35, 4800–4808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, M.-C.; Eid, P.; Wong, P.T.T.; Macgregor, R.B. High Pressure Fourier Transform Infrared Spectroscopy of Poly(DA)Poly(DT), Poly(DA) and Poly(DT). Biophys. Chem. 1999, 76, 87–94. [Google Scholar] [CrossRef]
- Takahashi, S.; Sugimoto, N. Effect of Pressure on the Stability of G-Quadruplex DNA: Thermodynamics under Crowding Conditions. Angew. Chem. Int. Ed. 2013, 52, 13774–13778. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.; Anders, C.; Erwin, N.; Winter, R. Osmolyte Effects on the Conformational Dynamics of a DNA Hairpin at Ambient and Extreme Environmental Conditions. Angew. Chem. Int. Ed. 2017, 56, 5045–5049. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.; Anders, C.; Schummel, P.H.; Winter, R. Antagonistic Effects of Natural Osmolyte Mixtures and Hydrostatic Pressure on the Conformational Dynamics of a DNA Hairpin Probed at the Single-Molecule Level. Phys. Chem. Chem. Phys. 2018, 20, 13159–13170. [Google Scholar] [CrossRef]
- Patra, S.; Schuabb, V.; Kiesel, I.; Knop, J.-M.; Oliva, R.; Winter, R. Exploring the Effects of Cosolutes and Crowding on the Volumetric and Kinetic Profile of the Conformational Dynamics of a Poly DA Loop DNA Hairpin: A Single-Molecule FRET Study. Nucleic Acids Res. 2019, 47, 981–996. [Google Scholar] [CrossRef]
- Hughes, F.; Steiner, R.F. Effects of Pressure on the Helix-Coil Transitions of the Poly A-Poly U System. Biopolymers 1966, 4, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Downey, C.D.; Crisman, R.L.; Randolph, T.W.; Pardi, A. Influence of Hydrostatic Pressure and Cosolutes on RNA Tertiary Structure. J. Am. Chem. Soc. 2007, 129, 9290–9291. [Google Scholar] [CrossRef]
- Schuabb, C.; Berghaus, M.; Rosin, C.; Winter, R. Exploring the Free Energy and Conformational Landscape of TRNA at High Temperature and Pressure. ChemPhysChem 2015, 16, 138–146. [Google Scholar] [CrossRef]
- Clifford, S.M.; Lasue, J.; Heggy, E.; Boisson, J.; McGovern, P.; Max, M.D. Depth of the Martian Cryosphere: Revised Estimates and Implications for the Existence and Detection of Subpermafrost Groundwater. J. Geophys. Res. 2010, 115, E07001. [Google Scholar] [CrossRef]
- Osterloo, M.M.; Hamilton, V.E.; Bandfield, J.L.; Glotch, T.D.; Baldridge, A.M.; Christensen, P.R.; Tornabene, L.L.; Anderson, F.S. Chloride-Bearing Materials in the Southern Highlands of Mars. Science 2008, 319, 1651–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gendrin, A. Sulfates in Martian Layered Terrains: The OMEGA/Mars Express View. Science 2005, 307, 1587–1591. [Google Scholar] [CrossRef] [Green Version]
- Hecht, M.H.; Kounaves, S.P.; Quinn, R.C.; West, S.J.; Young, S.M.M.; Ming, D.W.; Catling, D.C.; Clark, B.C.; Boynton, W.V.; Hoffman, J.; et al. Detection of Perchlorate and the Soluble Chemistry of Martian Soil at the Phoenix Lander Site. Science 2009, 325, 64–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, T.H.; Hodyss, R.; Choukroun, M.; Johnson, P.V. Chemistry of frozen sodium–magnesium–sulfate–chloride brines: Implications for surface expression of europa’s ocean composition. Astrophys. J. Lett. 2016, 816, L26. [Google Scholar] [CrossRef]
- Zolotov, M.Y. Aqueous Origins of Bright Salt Deposits on Ceres. Icarus 2017, 296, 289–304. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Gatto, G.J.; Stryer, L. Biochemistry, 8th ed.; W.H. Freeman & Company, a Macmillan Education Imprint: New York, NY, USA, 2015; ISBN 978-1-4641-2610-9. [Google Scholar]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-Binding Proteins: Modular Design for Efficient Function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Li, Q.; Xiang, J.; Yang, Q.; Sun, H.; Guan, A.; Wang, L.; Liu, Y.; Yu, L.; Shi, Y.; et al. Thioflavin T as an Efficient Fluorescence Sensor for Selective Recognition of RNA G-Quadruplexes. Sci. Rep. 2016, 6, 24793. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, S.; Arita-Morioka, K.; Mizunoe, Y.; Yamanaka, K.; Ogura, T. Thioflavin T as a Fluorescence Probe for Monitoring RNA Metabolism at Molecular and Cellular Levels. Nucleic Acids Res. 2015, 43, e92. [Google Scholar] [CrossRef] [Green Version]
- Warner, K.D.; Hajdin, C.E.; Weeks, K.M. Principles for Targeting RNA with Drug-like Small Molecules. Nat. Rev. Drug Discov. 2018, 17, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wilkinson, K.A.; Weeks, K.M. Complex Ligand-Induced Conformational Changes in TRNA Asp Revealed by Single-Nucleotide Resolution SHAPE Chemistry. Biochemistry 2008, 47, 3454–3461. [Google Scholar] [CrossRef]
- Agudelo, D.; Bourassa, P.; Beauregard, M.; Bérubé, G.; Tajmir-Riahi, H.-A. TRNA Binding to Antitumor Drug Doxorubicin and Its Analogue. PLoS ONE 2013, 8, e69248. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sánchez, L.; Lax, P.; Esquiva, G.; Martín-Nieto, J.; Pinilla, I.; Cuenca, N. Safranal, a Saffron Constituent, Attenuates Retinal Degeneration in P23H Rats. PLoS ONE 2012, 7, e43074. [Google Scholar] [CrossRef] [Green Version]
- Oliva, R.; Mukherjee, S.; Winter, R. Unraveling the Binding Characteristics of Small Ligands to Telomeric DNA by Pressure Modulation. Sci. Rep. 2021, 11, 9714. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Banerjee, S.; Cinar, H.; Ehrt, C.; Winter, R. Alteration of Protein Binding Affinities by Aqueous Two-Phase Systems Revealed by Pressure Perturbation. Sci. Rep. 2020, 10, 8074. [Google Scholar] [CrossRef]
- Jahmidi-Azizi, N.; Oliva, R.; Gault, S.; Cockell, C.S.; Winter, R. The Effects of Temperature and Pressure on Protein-Ligand Binding in the Presence of Mars-Relevant Salts. Biology 2021, 10, 687. [Google Scholar] [CrossRef]
- Pawlowska, R.; Janicka, M.; Jedrzejczyk, D.; Chworos, A. RNA Fragments Mimicking TRNA Analogs Interact with Cytochrome c. Mol. Biol. Rep. 2016, 43, 295–304. [Google Scholar] [CrossRef]
- Suga, K.; Tanabe, T.; Tomita, H.; Shimanouchi, T.; Umakoshi, H. Conformational Change of Single-Stranded RNAs Induced by Liposome Binding. Nucleic Acids Res. 2011, 39, 8891–8900. [Google Scholar] [CrossRef] [Green Version]
- Gray, D.M.; Ratliff, R.L.; Vaughan, M.R. Circular dichroism spectroscopy of DNA. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1992; Volume 211, pp. 389–406. ISBN 978-0-12-182112-8. [Google Scholar]
- Wells, B.D.; Yang, J.T. Computer Probe of the Circular Dichroic Bands of Nucleic Acids in the Ultraviolet Region. II. Double-Stranded Ribonucleic Acid and Deoxyribonucleic Acid. Biochemistry 1974, 13, 1317–1321. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.K.; Draper, D.E. On the Role of Magnesium Ions in RNA Stability. Biopolymers 1998, 48, 113–135. [Google Scholar] [CrossRef]
- Draper, D.E. RNA Folding: Thermodynamic and Molecular Descriptions of the Roles of Ions. Biophys. J. 2008, 95, 5489–5495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-León, S.; Schwierz, N. Hofmeister Series for Metal-Cation–RNA Interactions: The Interplay of Binding Affinity and Exchange Kinetics. Langmuir 2020, 36, 5979–5989. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Jahmidi-Azizi, N.; Mukherjee, S.; Winter, R. Harnessing Pressure Modulation for Exploring Ligand Binding Reactions in Cosolvent Solutions. J. Phys. Chem. B 2021, 125, 539–546. [Google Scholar] [CrossRef]
- Lavelle, L.; Fresco, J.R. Stabilization of Nucleic Acid Triplexes by High Concentrations of Sodium and Ammonium Salts Follows the Hofmeister Series. Biophys. Chem. 2003, 105, 681–699. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Chalikian, T.V.; Macgregor, R.B. On Empirical Decomposition of Volumetric Data. Biophys. Chem. 2019, 246, 8–15. [Google Scholar] [CrossRef]
- Chalikian, T.V.; Macgregor, R.B. Volumetric Properties of Four-Stranded DNA Structures. Biology 2021, 10, 813. [Google Scholar] [CrossRef]
- Gabelica, V.; Maeda, R.; Fujimoto, T.; Yaku, H.; Murashima, T.; Sugimoto, N.; Miyoshi, D. Multiple and Cooperative Binding of Fluorescence Light-up Probe Thioflavin T with Human Telomere DNA G-Quadruplex. Biochemistry 2013, 52, 5620–5628. [Google Scholar] [CrossRef]






| Solution Conditions | Kb/106 M−1 |
|---|---|
| Tris-HCl 20 mM, 40 mM KCl, pH 6.9 | 0.16 ± 0.01 |
| +150 mM NaCl | 0.12 ± 0.01 |
| +1 mM MgCl2 | 0.078 ± 0.010 |
| +1 mM MgSO4 | 0.009 ± 0.002 |
| +1 mM Mg(ClO4)2 | 0.053 ± 0.019 |
| Solution Conditions | p/bar | Kb/106 M−1 |
|---|---|---|
| Tris-HCl 20 mM, 40 mM KCl, pH 6.9 | 1 | 0.16 ± 0.01 |
| 500 | 0.18 ± 0.01 | |
| 1000 | 0.25 ± 0.03 | |
| 1500 | 0.36 ± 0.01 | |
| 2000 | 0.50 ± 0.05 | |
| +150 mM NaCl | 1 | 0.12 ± 0.01 |
| 500 | 0.15 ± 0.03 | |
| 1000 | 0.23 ± 0.05 | |
| 1500 | 0.31 ± 0.09 | |
| 2000 | 0.43 ± 0.11 | |
| +1 mM MgCl2 | 1 | 0.078 ± 0.007 |
| 500 | 0.081 ± 0.007 | |
| 1000 | 0.13 ± 0.02 | |
| 1500 | 0.18 ± 0.01 | |
| 2000 | 0.25 ± 0.01 | |
| +1 mM MgSO4 | 1 | 0.009 ± 0.002 |
| 500 | 0.011 ± 0.002 | |
| 1000 | 0.017 ± 0.001 | |
| 1500 | 0.030 ± 0.007 | |
| 2000 | 0.039 ± 0.002 | |
| +1 mM Mg(ClO4)2 | 1 | 0.053 ± 0.019 |
| 500 | 0.056 ± 0.020 | |
| 1000 | 0.074 ± 0.023 | |
| 1500 | 0.078 ± 0.025 | |
| 2000 | 0.091 ± 0.024 |
| Solution Conditions | Kb/106 M−1 | ΔG°b/kJ mol−1 | ΔH°b/kJ mol−1 | TΔS°b/kJ mol−1 | ΔVb/mL mol−1 |
|---|---|---|---|---|---|
| Tris-HCl 20 mM, 40 mM KCl, pH 6.9 | 0.16 ± 0.01 | −29.7 ± 0.2 | −4.1 ± 0.2 | 25.6 ± 0.2 | −14.9 ± 1.3 |
| +150 mM NaCl | 0.12 ± 0.01 | −29.0 ± 0.2 | N.D. * | N.D. * | −16.6 ± 0.8 |
| +1 mM MgCl2 | 0.078 ± 0.007 | −27.9 ± 0.2 | 40.5 ± 2.2 | 68.4 ± 2.2 | −15.2 ± 2.1 |
| +1 mM MgSO4 | 0.009 ± 0.002 | −22.6 ± 0.6 | 14.2 ± 2.0 | 36.7 ± 2.1 | −19.9 ± 1.9 |
| +1 mM Mg(ClO4)2 | 0.053 ± 0.019 | −27.0 ± 0.9 | 42.6 ± 6.7 | 69.6 ± 6.7 | −7.0 ± 0.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahmidi-Azizi, N.; Gault, S.; Cockell, C.S.; Oliva, R.; Winter, R. Ions in the Deep Subsurface of Earth, Mars, and Icy Moons: Their Effects in Combination with Temperature and Pressure on tRNA–Ligand Binding. Int. J. Mol. Sci. 2021, 22, 10861. https://doi.org/10.3390/ijms221910861
Jahmidi-Azizi N, Gault S, Cockell CS, Oliva R, Winter R. Ions in the Deep Subsurface of Earth, Mars, and Icy Moons: Their Effects in Combination with Temperature and Pressure on tRNA–Ligand Binding. International Journal of Molecular Sciences. 2021; 22(19):10861. https://doi.org/10.3390/ijms221910861
Chicago/Turabian StyleJahmidi-Azizi, Nisrine, Stewart Gault, Charles S. Cockell, Rosario Oliva, and Roland Winter. 2021. "Ions in the Deep Subsurface of Earth, Mars, and Icy Moons: Their Effects in Combination with Temperature and Pressure on tRNA–Ligand Binding" International Journal of Molecular Sciences 22, no. 19: 10861. https://doi.org/10.3390/ijms221910861
APA StyleJahmidi-Azizi, N., Gault, S., Cockell, C. S., Oliva, R., & Winter, R. (2021). Ions in the Deep Subsurface of Earth, Mars, and Icy Moons: Their Effects in Combination with Temperature and Pressure on tRNA–Ligand Binding. International Journal of Molecular Sciences, 22(19), 10861. https://doi.org/10.3390/ijms221910861






